Abstract
In this paper we consider a basic clustering problem that has uses in bioinformatics. A structural fragment is a sequence of ℓ points in a 3D space, where ℓ is a fixed natural number. Two structural fragments and are equivalent if and only if under some rotation R and translation τ. We consider the distance between two structural fragments to be the sum of the squared Euclidean distance between all corresponding points of the structural fragments. Given a set of n structural fragments, we consider the problem of finding k (or fewer) structural fragments , so as to minimize the sum of the distances between each of to its nearest structural fragment in . In this paper we show a polynomial-time approximation scheme (PTAS) for the problem through a simple sampling strategy.
1. Introduction
In this paper we consider the problem of clustering similar sequences of 3D points. Two such sequences of points are considered the same if they are equivalent under rotation and translation. The scenario which we consider is as follows. Suppose there is an original sequence of points that gave rise to a few variations of itself, through slight changes in some or all of its points. Now given these variations of the sequence, we are to reconstruct the original sequence. A likely candidate for such an original sequence would be a sequence which is “nearest" in terms of some distance measure, to the variations.
A more complicated scenario involves k original sequences of the same length. Formally, we formulate the problem as follows. Given n sequences of points , we are to find a set of k sequences , such that the sum of distances
is minimized. In this paper we consider the case where is the minimum sum of squared Euclidean distances between each of the points in the two sequences and , under all possible rigid transformations on the sequences of points. A cost function in the form of the squared Euclidean distance is used in many techniques for clustering 3D points []. Since our clustering problem is quite different from those previously studied, it calls for a new technique. (The “square" in the distance measure is to fulfill a condition needed by the method in this paper. The method does not work, for example, in the case of the root mean squared Euclidean distance. On the other hand, the method easily adapts to other distance measures that fulfill the required condition.)
Such a problem has potential use in clustering protein structures. A protein structure is typically given as a sequence of points in 3D space, and for various reasons, there are typically minor variations in their measured structures. The problem can be considered a model of the situation where we have a set of measurements of a few protein structures, and are to reconstruct the original structures.
In this paper, we show that there is a polynomial-time approximation scheme (PTAS) for the problem, through a sampling strategy. More precisely, we show that an optimal solution obtained by sampling smaller subsets of the input suffices to give us an approximate solution, and the approximation ratio improves as we increase the size of the subsets we sample.
2. Preliminaries
Throughout this paper we let ℓ be a fixed non-zero natural number. A structural fragment is a sequence of ℓ 3D-points. The mean square distance () between two structural fragments and , is defined to be
where is the set of all rotation matrices, the set of all translation vectors, and is the Euclidean distance between .
The root of the measure, is a measure that has been extensively studied. Note that , that minimize to give us will also give us , and vice versa. Since given any f and g, there are closed form equations [,] for finding R and τ that give , can be computed efficiently for any f and g.
Furthermore, it is known that to minimize , the centroid of f and g must coincide []. Due to this, without loss of generality we assume that all structural fragments have centroids at the origin. Such transformations can be done in time. After such transformations, in computing , only the parameter need to be considered, that is,
Suppose that given a set of n structural fragments , we are to find k structural fragments , such that each structural fragment is “near", in terms of the , to at least one of the structural fragments in . We formulate such a problem as follows:
| k-Consensus Structural Fragments Problem Under | |
| Input: | n structural fragments , and a non-zero natural |
| number . | |
| Output: | k structural fragments , minimizing the cost |
| . |
In this paper we will demonstrate that there is a PTAS for the problem.
We use the following notations: Cardinality of a set A is written . For a set A and non-zero natural number n, denotes the set of all length n sequences of elements of A. Let elements in a set A be indexed, say , then denotes the set of all the length m sequences , where . For a sequence S, denotes the i-th element in S, and denotes its length.
3. PTAS for the k-Consensus Structural Fragments
The following lemma, from [], is central to the method.
Lemma 1 ([]) Let be a sequence of real numbers and let , . Then the following equation holds:
Let be a sequence of 3D points.
One can similarly extend the equation for structural fragments. Let be n structural fragments, the equation becomes:
The equation says that there exists a sequence of r structural fragments such that
Our strategy uses this fact —in essentially the same way as in []— to approximate the optimal solution for the k-consensus structural fragments problem. That is, by exhaustively sampling every combination of k sequences, each of r elements from the space , where is the input and is a fixed selected set of rotations, which we next discuss.
3.1. Discretized Rotation Space
Any rotation can be represented by a normalized vector u and a rotation angle θ, where u is the axis about which an object is rotated by θ. If we apply to a vector v, we obtain vector , which is:
where · represents dot product, and × represent cross product.
By the equation, one can verify that a change of ϵ in u will result in a change of at most in for some computable ; and a change of ϵ in θ will result in a change of at most in for some computable . Now any rotation along an axis through the origin can be written in the form , where are respectively a rotation along each of the axes. Similarly, changes of ϵ in , and will result in a change of at most , for some computable .
