A Novel Coupling Algorithm Based on Glowworm Swarm Optimization and Bacterial Foraging Algorithm for Solving Multi-Objective Optimization Problems
Abstract
:1. Introduction
2. Basic Concepts
2.1. The Multi-Objective Optimization Problems
2.2. Standard Glowworm Swarm Optimization Algorithm (GSO)
- (1)
- Initialization
- (2)
- Updating luciferin
- (3)
- Roulette selection
- Glowworm needs to be within the perceived radius of glowworm ;
- The luciferin of glowworm is brighter than that of glowworm .
- (4)
- Neighborhood range update rule
2.3. Standard Bacterial Foraging Algorithm (BFO)
- ➢
- The Chemotaxis
- ➢
- The Swarming
- ➢
- The Reproduction
- ➢
- The Elimination and Dispersal Operation
3. The MGSOBFO Algorithm
3.1. Fast Non-Dominated Sorting Approach and Crowding Distance
- (1)
- Fast Non-dominated Sorting Approach
| Algorithm 1: Fast non-dominated sort approach |
| for each , for each if // if p dominated q then |
| else if then end if then // p belong to the first front |
| end |
| i = 1 //Initialize the front counter |
| While , //Q represents the next front for store |
| For each if //q belong to the next front then , |
i=i+1 |
| end |
- (2)
- Crowding-distance calculation approach
3.2. The Self-Adaptive for Chemotaxis
- (a)
- if , that means is better than ;
- (b)
- if , that means is better than ;
- (c)
- If there is no dominant relationship between and , normalization is carried out for different fitness values. The process is as follows:
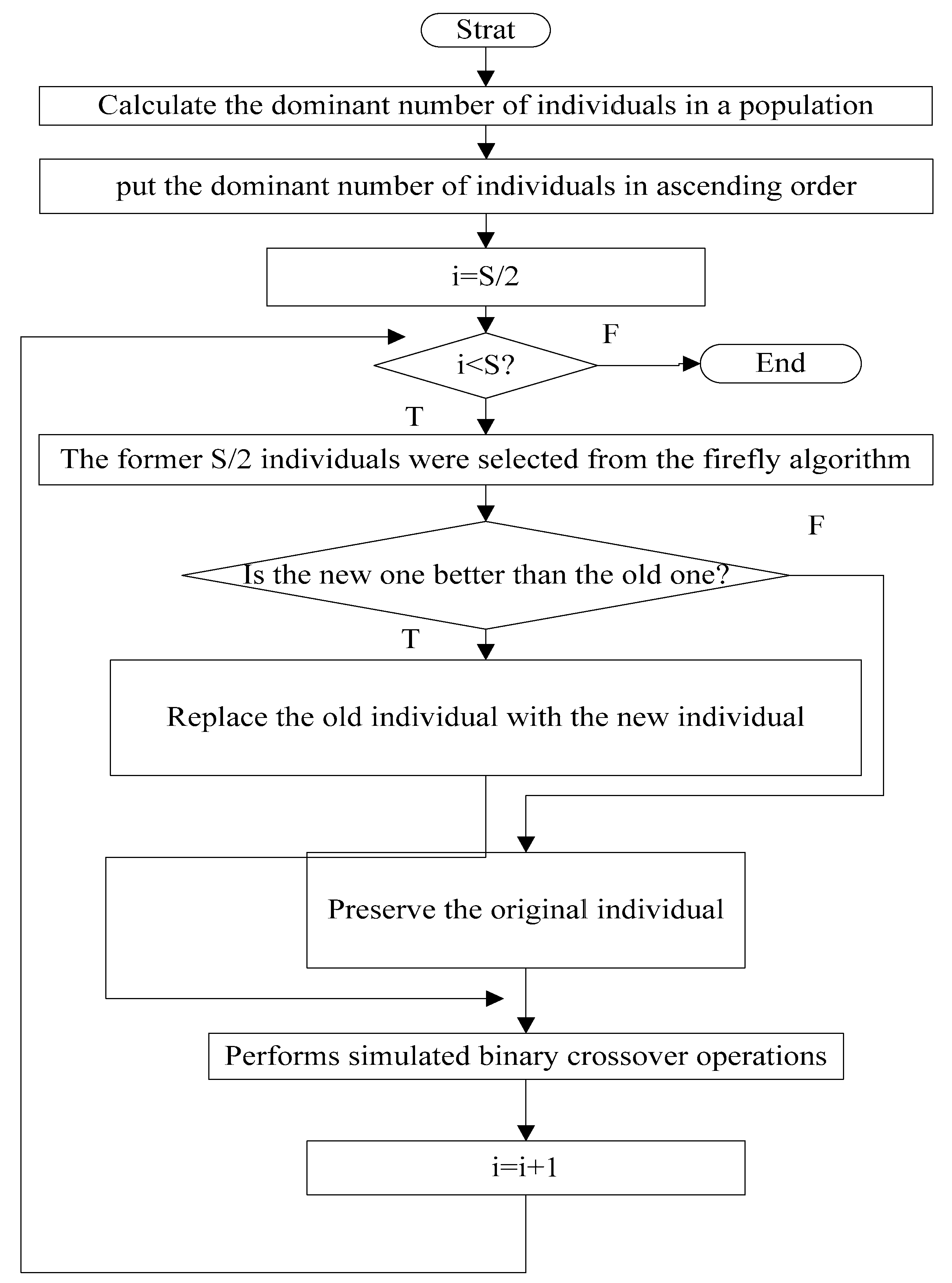
3.3. The Replication Operations Based on Crossover
3.4. The Elimination and Dispersal Operations Based on Mutation
3.5. The Flow Chart of MGSO-BFO
| Algorithm 2: The MGSO-BFO |
| Step 1: Create a random population N of size S, and initialize the required parameters; Step 2: Randomly generate the initial population Step 3: Elimination and Dispersal Operations loop: let j=0, j = j + 1, (the number of Elimination and Dispersal Operations step); Step 4: The replication operations loop: let k = 0, k = k + 1; (the number of replication step) Step 5: Chemotactic loop: let L = 0, L = L + 1; (the number of chemotactic step) Take the chemotactic step for the ith bacterium