Quantification of Bacterial DNA from Infected Human Root Canals Using qPCR and DAPI after Disinfection with Established and Novel Irrigation Protocols
Abstract
:1. Introduction
2. Materials and Methods
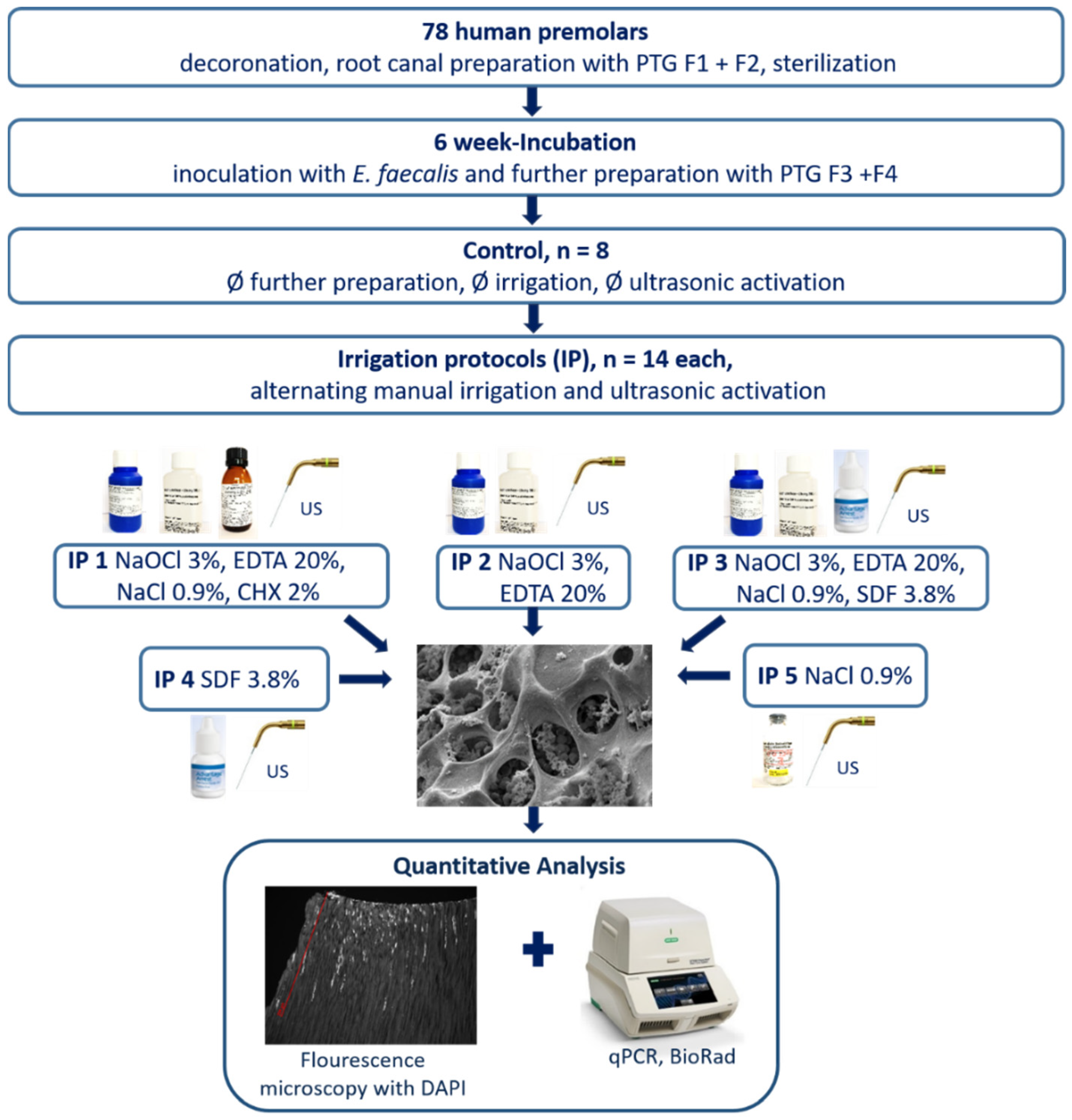
2.1. Chemicals, Bacterial Strains, and Teeth
2.2. Inoculation with E. faecalis
2.3. Application of the Irrigation Protocols
2.4. Preparation of Specimens for DAPI Staining and Microscopical Analyses
2.5. Grinding of Teeth and Purification of DNA and qPCR
2.6. Quantitative Real-Time PCR and Quantification of Bacterial Colonization
2.7. Statistical Analyses
3. Results
3.1. Outline of the Procedure for Evaluation of Novel and Established Irrigation Protocols
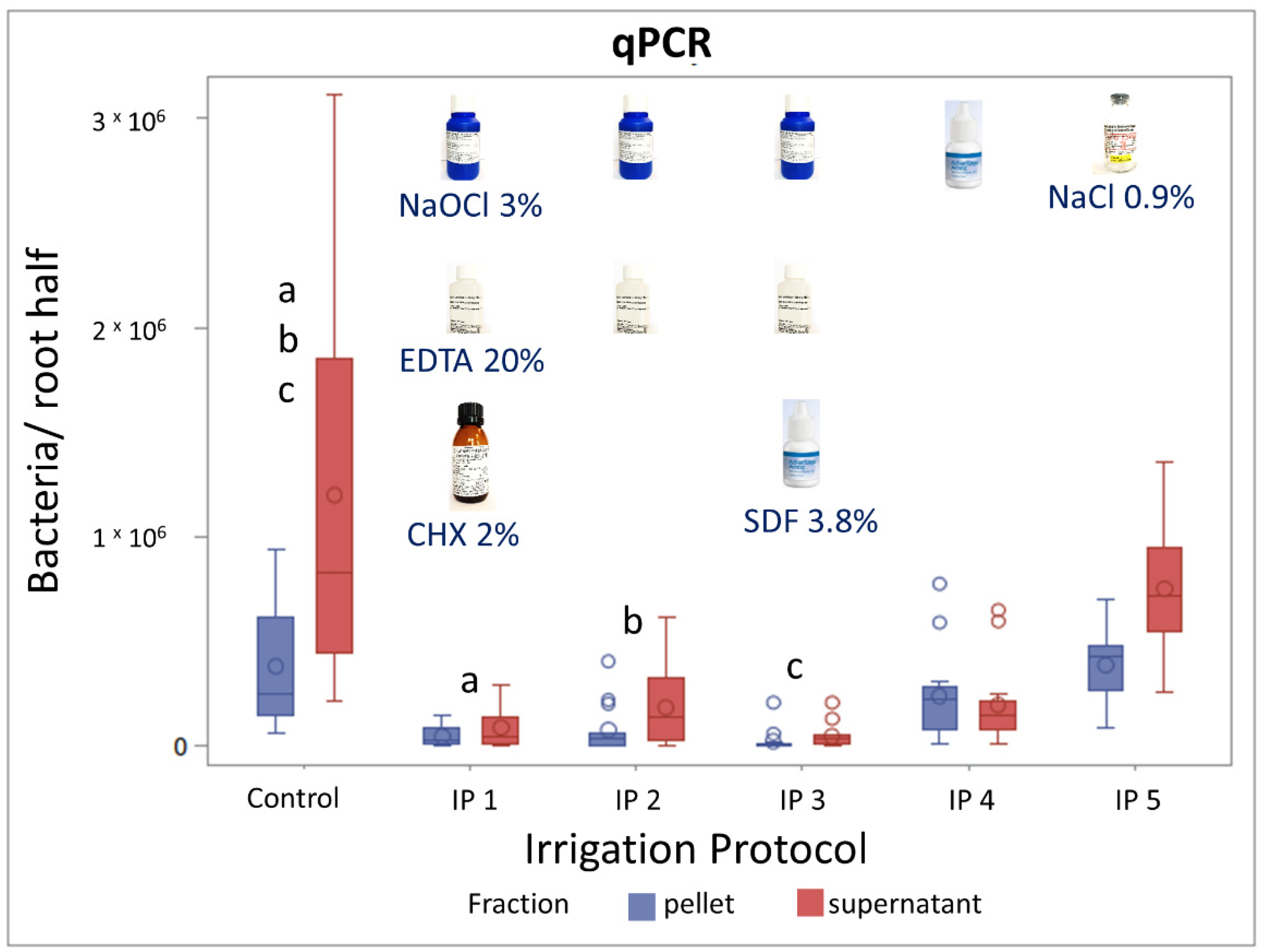
3.2. Evaluation of Irrigation Protocols by qPCR
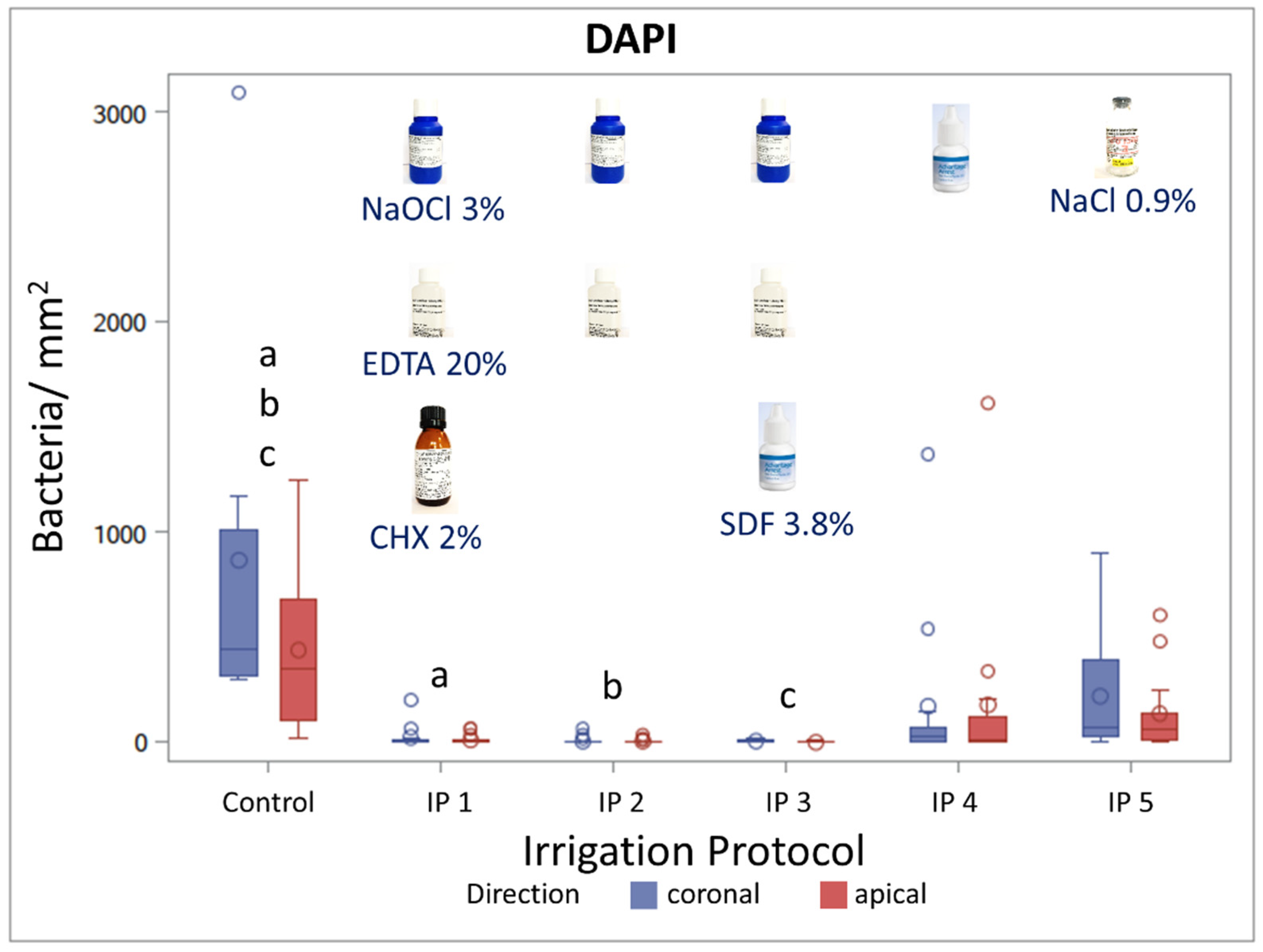
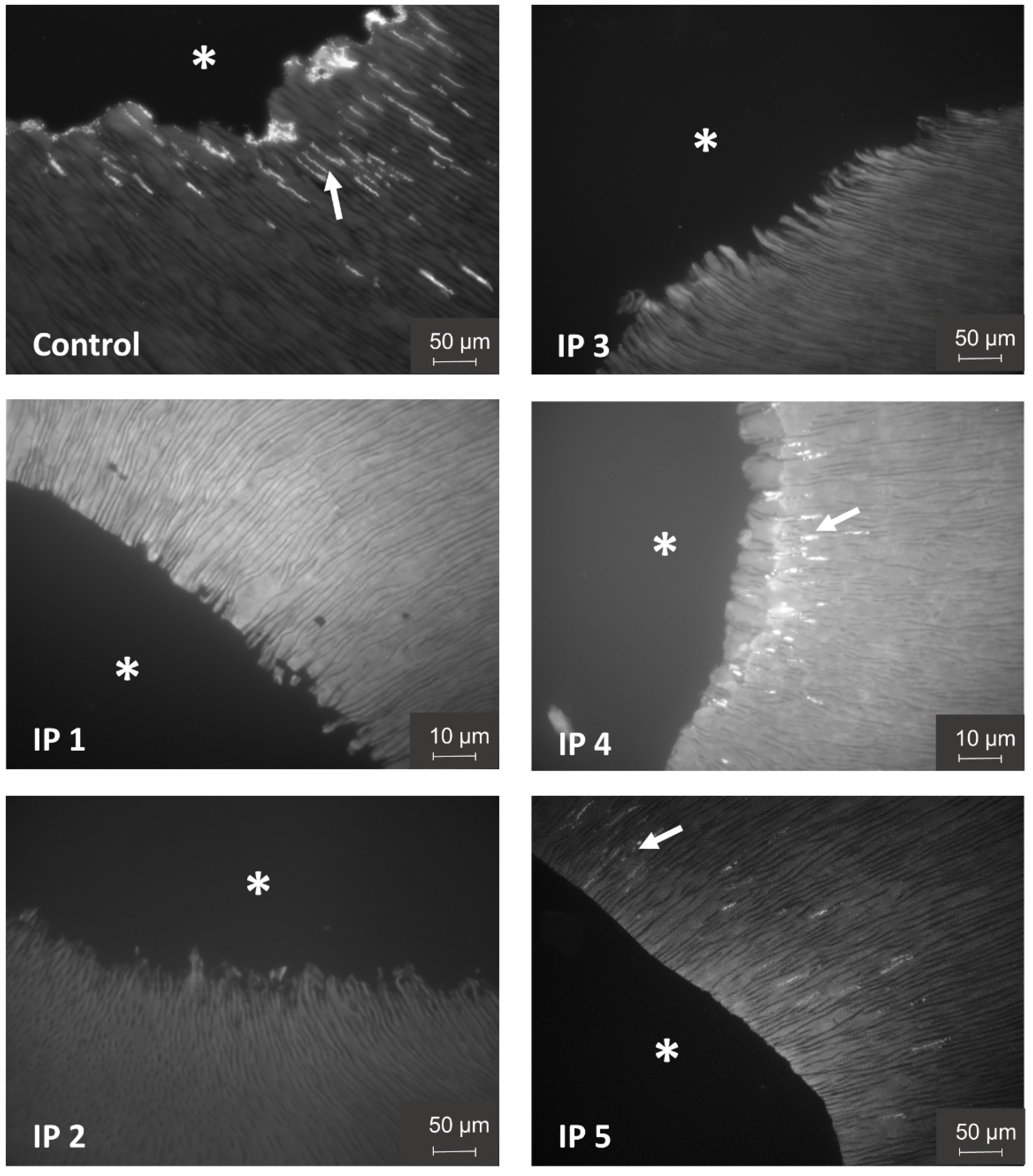
3.3. Evaluation of Irrigation Protocols by DAPI Staining
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tzanetakis, G.N.; Azcarate-Peril, A.M.; Zachaki, S.; Panopoulos, P.; Kontakiotis, E.G.; Madianos, P.N.; Divaris, K. Comparison of Bacterial Community Composition of Primary and Persistent Endodontic Infections Using Pyrosequencing. J. Endod. 2015, 41, 1226–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzanetakis, G.N.; Giannakoulas, D.G.; Papanakou, S.; Gizani, S.; Lygidakis, N. Regenerative endodontic therapy of immature permanent molars with pulp necrosis: A cases series and a literature review. Eur. Arch. Paediatr. Dent. 2021, 22, 515–525. [Google Scholar] [CrossRef]