We discretize the values that each , may take within the range into a series of angles of angular difference ϑ. There are hence at most of such values for each , . Let denote the set of all possible discretized rotations . Note that is of order .
Let be the diameter of a ball that is able to encapsulate each of . Hence any distance between two points among is at most . In this paper we assume d to be constant with respect to the input size. Note that for a protein structure, is of order []. For any , we can choose ϑ so small that for any rotation R and any point , there exists such that .
3.2. A Polynomial-time Algorithm With Cost
Our algorithm for the k-consensus structural fragments problem is summarized in Table 1.
This is what the algorithm does: In (2), we explore m distinct subsets from , in the hope that each subset is from a distinct cluster in the optimal clustering. Since we explore all possible such subsets this is bound to happen. We then try to evaluate the score of each subset by sampling up to r structural fragments (allowing repeats) from it (from (2.1) onwards). Such an evaluation is possible due to Equation 7. The evaluation also requires us to exhaustively try out all possible transformations in , which is what we try to do in (2.2). Each of these samplings of produces a consensus structural fragment for in (2.3), the score of which is evaluated in (2.4). Finally in (3), we output the consensus patterns which give us the best score.
We now analyze the runtime complexity of the algorithm. Consider the number of in (2.1) that are possible. Let each be represented by a length r string of symbols, n of which each represents one of , while the remaining symbol represents “nothing". It is clear that for any , any , or (where ), can be represented by one such string. Furthermore, any can be completely represented by k such strings — that is, to represent the case where , strings can be set to “nothing" completely. From this, we can see that there are at most possible combinations of .
For each of these combinations, there are possible combinations of at (2.2), hence resulting in iterations to run for (2.3) to (2.5). Since (2.3) can be done in , (2.4) in , and (2.5) in time, the algorithm completes in time.
We argue that eventually is at most of the optimal solution plus a factor. Suppose the optimal solution results in the disjoint clusters .
For each , , let be a structural fragment which minimizes . Furthermore, for each , let be a rotation where
and let

Table 1.
Polynomial-time algorithm for the problem.
By the property of the measure, it can be shown that is the average of . For each where , by Equation 6,
For each such , let be such that
Without loss of generality assume that each . Let
Then we may write,
For each rotation , let be a closest rotation to within . Also, let
Since we exhaustively sample all possible for all possible and for all , it is clear that:
Hence by Equation 14, is at most of the optimal solution plus a factor . Let ,
Theorem 2 For any , a -approximation solution for the k-consensus structural fragments problem can be computed in
time.
The factor c in Theorem 2 is due to error introduced by the use of discretization in rotations. If we are able to estimate a lower bound of , we can scale this error by refining the discretization such that c is an arbitrarily small factor of . To do so, in the next section we show a lower bound to .
3.3. A Polynomial-time 4-approximation Algorithm
We now show a 4-approximation algorithm for the k-consensus structural fragments problem. We first show the case for , and then generalizes the result to all .
Let the input n structural fragments be , , …, . Let , be the structural fragment where
is minimized. Note that can be found in time , since for any , (more precisely, ) can be computed in time using closed form equations from [].
We argue that is a 4-approximation. Let the optimal structural fragment be , the corresponding distance , and let () be the fragment where is minimized.
We first note that the cost of using as solution, . To continue we first establish the following claim.
Claim 1 .
