Abstract
Over the past few years, the popularity of graphene as a potential 2D material has increased since graphene-based materials have applications in a variety of fields, including medicine, engineering, energy, and the environment. A large number of graphene sheets as well as an understanding of graphene’s structural hierarchy are critical to the development of graphene-based materials. For a variety of purposes, it is essential to understand the fundamental structural properties of graphene. Molecular descriptors were used in this study to investigate graphene sheets’ structural behaviour. Based on our findings, reverse degree-based molecular descriptors can significantly affect the exchange-correlation energy prediction. For the exchange-correlation energy of graphene sheets, a linear regression analysis was conducted using the reverse general inverse sum indeg descriptor, . From , a set of reverse topological descriptors can be obtained all at once as a special case, resulting in a model with a high correlation coefficient (R between 0.896 and 0.998). Used together, these reverse descriptors are graphed in relation to their response to graphene. Based on this study’s findings, it is possible to predict the exchange correlation energy as well as the geometric structures of graphene sheets with very little computational cost.
1. Introduction
Carbon is a widely studied and influential element across many scientific disciplines. Many allotropes of carbon exist, each with special properties, such as graphite, diamond, and amorphous carbon as well as fullerenes, carbon nanotubes (CNTs), and graphene [1,2,3,4,5,6]. Graphene is at the forefront of research in fields such as physics, chemistry, and materials science, among many others. Researchers have been intrigued by graphene due to its great mechanical, transportable, optical, and thermal properties as well as its thermal stability and unique electronic structures [7,8,9]. Graphene is packed in a unique two-dimensional nano-carbon hexagonal lattice [10,11]. Graphene’s unique combination of characteristics strongly qualifies it for use in multiple applications, such as biosensors [9], membranes [12], drug delivery, tissue engineering, sensing applications [13], photodetectors [14], electrochemical sensors [15], and hydrogen-based energy storage [16].
A nanostructure is composed of distinct and measurable elements, known as nano-patterns. In contrast to random patterns, these patterns follow the order of chemical and physical laws. Physical and chemical laws determine how atoms and molecules form discrete and measurable geometric structures, ranging from repeating lattices to complex shapes. Rules from the chemical graph theory can be used to analyze and predict the properties of these well-defined structures [17]. In the chemical graph theory, a chemical structure is represented by a corresponding molecular graph, where vertices represent atoms and edges represent bonds [18]. Molecular descriptors are commonly used in the chemical graph theory to predict various properties of chemical structures. Among the many molecular descriptors available, the topological molecular descriptors are a prominent [19,20]. Topological molecular descriptors are used to transform molecular graphs into mathematical models as well as encrypt significant amounts of information about the molecular structure. Topological molecular descriptors can be classified into a number of groups according to their graph parameters. Some of the well-known topological descriptors include distance [21], degree [22], eccentricity [23], and spectrum-based descriptors [24]. Researchers often prefer degree-based topological descriptors due to their simplicity, and some of the most popular degree-based topological descriptors are the first and second Zagreb [25], Randić [26], sum−connectivity [27], and geometric−arithmetic descriptors [28], etc. Wei et al. [29] recently introduced many reverse degree-based topological descriptors, inspired by their work on degree-based topological descriptors.
In this article, molecular graphs are represented by . denotes the degree of a vertex , and is the maximum degree of the graph The reverse degree [30] of a vertex is defined as .
To derive a set of reverse degree-based topological descriptors, we first define the reverse general inverse sum indeg descriptor, denoted by , as follows: where and are any real numbers.
Table 1 Some reverse degree-based topological descriptors derived from the reverse general inverse sum indeg descriptor by assigning specific values to the parameters and .

Table 1.
Some reverse degree-based topological descriptors derived from the reverse general inverse sum indeg descriptor.
Our main objective is to offer an alternate method, with high accuracy, for computing the exchange-correlation energies of graphene sheets. The DFT calculations of the exchange-correlation energies of graphene sheets have the advantage of being accurate, but they also have the disadvantage of being computationally expensive. Therefore, Section 2 provides a relationship between the exchange-correlation energy of graphene sheets and reverse degree-based topological descriptors. Section 3 contains detailed analytical results for graphene using reverse degree topological descriptors and polynomial as well as numerical comparisons.