- Sørensen, L.H.; Kirkevang, L.L. Establishment of a Danish endodontic practice-based research network: Baseline data. Acta Odontol. Scand. 2021, 79, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Rôças, I.N.; Siqueira, J.F. Root canal microbiota of teeth with chronic apical periodontitis. J. Clin. Microbiol. 2008, 46, 3599–3606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plotino, G.; Cortese, T.; Grande, N.M.; Leonardi, D.P.; Di Giorgio, G.; Testarelli, L.; Gambarini, G. New Technologies to Improve Root Canal Disinfection. Braz. Dent. J. 2016, 27, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Nair, P.N.R.; Sjögren, U.; Krey, G.; Kahnberg, K.-E.; Sundqvist, G. Intraradicular Bacteria and Fungi in Root-filled, Asymptomatic Human Teeth with Therapy-resistant Periapical Lesions: A Long-term Light and Electron Microscopic Follow-up Study. J. Endod. 1990, 16, 580–588. [Google Scholar] [CrossRef]
- Siqueira, J.F.; De Uzeda, M. Disinfection by calcium hydroxide pastes of dentinal tubules infected with two obligate and one facultative anaerobic bacteria. J. Endod. 1996, 22, 674–676. [Google Scholar] [CrossRef]
- Siqueira, J.F.; Rôças, I.N. Clinical Implications and Microbiology of Bacterial Persistence after Treatment Procedures. J. Endod. 2008, 34, 1291–1301. [Google Scholar] [CrossRef]
- Peters, O.A.; Laib, A.; Göhring, T.N.; Barbakow, F. Changes in root canal geometry after preparation assessed by high-resolution computed tomography. J. Endod. 2001, 27, 1–6. [Google Scholar] [CrossRef]