PROOF. In [], it is shown that
Squaring both sides gives
Since
we have . ▮
By the above claim,
Hence . We now extend this to k structural fragments. 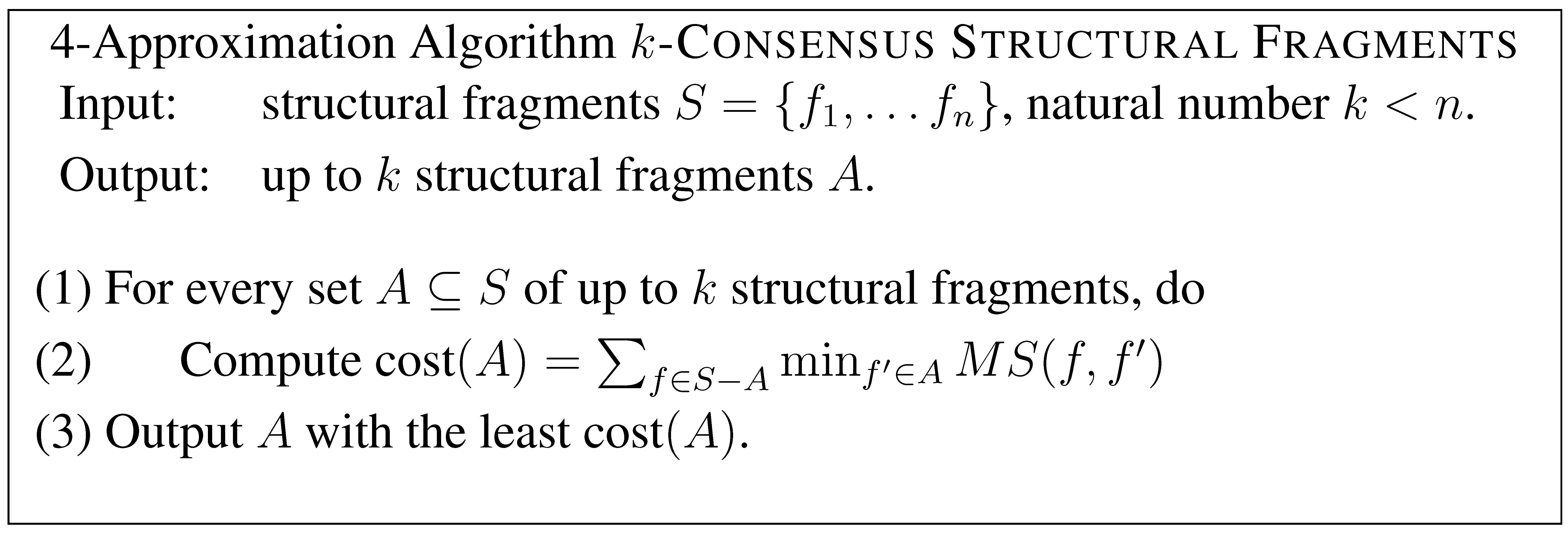

We first pre-compute for every pair of , which takes time . Then, at step (1), there are at most combinations of A, each which takes time to compute at step (2). Hence in total we can perform the computation in time. To see that the solution is a 4-approximation, let where be an optimal clustering. Then, by our earlier argument, there exists , , …, such that each is a 4-approximation for , and hence is a 4-approximation for the k-consensus structural fragments problem. Since the algorithm exhaustively search for every combination of up to k fragments, it gives a solution at least as good as , and hence is a 4-approximation algorithm.
Theorem 3 A 4-approximation solution for the k-consensus structural fragments problem can be computed in time.
3.4. A Polynomial-time Approximation Scheme
Recall that the algorithm in Section 3.2 has cost where . From Section 3.3 we have a lower bound D of . We want . To do so, it suffices that we set . This results in an of order . Substituting this in Theorem 2, and combining with Theorem 3, we get the following.
Theorem 4 For any , a -approximation solution for the k-consensus structural fragments problem can be computed in
time, where .
4. Discussions
The method in this paper depends on Lemma 1. For this reason, the technique does not extend to the problem under distance measures where Lemma 1 cannot be applied, for example, the measure. However, should Lemma 1 apply to a distance measure, it should be easy to adapt the method here to solve the problem for that distance measure.
One can also formulate variations of the k-consensus structural fragments problem. For example, 

While the cost function of the k-consensus structural fragments problem resembles that of the k-means problem, the cost function of the k-closest structural fragments resembles that of the (absolute) k-center problem. One interesting problem for future study is whether this problem has a PTAS or not. It is not clear how to generalize the technique employed in this paper to k-closest structural fragments problem under .
References
- Jain, A. K.; Murty, M. N.; Flynn, P. J. Data clustering: a review. ACM Computing Surveys 1999, 31(3), 264–323. [Google Scholar] [CrossRef]
- Arun, K. S.; Huang, T. S.; Blostein, S. D. Least-squares fitting of two 3-d point sets. IEEE Trans. Pattern Anal. Mach. Intell. 1987, 9(5), 698–700. [Google Scholar] [CrossRef] [PubMed]
- Umeyama, S. Least-squares estimation of transformation parameters between two point patterns. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13(4), 376–380. [Google Scholar] [CrossRef]
- Qian, J.; Li, S. C.; Bu, D.; Li, M.; Xu, J. Finding compact structural motifs. In Combinatorial Pattern Matching, 18th Annual Symposium, CPM 2007, London, Canada, July 9-11, 2007, Proceedings; Ma, B., Zhang, K.Z., Eds.; Springer, 2007; Vol. 4580 of Lecture Notes in Computer Science, pp. 142–149. [Google Scholar]
- Hao, M.; Rackovsky, S.; Liwo, A.; Pincus, M.; Scheraga, H. Effects of compact volume and chain stiffness on the conformations of native proteins. Proc. Natl. Acad. Sci. 1992, 89, 6614–6618. [Google Scholar] [CrossRef] [PubMed]
- Boris, S. A revised proof of the metric properties of optimally superimposed vector sets. Acta Crystallographica Section A 2002, 58(5), 506. [Google Scholar]
© 2008 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