2. Relationship between the Exchange-Correlation Energy and the Reverse General Inverse Sum Indeg Descriptor of Graphene Sheets
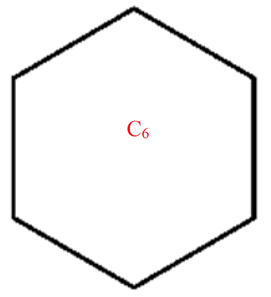
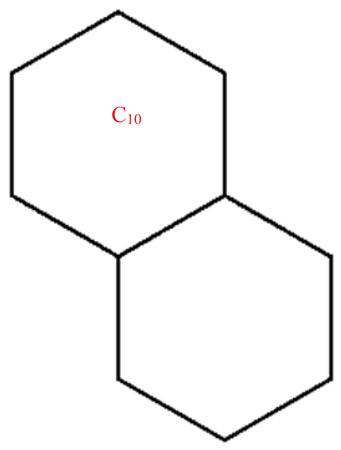
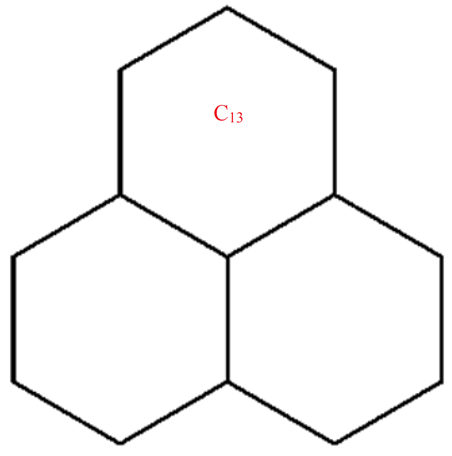
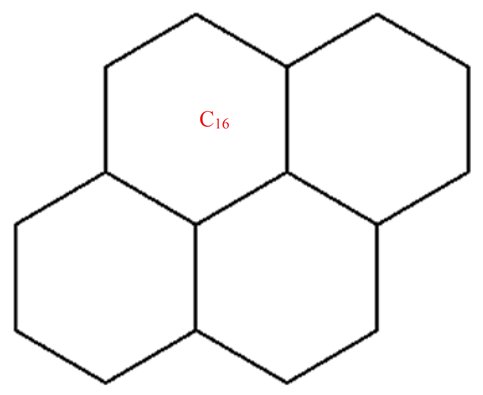
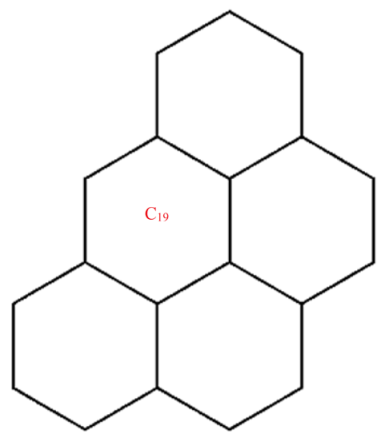
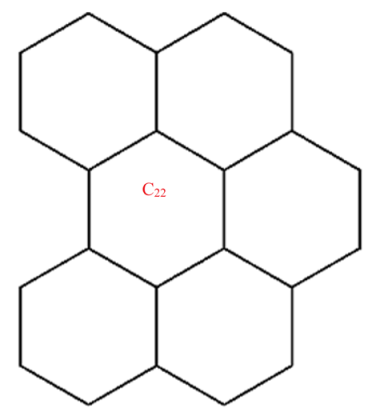
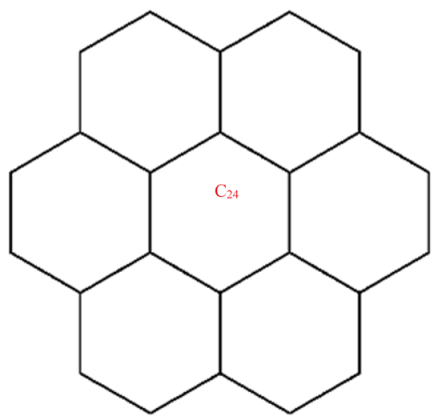
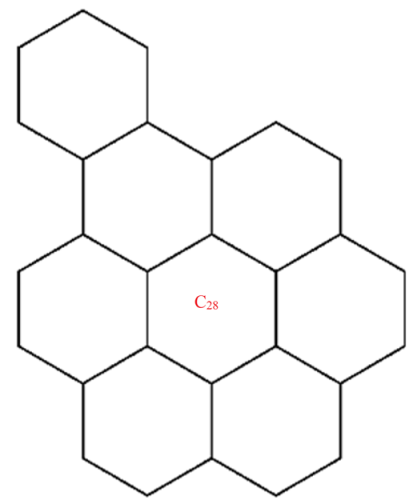
A wide range of molecular descriptors have been proposed in the current literature, but many of them show little evidence that they correlate with any of the physical or chemical properties of the chemical structure. This section highlights the inquiry that was undertaken to determine whether reverse general inverse sum indeg descriptors possess any predictive power and whether or not they should be used in any chemical applications. In order to achieve this, we selected ten graphene sheets from one cycle to ten cycles. The molecular structures of these graphene sheets are provided in Table 2. The exchange-correlation energies (ECE) of these graphene sheets were obtained from the literature [31] and have been listed in Table 2.