as follows: I. Calculate fitness function for all objective functions. II. let , (save a better fitness may be found so far) III. Tumble: create a random vector IV. Make movement with a self-adaption step(Specific see Formula (16)) for ith bacterium in direction. V. Computer for all objective functions. VI. Swim: Let m = 0 (m respect the swim length counter) While (1) Let m = m + 1 (2) If ( dominated ), let (3) ues to computer the new . VII. If , go to process the next bacterium. Step 6: if , go to the Step5 Step 7: The replication operations: I. Perform non-dominated sorting of BFO and GSO, and identify different front II. Prepare composite population by combining the BFO (s/2) with GSO (s/2). III. Performs simulated binary crossover operations IV. Select a new population of size S from the composite population. IV. if , go to the Step4 Step 8: Elimination and Dispersal Operations I. For each bacterium , if , use the mutation process shown in Section 3.4. II. if , go to the Step 3, else go to Step 9. Step 9: End. |
4. Experimental and Discussion
4.1. Test Set and Performance Measures
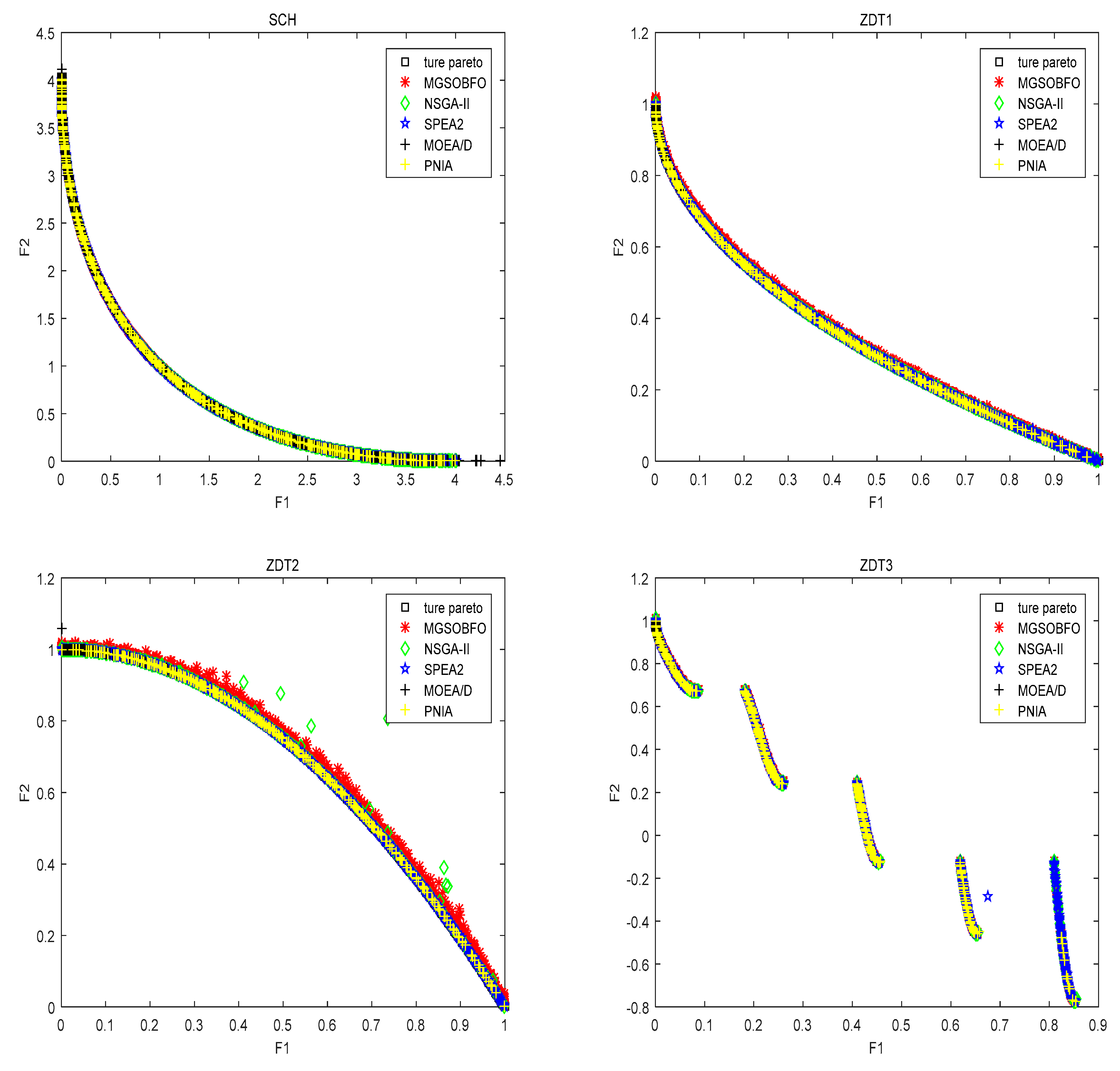
4.2. Comparison with State-of-the-Art Algorithm
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Problems | Metrics | MOEA/D | SPEA2 | PNIA | NSGA-II | MGSO-BFO |
|---|---|---|---|---|---|---|
| SCH | Mean (GD) | 4.83 × 10−2 | 4.41 × 10−2 | 4.81 × 10−2 | 6.17 × 10−2 | 4.75 × 10−2 |
| std (GD) | 4.51 × 10−2 | 1.14 × 10−2 | 1.93 × 10−3 | 2.96 × 10−3 | 3.41 × 10−2 | |
| Mean (SP) | 1.71 × 10−2 | 7.01 × 10−3 | 4.96 × 10−3 | 1.75 × 10−2 | 1.63 × 10−2 | |
| std (SP) | 1.64 × 10−2 | 1.61 × 10−3 | 3.66 × 10−3 | 1.02 × 10−2 | 2.14 × 10−2 | |
| ZDT1 | mean (GD) | 3.67 × 10−8 | 2.80 × 10−5 | 6.56 × 10−4 | 2.58 × 10−4 | 3.97 × 10−15 |
| std (GD) | 1.81 × 10−6 | 1.34 × 10−5 | 1.64 × 10−4 | 1.17 × 10−4 | 1.17 × 10−14 | |
| mean (SP) | 3.22 × 10−3 | 1.94 × 10−3 | 1.04 × 10−3 | 5.09 × 10−3 | 4.40 × 10−3 | |
| std (SP) | 4.21 × 10−3 | 3.38 × 10−4 | 1.06 × 10−3 | 3.92 × 10−3 | 7.30 × 10−3 | |
| ZDT2 | mean (GD) | 1.78 × 10−6 | 1.27 × 10−5 | 8.76 × 10−4 | 8.42 × 10−5 | 2.31 × 10−14 |
| std (GD) | 2.61 × 10−4 | 1.21 × 10−5 | 1.17 × 10−3 | 1.32 × 10−4 | 4.10 × 10−14 | |