- Kirsch, J.; Basche, S.; Neunzehn, J.; Dede, M.; Dannemann, M.; Hannig, C.; Weber, M.T. Is it really penetration? Locomotion of devitalized Enterococcus faecalis cells within dentinal tubules of bovine teeth. Arch. Oral Biol. 2017, 83, 289–296. [Google Scholar] [CrossRef]
- Kirsch, J.; Basche, S.; Neunzehn, J.; Dede, M.; Dannemann, M.; Hannig, C.; Weber, M.T. Is it really penetration? Part 2. Locomotion of Enterococcus faecalis cells within dentinal tubules of bovine teeth. Clin. Oral Investig. 2019, 23, 4325–4334. [Google Scholar] [CrossRef] [PubMed]
- Love, R.M.; Jenkinson, H.F. Invasion of dentinal tubules by oral bacteria. Crit. Rev. Oral Biol. Med. Off. Publ. Am. Assoc. Oral Biol. 2002, 13, 171–183. [Google Scholar] [CrossRef] [Green Version]
- Molander, A.; Reit, C.; Dahlén, G.; Kvist, T. Microbiological status of root-filled teeth with apical periodontitis. Int. Endod. J. 1998, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rocas, I.; Siquera, J.; Santos, K. Association of Enterococcus faecalis with different forms of periradicular. J. Endod. 2004, 30, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.F.; Rôças, I.N. Polymerase chain reaction-based analysis of microorganisms associated with failed endodontic treatment. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2004, 97, 85–94. [Google Scholar] [CrossRef]
- Love, R.M. Enterococcus faecalis-A mechanism for its role in endodontic failure. Int. Endod. J. 2001, 34, 399–405. [Google Scholar] [CrossRef]