Table 2.
Graphene sheets from C6 to C32 with their exchange-correlation energy and reverse general inverse sum indeg descriptors.
The reverse general inverse sum indeg descriptors of these graphene sheets were obtained through direct calculations using the edge partition technique. For example, the reverse general inverse sum indeg descriptor for the graphene sheet C24 (), shown in Table 2, was obtained by the following way: The molecular graph of C24 had 24 vertices, 30 edges, and . Based on the reverse degrees of each of the vertices, the edges set of was partitioned into three sets: with cardinalities . From the definition of a reverse general inverse sum indeg descriptor, we used the following:
After simplification, we arrived at .
Table 3 lists 11 reverse topological descriptors: the reverse first and second Zagreb descriptor, the reverse Randić descriptor, the reverse sum−connectivity descriptor, the reverse harmonic descriptor, the reverse hyper Zagreb descriptor, the reverse geometric−arithmetic descriptor, the reverse arithmetic−geometric descriptor, the reverse inverse sum indeg descriptor, the reverse redefined first Zagreb descriptor, and the reverse redefined third Zagreb descriptor. These descriptors were obtained by setting specific values of p and q, such as the following:
in the reverse general inverse sum indeg descriptors (Table 1) for each graphene sheet from C6 to C24.


Table 3.
Values of the reverse topological descriptors from C6 to C32.
Table 3.
Values of the reverse topological descriptors from C6 to C32.
| Graphene Sheets | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C6 | 12 | 6 | 6 | 4.2426 | 6 | 24 | 6 | 6 | 3 | 12 | 12 |
| C10 | 38 | 33 | 6.8284 | 6.0165 | 6.6667 | 136 | 10.771 | 11.243 | 9.1667 | 14 | 122 |
| C13 | 48 | 39 | 10.243 | 8.5855 | 10 | 162 | 14.657 | 15.364 | 11.5 | 21 | 138 |
| C16 | 58 | 45 | 13.657 | 11.154 | 13.333 | 188 | 18.542 | 19.485 | 13.833 | 28 | 154 |
| C19 | 68 | 51 | 17.071 | 13.723 | 16.667 | 214 | 22.428 | 23.606 | 16.167 | 35 | 170 |
| C22 | 79 | 60 | 20.364 | 16.267 | 20 | 249 | 26.485 | 27.546 | 19 | 41.5 | 202 |
| C24 | 84 | 60 | 23.485 | 18.414 | 23 | 252 | 29.314 | 30.728 | 20 | 48 | 192 |
| C28 | 98 | 71 | 27.485 | 21.534 | 27 | 296 | 34.314 | 35.728 | 23.5 | 56 | 230 |
| C30 | 104 | 73 | 30.399 | 24.104 | 30.33 | 306 | 37.199 | 38.849 | 24.833 | 62 | 230 |
| C32 | 110 | 75 | 33.314 | 25.673 | 32.667 | 316 | 40.97 | 41.97 | 26.167 | 68 | 230 |
To predict the exchange-correlation energy of the graphene sheets, the following linear regression model was used:
where is the exchange-correlation energy of the graphene sheets from C6 to C32, is the regression model constant, is the reverse topological descriptor coefficient, and is any predictor from Table 1. This linear regression model was used in compiling Table 4, which used SPSS software to show the regression equations of the 11 reverse topological descriptors, the correlation coefficient between the exchange-correlation energy of the graphene sheets, and the reverse topological descriptors from the data obtained from Table 2 and Table 3. Statistical quantities, such as the standard error (SE) and the F-test, were used to check the reliability of the predictive models listed in Table 4. Based on Table 2 and Table 3, we found that the reverse topological descriptors and the exchange-correlation energy exhibit similar trends and Figure 1 and Figure 2 illustrates this similarity. Figure 3 shows the linear relationship between the exchange-correlation energy while Figure 4 graphically depicts the predictive potential of the reverse topological descriptors via the square of the correlation with the help of the reverse topological descriptors of the studied graphene sheets using the regression model presented in Table 4.

Table 4.
Linear prediction models with statistical parameters of the exchange-correlation energy of graphene sheets from C6 to C32.