| mean (SP) | 8.25 × 10−4 | 2.13 × 10−11 | 2.36 × 10−3 | 2.81 × 10−3 | 8.35 × 10−4 | |
| std (SP) | 1.30 × 10−3 | 8.83 × 10−4 | 3.17 × 10−3 | 4.36 × 10−3 | 1.30 × 10−3 | |
| ZDT3 | mean (GD) | 3.20 × 10−8 | 4.43 × 10−6 | 1.88 × 10−4 | 1.40 × 10−4 | 1.71 × 10−14 |
| std (GD) | 2.04 × 10−6 | 1.71 × 10−6 | 1.01 × 10−4 | 9.98 × 10−5 | 2.90 × 10−14 | |
| mean (SP) | 1.65 × 10−4 | 1.89 × 10−3 | 4.08 × 10−4 | 5.56 × 10−3 | 1.00 × 10−3 | |
| std (SP) | 4.34 × 10−3 | 5.99 × 10−4 | 5.47 × 10−4 | 3.81 × 10−3 | 1.70 × 10−3 | |
| ZDT4 | mean (GD) | 4.23 × 10−3 | 6.95 × 10−2 | 1.25 × 10−2 | 2.68 × 10−1 | 1.18 × 10−4 |
| std (GD) | 2.21 × 10−2 | 4.19 × 10−2 | 1.03 × 10−2 | 1.30 × 10−1 | 2.39 × 10−2 | |
| mean (SP) | 2.10 × 10−2 | 4.37 × 10−3 | 1.46 × 10−3 | 5.09 × 10−3 | 1.70 × 10−3 | |
| std (SP) | 3.12 × 10−3 | 4.64 × 10−3 | 3.20 × 10−3 | 1.11 × 10−2 | 1.00 × 10−3 | |
| ZDT6 | mean (GD) | 6.22 × 10−3 | 1.33 × 10−3 | 1.78 × 10−3 | 5.49 × 10−3 | 7.5 × 10−3 |
| std (GD) | 2.78 × 10−3 | 1.84 × 10−4 | 2.31 × 10−4 | 1.27 × 10−3 | 2.43 × 10−3 | |
| mean (SP) | 4.20 × 10−3 | 1.45 × 10−3 | 1.35 × 10−3 | 4.43 × 10−3 | 4.89 × 10−4 | |
| std (SP) | 2.44 × 10−4 | 5.74 × 10−4 | 9.87 × 10−4 | 2.05 × 10−3 | 2.30 × 10−3 |
Appendix B
| Problems | Metrics | MOEA/D | SPEA2 | PNIA | NSGA-II | MGSO-BFO |
|---|---|---|---|---|---|---|
| SCH | mean (IGD) | 1.6237 | 2.7448 | 1.5049 | 2.7305 | 1.4388 |
| Std (IGD) | 7.2145 | 12.2751 | 8.2428 | 12.2112 | 7.8809 | |
| ZDT1 | mean (IGD) | 0.4378 | 0.6175 | 0.5368 | 0.5861 | 0.4331 |
| Std (IGD) | 2.7503 | 2.7615 | 2.9403 | 2.6210 | 2.3720 | |
| ZDT2 | mean (IGD) | 0.6734 | 0.7002 | 0.5396 | 0.7706 | 0.4595 |
| Std (IGD) | 2.9145 | 3.1314 | 2.9515 | 3.4461 | 2.3168 | |
| ZDT3 | mean (IGD) | 0.2165 | 0.2384 | 0.5388 | 0.2445 | 0.3068 |
| Std (IGD) | 1.1362 | 1.0661 | 2.9512 | 1.0981 | 2.7706 | |
| ZDT4 | mean (IGD) | 1.1436 | 2.1738 | 1.2983 | 3.4946 | 0.4898 |
| Std (IGD) | 4.2264 | 9.7216 | 7.1113 | 15.6284 | 2.6827 | |
| ZDT6 | mean (IGD) | 0.4360 | 0.6646 | 0.5026 | 0.6357 | 0.4014 |
| Std (IGD) | 2.1065 | 2.9720 | 2.7527 | 2.8431 | 2.1985 |
Appendix C
| Problems | MOEA/D | SPEA2 | PNIA | NSGA-II | MGSO-BFO |