- Kayaoglu, G.; Erten, H.; Ørstavik, D. Possible role of the adhesin ace and collagen adherence in conveying resistance to disinfectants on Enterococcus faecalis. Oral Microbiol. Immunol. 2008, 23, 449–454. [Google Scholar] [CrossRef]
- Boccella, M.; Santella, B.; Pagliano, P.; De Filippis, A.; Casolaro, V.; Galdiero, M.; Borrelli, A.; Capunzo, M.; Boccia, G.; Franci, G. Prevalence and antimicrobial resistance of enterococcus species: A retrospective cohort study in Italy. Antibiotics 2021, 10, 1552. [Google Scholar] [CrossRef]
- Santella, B.; Folliero, V.; Della Rocca, M.T.; Zannella, C.; Pignataro, D.; Montella, F.; Folgore, A.; Greco, G.; Galdiero, M.; Galdiero, M.; et al. Distribution of antibiotic resistance among Enterococcus spp. isolated from 2017 to 2018 at the University Hospital “Luigi Vanvitelli” of Naples, Italy. Int. J. Mol. Clin. Microbiol. 2019, 9, 1197–1204. [Google Scholar]
- Buetti, N.; Wassilew, N.; Rion, V.; Senn, L.; Gardiol, C.; Widmer, A.; Marschall, J. Emergence of vancomycin-resistant enterococci in Switzerland: A nation-wide survey. Antimicrob. Resist. Infect. Control 2019, 8, 16. [Google Scholar] [CrossRef] [Green Version]
- Hiraishi, N.; Yiu, C.K.Y.; King, N.M.; Tagami, J.; Tay, F.R. Antimicrobial efficacy of 3.8% silver diamine fluoride and its effect on root dentin. J. Endod. 2010, 36, 1026–1029. [Google Scholar] [CrossRef] [PubMed]