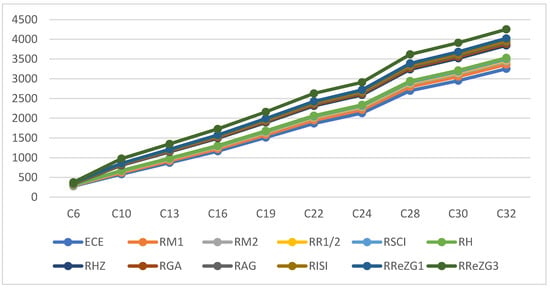
Figure 1.
Variation of reverse topological descriptors and ECE.
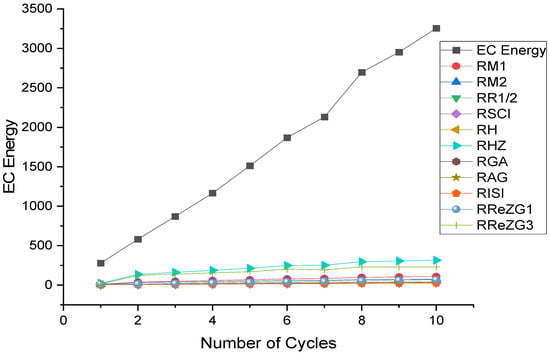
Figure 2.
ECE and reverse descriptors.
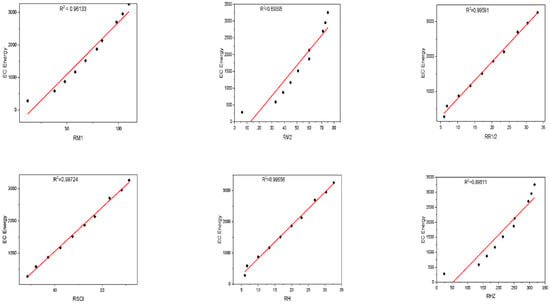
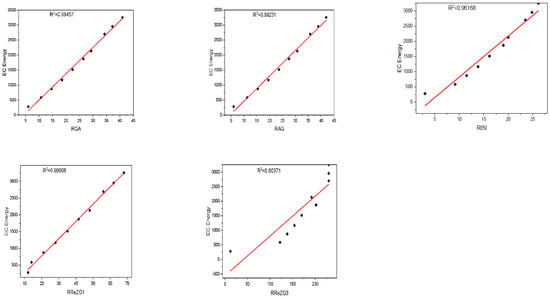
Figure 3.
Plot between the reverse topological descriptors and ECE of graphene sheets from C6 to C32.
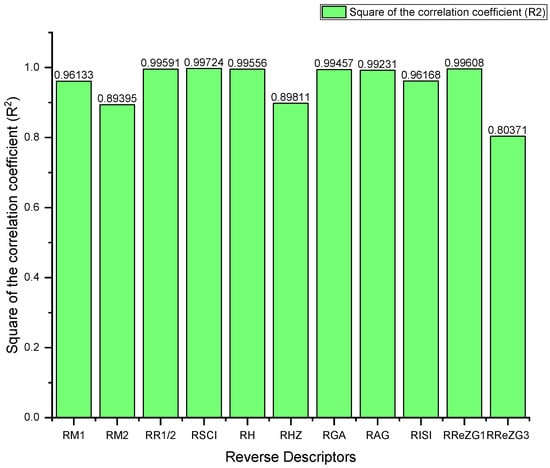
Figure 4.
Predictive potential of the reverse topological descriptors via square of the correlation coefficient .