|---|---|---|---|---|---|
| SCH | 1.14 × 102 | 2.74 × 102 | 1.50 × 102 | 2.73 × 102 | 1.13 × 102 |
| ZDT1 | 2.23 × 102 | 3.56 × 102 | 3.74 × 102 | 2.40 × 102 | 1.12 × 102 |
| ZDT2 | 2.74 × 102 | 2.24 × 102 | 2.02 × 102 | 2.14 × 102 | 1.44 × 101 |
| ZDT3 | 2.45 × 102 | 2.52 × 102 | 3.88 × 102 | 2.38 × 102 | 2.55 × 102 |
| ZDT4 | 3.02 × 103 | 3.45 × 103 | 2.96 × 102 | 3.10 × 103 | 2.68 × 102 |
| ZDT6 | 2.74 × 102 | 2.97 × 102 | 2.75 × 103 | 2.82 × 103 | 2.19 × 102 |
References
- Heller, L.; Sack, A. Unexpected failure of a Greedy choice Algorithm Proposed by Hoffman. Int. J. Math. Comput. Sci. 2017, 12, 117–126. [Google Scholar]
- Pisut, P.; Voratas, K. A two-level particle swarm optimization algorithm for open-shop scheduling problem. Int. J. Comput. Sci. Math. 2016, 7, 575–585. [Google Scholar]
- Zhu, H.; He, Y.; Wang, X.; Tsang, E.C. Discrete differential evolutions for the discounted {0-1} knapsack problem. Int. J. Bio-Inspir. Comput. 2017, 10, 219–238. [Google Scholar] [CrossRef]
- Fourman, M.P. Compaction of Symbolic Layout using Genetic Algorithms. In Genetic Algorithms and Their Applications. In Proceedings of the First Internation Conference on Genetic Algorithms, Pittsburg, PA, USA, 24–26 July 1985; pp. 141–153. [Google Scholar]
- Das, D.; Suganthan, P.N. Differential Evolution: A Survey of the State-of-the-Art. IEEE Trans. Evolut. Comput. 2011, 15, 4–31. [Google Scholar] [CrossRef]
- Figueiredo, E.M.N.; Carvalho, D.F.; BastosFilho, C.J.A. Many Objective Particle Swarm Optimization. Inf. Sci. 2016, 374, 115–134. [Google Scholar] [CrossRef]
- Cortés, P.; Muñuzuri, J.; Onieva, L.; Guadix, J. A discrete particle swarm optimisation algorithm to operate distributed energy generation networks efficiently. Int. J. Bio-Inspir. Comput. 2018, 12, 226–235. [Google Scholar] [CrossRef]
- Ning, J.; Zhang, Q.; Zhang, C.; Zhang, B. A best-path-updating information-guided ant colony optimization algorithm. Inf. Sci. 2018, 433–434, 142–162. [Google Scholar] [CrossRef]
- Wang, H.; Wu, Z.; Rahnamayan, S.; Sun, H.; Liu, Y.; Pan, J.S. Multi-strategy ensemble artificial bee colony algorithm. Inf. Sci. 2014, 279, 587–603. [Google Scholar] [CrossRef]