- Mathew, V.B.; Madhusudhana, K.; Sivakumar, N.; Venugopal, T.; Reddy, R.K. Anti-microbial efficiency of silver diamine fluoride as an endodontic medicament-An ex vivo study. Contemp. Clin. Dent. 2012, 3, 262–264. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.J.-Y.; Botelho, M.G.; Matinlinna, J.P. Silver compounds used in dentistry for caries management: A review. J. Dent. 2012, 40, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; McGrath, C.; Lo, E.C.M.; Li, J.Y. Silver diamine fluoride and education to prevent and arrest root caries among community-dwelling elders. Caries Res. 2013, 47, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, V.E.; Filho, A.V.; Ribeiro Targino, A.G.; Pelagio Flores, M.A.; Galembeck, A.; Caldas, A.F.; Rosenblatt, A. A new “silver-Bullet” to treat caries in children-Nano Silver Fluoride: A randomised clinical trial. J. Dent. 2014, 42, 945–951. [Google Scholar] [CrossRef] [Green Version]
- Urquhart, O.; Tampi, M.P.; Pilcher, L.; Slayton, R.L.; Araujo, M.W.B.; Fontana, M.; Guzmán-Armstrong, S.; Nascimento, M.M.; Nový, B.B.; Tinanoff, N.; et al. Nonrestorative Treatments for Caries: Systematic Review and Network Meta-analysis. J. Dent. Res. 2019, 98, 14–26. [Google Scholar] [CrossRef]
- Al-Madi, E.M.; Al-Jamie, M.A.; Al-Owaid, N.M.; Almohaimede, A.A.; Al-Owid, A.M. Antibacterial efficacy of silver diamine fluoride as a root canal irrigant. Clin. Exp. Dent. Res. 2019, 5, 551–556. [Google Scholar] [CrossRef] [Green Version]
- Görduysus, M.Ö.; Yilmaz, Z.; Görduysus, M.; Kaya, C.; Gülmez, D.; Emini, L.; Hoxha, V. Bacterial Reduction in Infected Root Canals Treated With Calcium Hydroxide Using Hand and Rotary Instrument: An In-Vivo Study. Clin. Dent. Res. 2012, 36, 15–21. [Google Scholar]