3. Reverse General Inverse Sum Indeg Descriptor of Graphene
This section covers graphene systems, which have gained a lot of research interest across a wide range of applications due to their fascinating properties. There are numerous studies [32,33,34,35,36,37,38,39,40] dedicated to the computation of topological descriptors of graphene systems in recent years. Most of these studies are devoted to obtaining an individual formula for each topological descriptor. This article presents a general reverse degree-based topological descriptor, namely, a reverse general inverse sum indeg descriptor from which 11 other reverse degree-based topological descriptors can be obtained. To compute the general reverse inverse sum indeg descriptor for the molecular structure of the graphene under study, we considered four different cases based on the number of rows (l) and the number of benzene rings in each row (k). Initially, the case in which the number of rows and the number of rings in each row were both greater than one was considered, as shown in Figure 5 and Figure 6 as 3D plots. For the second case, the graphene structure had only one row and more than one benzene ring. Figure 7 shows such a situation. In the third case, there was more than one row with only one benzene ring in each column, as shown in Figure 8 and Figure 9 as 3D plots. Figure 10 represents the last case where there was only one benzene ring. Using these four cases and edge partitioning as well as degree counting and graph structure analysis, the reverse general inverse sum indeg descriptor of graphene (Ǥ) was derived as follows:
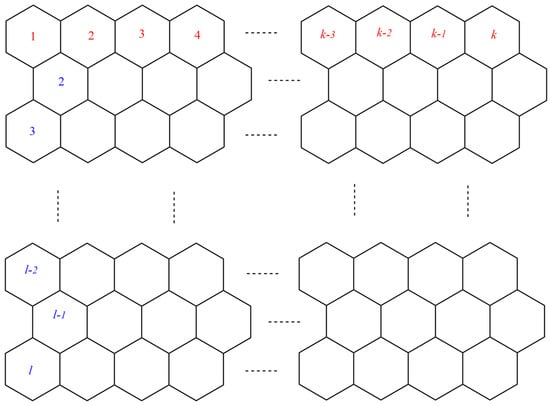
Figure 5.
Graphene structure with .
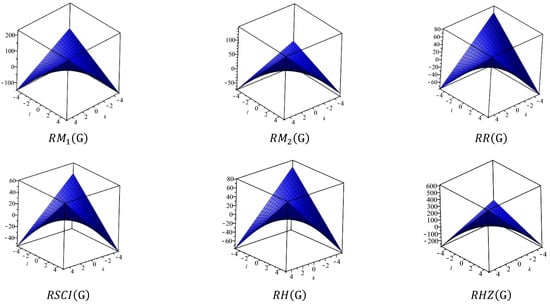
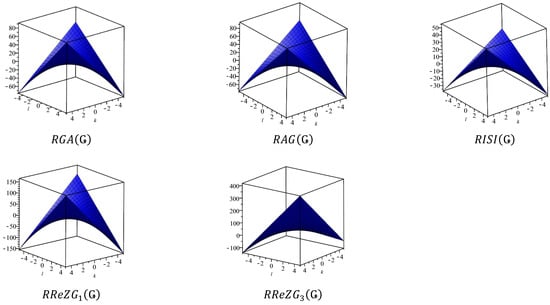
Figure 6.
3D Plots when .

Figure 7.
Graphene structure with .
Figure 8.
Graphene structure with .
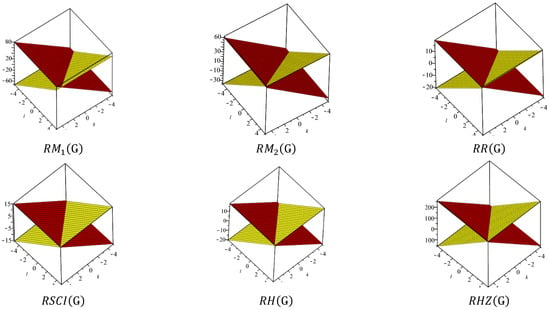
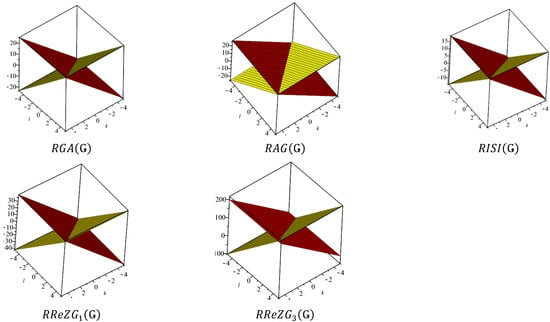
Figure 9.
3D plots when and
Figure 10.
Graphene structure with .
Theorem 1.
The reverse general inverse sum indeg descriptor of graphene is as follows:
if
Proof.
The proof was built by taking the four cases into account. □
Case 1.
From the graph structure analysis, the reverse edge partition of graphene whencontainededges, edges, andedges.
Then, applying the definition of the reverse general inverse sum indeg descriptor, , we arrived at
Using Table 1, in Equation (1), the following 11 reverse topological descriptors for the graphene when were obtained.
Remark 1.
- (i)
- (ii)
- (iii)
- (iv)
- (v)
- (vi)
- (vii)
- (viii)
- (ix)
- (x)
- (xi)
Case 2.
When the reverse edge partition of graphene contained edges, edges, and edges.
Applying the definition of the reverse general inverse sum indeg descriptor, ,
Using Table 1, in Equation (2), we noted the following 11 reverse topological descriptors for the graphene when
Remark 2.
- (i)
- (ii)
- (iii)
- (iv)
- (v)
- (vi)
- (vii)
- (viii)
- (ix)
- (x)
- (xi)
Case 3.