- Cui, Z.; Zhang, J.; Wang, Y.; Cao, Y.; Cai, X.; Zhang, W.; Chen, J. A pigeon-inspired optimization algorithm for many-objective optimization problems. Sci. China Inf. Sci. 2019. [Google Scholar] [CrossRef]
- Cai, X.; Gao, X.; Xue, Y. Improved bat algorithm with optimal forage strategy and random disturbance strategy. Int. J. Bio-Inspir. Comput. 2016, 8, 205–214. [Google Scholar] [CrossRef]
- Cui, Z.; Li, F.; Zhang, W. Bat algorithm with principal component analysis. Int. J. Mach. Learn. Cybern. 2018. [Google Scholar] [CrossRef]
- Yang, C.; Ji, J.; Liu, J.; Yin, B. Bacterial foraging optimization using novel chemotaxis and conjugation strategies. Inf. Sci. 2016, 363, 72–95. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, H.; Cui, Z.; Chen, J. Hybrid multi-objective cuckoo search with dynamical local search. Memet. Comput. 2018, 10, 199–208. [Google Scholar] [CrossRef]
- Cui, Z.; Sun, B.; Wang, G.; Xue, Y.; Chen, J. A novel oriented cuckoo search algorithm to improve DV-Hop performance for cyber-physical systems. J. Parallel Distrib. Comput. 2017, 103, 42–52. [Google Scholar] [CrossRef]
- Abdel-Baset, M.; Zhou, Y.; Ismail, M. An improved cuckoo search algorithm for integer programming problems. Int. J. Comput. Sci. Math. 2018, 9, 66–81. [Google Scholar] [CrossRef]
- Zhou, J.; Dong, S. Hybrid glowworm swarm optimization for task scheduling in the cloud environment. Eng. Optim. 2018, 50, 949–964. [Google Scholar] [CrossRef]
- Yu, G.; Feng, Y. Improving firefly algorithm using hybrid strategies. Int. J. Comput. Sci. Math. 2018, 9, 163–170. [Google Scholar] [CrossRef]
- Cui, Z.; Cao, Y.; Cai, X.; Cai, J.; Chen, J. Optimal LEACH protocol with modified bat algorithm for big data sensing systems in Internet of Things. J. Parallel Distrib. Comput. 2017. [Google Scholar] [CrossRef]
- Cai, X.; Wang, H.; Cui, Z.; Cai, J.; Xue, Y.; Wang, L. Bat algorithm with triangle-flipping strategy for numerical optimization. Int. J. Mach. Learn. Cybern. 2018, 9, 199–215. [Google Scholar] [CrossRef]
- Srinivas, N.; Deb, K. Muiltiobjective Optimization Using Nondominated Sorting in Genetic Algorithms. Evolut. Comput. 1994, 2, 221–248. [Google Scholar] [CrossRef]