Forthe reverse edge partition of the graphene containsedges, edges, andedges.
Using the definition of the reverse general inverse sum indeg descriptor, ,
Using Table 1, in Equation (3), we noted the following reverse topological descriptors for the graphene when
Remark 3.
- (i)
- (ii)
- (iii)
- (iv)
- (v)
- (vi)
- (vii)
- (viii)
- (ix)
- (x)
- (xi)
Case 4.
Whenthe reverse edge partition of the graphene contained onlyedges, and by the definition of reverse general inverse sum indeg descriptor, ,
In this case, we noted the following 11 reverse topological descriptors for the graphene as follows:
Remark 4.
- (i)
- (ii).
- (ii)
- (iv).
- (iii)
- (vi).
- (iv)
- (viii).
- (v)
- (x).
- (vi)
In Table 5, the numerical values of the 11 reverse topological descriptors calculated with graphene’s analytical expressions when are presented. From Table 5, it is possible to see how individual reverse topological descriptor differ and how they are similar. The computational results show that reverse topological descriptors are highly dependent on the values of l and k. As these values increase, the magnitude of all reverse descriptors also increases, and this can be visualized by the 3D graphical representation in Figure 11.

Table 5.
Numerical values of the graphene for .
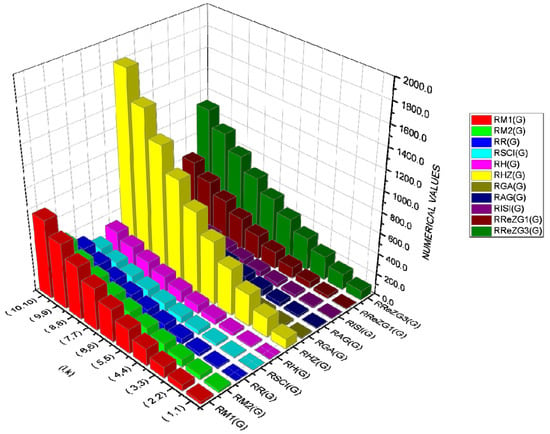
Figure 11.
An interactive visualization of Table 5.
4. Conclusions
In this paper, we presented a reverse general inverse sum inverse degree descriptor from which one can derive a set of reverse degree-based topological descriptors. In order to assess the predictive potential of , we selected the exchange-correlation energy of the graphene sheets as a data example. Based on the results obtained in this article, we can summarize them as follows:
- The regression models (Table 4) derived from reverse topological descriptors in the present article were extremely accurate for predicting the exchange-correlation energies in the graphene sheets.
- The reverse sum−connectivity descriptor with was the best predictor among the 11 studied descriptors. Meanwhile, the reverse redefined first Zagreb descriptor performed poorly.
- The density functional theory (DFT) calculations of the electronic structure, such as the exchange-correlation energies of the graphene sheets, were precise; however, they were computationally expensive while the reverse topological descriptors models presented herein required minimal computations and provided high levels of accuracy.
- Analytical expressions of the reverse first and second Zagreb descriptor, reverse Randić descriptor, reverse sum−connectivity descriptor, reverse harmonic descriptor, reverse hyper Zagreb descriptor, reverse geometric−arithmetic descriptor, reverse inverse sum indeg descriptor, and reverse redefined first and third Zagreb descriptors have been obtained for graphene structures.
- Researchers who are trying to better understand the behaviour of graphene are likely to find the numerical values and graphical representations presented in this article helpful.
Author Contributions
Conceptualization, M.A., P.A., W.H.E.-G. and H.A.E.-H.; methodology, M.A., P.A., W.H.E.-G. and H.A.E.-H.; software, M.A. and P.A.; validation, W.H.E.-G. and H.A.E.-H.; formal analysis, P.A. and H.A.E.-H.; investigation, P.A., W.H.E.-G. and H.A.E.-H.; data curation, M.A., P.A., W.H.E.-G. and H.A.E.-H.; writing—original draft preparation, P.A.; writing—review and editing, M.A. and W.H.E.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the raw data supporting the conclusion of this paper were provided by the authors.
Acknowledgments
Authors would like to thank the Deanship of Scientific Research, Qassim University, for funding publication of this project.
Conflicts of Interest
The authors declare that they have no competing interest.
References
- Iijima, S. Helical microtubules of graphitic carbon. Nature 1991, 354, 56–58. [Google Scholar] [CrossRef]
- Kroto, H.W. JR HEATH, SC O’BRIEN, RF CURL, AND RE SMALLEy. Astrophys. J. 1987, 314, 352. [Google Scholar] [CrossRef]
- Novoselov, K.S.; Geim, A.K.; Morozov, S.V.; Jiang, D.; Zhang, Y.; Dubonos, S.V.; Grigorieva, I.V.; Firsov, A.A. Electric field effect in atomically thin carbon films. Science 2004, 306, 666–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, D.D.L. Review graphite. J. Mater. Sci. 2002, 37, 1475–1489. [Google Scholar] [CrossRef]
- Robertson, J. Amorphous carbon. Adv. Phys. 1986, 35, 317–374. [Google Scholar] [CrossRef]
- Spitsyn, B.; Bouilov, L.; Derjaguin, B. Vapor growth of diamond on diamond and other surfaces. J. Cryst. Growth 1981, 52, 219–226. [Google Scholar] [CrossRef]
- Kalman, J.; Torrent, F.; Navas, J.M. Cytotoxicity of three graphene-related materials in rainbow trout primary hepatocytes is not associated to cellular internalization. Ecotoxicol. Environ. Saf. 2022, 231, 113227. [Google Scholar] [CrossRef]
- Kobyliukh, A.; Olszowska, K.; Godzierz, M.; Kordyka, A.; Kubacki, J.; Mamunya, Y.; Pusz, S.; Stoycheva, I.; Szeluga, U. Effect of graphene material structure and iron oxides deposition method on morphology and properties of graphene/iron oxide hybrids. Appl. Surf. Sci. 2021, 573, 151567. [Google Scholar] [CrossRef]
- Yildiz, G.; Bolton-Warberg, M.; Awaja, F. Graphene and graphene oxide for bio-sensing: General properties and the effects of graphene ripples. Acta Biomater. 2021, 131, 62–79. [Google Scholar] [CrossRef]
- Clapa, M.; Gaj, J. Behavior of graphene under glow discharge plasma. Sens. Actuators A Phys. 2021, 332, 113069. [Google Scholar] [CrossRef]
- Park, S.; Ji, S.G.; Yoon, Y.; Kim, S.K.; Song, W.; Myung, S.; Lim, J.; Jung, H.-K.; Lee, S.S.; An, K.-S. Fabrication of fanlike L-shaped graphene nanostructures with enhanced thermal/electrochemical properties via laser irradiation. Carbon 2021, 182, 691–699. [Google Scholar] [CrossRef]
- Alam Amin, M.S.; Stüber, F.; Giralt, J.; Fortuny, A.; Fabregat, A.; Font, J. Ceramic-supported graphene oxide membrane bioreactor for the anaerobic decolorization of azo dyes. J. Water Process Eng. 2021, 45, 102499. [Google Scholar] [CrossRef]
- Pinelli, F.; Nespoli, T.; Fiorati, A.; Farè, S.; Magagnin, L.; Rossi, F. Graphene nanoplatelets can improve the performances of graphene oxide—Polyaniline composite gas sensing aerogels. Carbon Trends 2021, 5, 100123. [Google Scholar] [CrossRef]
- Shimomura, K.; Imai, K.; Nakagawa, K.; Kawai, A.; Hashimoto, K.; Ideguchi, T.; Maki, H. Graphene photodetectors with asymmetric device structures on silicon chips. Carbon Trends 2021, 5, 100100. [Google Scholar] [CrossRef]
- Kamsong, W.; Primpray, V.; Pasakon, P.; Sriprachuabwong, C.; Pakapongpan, S.; Mensing, J.P.; Wisitsoraat, A.; Tuantranont, A.; Karuwan, C. Highly sensitive and disposable screen-printed ionic liquid/graphene based electrochemical sensors. Electrochem. Commun. 2022, 135, 107209. [Google Scholar] [CrossRef]
- Tarasova, B.P.; Arbuzova, A.A.; Volodina, A.A.; Fursikova, P.V.; Mozhzhuhina, S.A.; Lototskyyd, M.V.; Yartyse, V.A. Metal hydride–Graphene composites for hydrogen based energy storage. J. Alloys Compd. 2022, 896, 162881. [Google Scholar] [CrossRef]
- Ivanciuc, O.; Balaban, A.T. The graph description of chemical structures. In Topological Indices and Related Descriptors in QSAR and QSPAR; CRC Press: Boca Raton, FL, USA, 2000; pp. 69–178. [Google Scholar]
- Trinajstic, N. Chemical graphs. In Chemical Graph Theory; CRC Press: Boca Raton, USA, 2018; pp. 45–60. [Google Scholar]