- Deb, K.; Agrawal, S.; Pratap, A.; Meyarivan, T. A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evolut. Comput. 2002, 6, 182–197. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, H. MOEA/D: A Multiobjective Evolutionary Algorithm Based on Decomposition. IEEE Trans. Evolut. Comput. 2007, 11, 712–731. [Google Scholar] [CrossRef]
- Horn, J.; Nafpliotis, N.; Goldberg, D.E. A niched Pareto genetic algorithm for multiobjective optimization. In Proceedings of the IEEE Conference on Evolutionary Computation IEEE World Congress on Computational Intelligence, Honolulu, HI, USA, 12–17 May 2002. [Google Scholar]
- Zitzler, E.; Laumanns, M.; Thiele, L. SPEA2: Improving the Strength Pareto Evolutionary Algorithm for Multiobjective Optimization. In Evolutionary Methods for Design, Optimization and Control with Applications To Industrial Problems, Proceedings of the Eurogen 2001, Athens, Greece, 19–21 September2001; International Center for Numerical Methods in Engineering: Barcelona, Spain, 2002. [Google Scholar]
- Yuan, J.; Gang, X.; Zhen, Z.; Chen, B. The pareto optimal control of inverter based on multi-objective immune algorithm. In Proceedings of theInternational Conference on Power Electronics & Ecce Asia, Seoul, Korea, 1–5 June2015. [Google Scholar]
- Wolpert, D.H.; Macready, W.G. No free lunch theorems for optimization. IEEE Trans. Evolut. Comput. 1997, 1, 67–82. [Google Scholar] [CrossRef]
- Qu, B.Y.; Suganthan, P.N. Constrained Multi-Objective Optimization Algorithm with Ensemble of Constraint Handling Methods. Eng. Optim. 2011, 43, 403. [Google Scholar] [CrossRef]
- Jin, Y.; Branke, J. Evolutionary optimization in uncertain environments-a survey. IEEE Trans. Evolut. Comput. 2005, 9, 303–317. [Google Scholar] [CrossRef]
- Yu, X.; Tang, K.; Chen, T.; Yao, X. Empirical analysis of evolutionary algorithms with immigrants schemes for dynamic optimization. Memet. Comput. 2009, 1, 3–24. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, Z.; Cui, Z.; Cai, X. NSGA-II with local perturbation. In Proceedings of the Control & Decision Conference, Chongqing, China, 28–30 May 2017. [Google Scholar]