- Todeschini, R.; Consonni, V. Handbook of Molecular Descriptors; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Mauri, A.; Consonni, V.; Todeschini, R. Molecular descriptors. In Handbook of Computational Chemistry; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Wiener, H. Correlation of heats of isomerization, and differences in heats of vaporization of isomers, among the paraffin hydrocarbons. J. Am. Chem. Soc. 1947, 69, 2636–2638. [Google Scholar] [CrossRef]
- Gutman, I.; Milovanović, E.; Milovanović, I. Beyond the Zagreb indices. AKCE Int. J. Graphs Comb. 2020, 17, 74–85. [Google Scholar] [CrossRef]
- Gupta, S.; Singh, M.; Madan, A.K. Connective eccentricity index: A novel topological descriptor for predicting biological activity. J. Mol. Graph. Model. 2000, 18, 18–25. [Google Scholar] [CrossRef]
- Redžepović, I.; Furtula, B. Predictive potential of eigenvalue-based topological molecular descriptors. J. Comput.-Aided Mol. Des. 2020, 34, 975–982. [Google Scholar] [CrossRef]
- Gutman, I.; Trinajstić, N. Graph theory and molecular orbitals. Total φ-electron energy of alternant hydrocarbons. Chem. Phys. Lett. 1972, 17, 535–538. [Google Scholar] [CrossRef]
- Randic, M. Characterization of molecular branching. J. Am. Chem. Soc. 1975, 97, 6609–6615. [Google Scholar] [CrossRef]
- Zhou, B.; Trinajstić, N. On general sum-connectivity index. J. Math. Chem. 2010, 47, 210–218. [Google Scholar] [CrossRef]
- Vukičević, D. Bond additive modeling 2. Mathematical properties of max-min rodeg index. Croat. Chem. Acta 2010, 83, 261–273. [Google Scholar]
- Wei, J.; Cancan, M.; Rehman, A.U.; Siddiqui, M.K.; Nasir, M.; Younas, M.T.; Hanif, M.F. On topological indices of remdesivir compound used in treatment of Corona virus (COVID 19). Polycycl. Aromat. Compd. 2021. [Google Scholar] [CrossRef]
- Kulli, V.R. On the sum connectivity reverse index of oxide and honeycomb networks. J. Comput. Math. Sci. 2017, 8, 408–413. [Google Scholar]
- Bakhshi, K.; Mojallal, S.A. Relation Between Topological Indices and Exchange-Correlation Energy for Graphene Nanosurface: A DFT Study. Fuller. Nanotub. Carbon Nanostructures 2013, 21, 617–623. [Google Scholar] [CrossRef]
- Sridhara, G.; Kanna, M.R.R.; Indumathi, R.S. Computation of Topological Indices of Graphene. J. Nanomater. 2015, 2015, 969348. [Google Scholar] [CrossRef] [Green Version]
- Shigehalli, V.S.; Kanabur, R. Computation of New Degree-Based Topological Indices of Graphene. J. Math. 2016, 2016, 4341919. [Google Scholar] [CrossRef]
- Jagadeesh, R.; Kanna, M.R.; Indumathi, R.S. Some results on topological indices of graphene. Nanomater. Nanotechnol 2016. [Google Scholar] [CrossRef] [Green Version]
- Berhe, M.; Wang, C. Computation of certain topological coindices of graphene sheet and () nanotubes and nanotorus. Appl. Math. Nonlinear Sci. 2019, 4, 455–468. [Google Scholar] [CrossRef] [Green Version]
- Mondal, S.; De, N.; Pal, A. Topological properties of Graphene using some novel neighborhood degree-based topological indices. Int. J. Math. Ind. 2019, 11, 1950006. [Google Scholar] [CrossRef] [Green Version]
- Parashivamurthy, H.L.; Kanna, M.R.R.; Jagadeesh, R. Some Topological Indices and Their Polynomials of Graphene. Orient. J. Chem. 2019, 35, 1514–1518. [Google Scholar] [CrossRef]
- Rada, J. Vertex-Degree Based Topological Indices of Graphene. Polycycl. Aromat. Compd. 2020. [Google Scholar] [CrossRef]
- Sudhakara, K.B.; Guruprasad, P.S.; Sriraj, M.A. Some edge degree based topological indices of Graphene. Proc. Jangjeon Math. Soc. 2020, 23, 253–270. [Google Scholar]
- Shanmukha, M.C.; Usha, A.; Basavarajappa, N.S.; Shilpa, K.C. Graph entropies of porous graphene using topological indices. Comput. Theor. Chem. 2021, 1197, 113142. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).