- Zitzle, E.; Deb, K.; Thiele, L. Comparison of Multiobjective Evolutionary Algorithm: Empirical Results. Evolut. Comput. 2000, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, J.D. Multiple objective optimization with vector evaluated genetic algorithms. In Proceedings of the First International Conference on Genetic Algorithms and Their Applications; L. Erlbaum Associates Inc.: Mahwah, NJ, USA, 1985; pp. 93–100. [Google Scholar]
- Schott, J.R. Fault tolerant design using single and multicriteria genetic algorithm optimization. Cell. Immunol. 1995, 37, 1–13. [Google Scholar]
- Mohammadi, A.; Omidvar, M.N.; Li, X. A new performance metric for user-preference based on multi-objective evolutionary algorithms. In Proceedings of the 2013 IEEE Congress on Evolutionary Computation (CEC), Cancun, Mexico, 20–23 June 2013; pp. 2825–2832. [Google Scholar]


| Function | Dimension | Range | Objective Function | Optimal Solution | Feature |
|---|---|---|---|---|---|
| SCH | 1 | convex | |||
| ZDT1 | 30 | [0, 1] | convex | ||
| ZDT2 | 30 | [0, 1] | Non-convex | ||
| ZDT3 | 30 | [0, 1] | convex discon-nected | ||
| ZDT4 | 10 | Non-convex | |||
| ZDT6 | 10 | [0, 1] | Non-convex non-uniformly |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Cui, Z.; Li, W. A Novel Coupling Algorithm Based on Glowworm Swarm Optimization and Bacterial Foraging Algorithm for Solving Multi-Objective Optimization Problems. Algorithms 2019, 12, 61. https://doi.org/10.3390/a12030061
Wang Y, Cui Z, Li W. A Novel Coupling Algorithm Based on Glowworm Swarm Optimization and Bacterial Foraging Algorithm for Solving Multi-Objective Optimization Problems. Algorithms. 2019; 12(3):61. https://doi.org/10.3390/a12030061
Chicago/Turabian StyleWang, Yechuang, Zhihua Cui, and Wuchao Li. 2019. "A Novel Coupling Algorithm Based on Glowworm Swarm Optimization and Bacterial Foraging Algorithm for Solving Multi-Objective Optimization Problems" Algorithms 12, no. 3: 61. https://doi.org/10.3390/a12030061
APA StyleWang, Y., Cui, Z., & Li, W. (2019). A Novel Coupling Algorithm Based on Glowworm Swarm Optimization and Bacterial Foraging Algorithm for Solving Multi-Objective Optimization Problems. Algorithms, 12(3), 61. https://doi.org/10.3390/a12030061