- Tan, K.S.; Yu, V.S.H.; Quah, S.Y.; Bergenholtz, G. Rapid method for the detection of root canal bacteria in endodontic therapy. J. Endod. 2015, 41, 447–450. [Google Scholar] [CrossRef]
- Herzog, D.B.; Hosny, N.A.; Niazi, S.A.; Koller, G.; Cook, R.J.; Foschi, F.; Watson, T.F.; Mannocci, F.; Festy, F. Rapid Bacterial Detection during Endodontic Treatment. J. Dent. Res. 2017, 96, 626–632. [Google Scholar] [CrossRef]
- Ruiz-Linares, M.; Aguado-Pérez, B.; Baca, P.; Arias-Moliz, M.T.; Ferrer-Luque, C.M. Efficacy of antimicrobial solutions against polymicrobial root canal biofilm. Int. Endod. J. 2017, 50, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Tsesis, I.; Elbahary, S.; Venezia, N.B.; Rosen, E. Bacterial colonization in the apical part of extracted human teeth following root-end resection and filling: A confocal laser scanning microscopy study. Clin. Oral Investig. 2018, 22, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Mohmmed, S.A.; Vianna, M.E.; Penny, M.R.; Hilton, S.T.; Mordan, N.; Knowles, J.C. Confocal laser scanning, scanning electron, and transmission electron microscopy investigation of Enterococcus faecalis biofilm degradation using passive and active sodium hypochlorite irrigation within a simulated root canal model. Microbiologyopen 2017, 6, e455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sterzenbach, T.; Pioch, A.; Dannemann, M.; Hannig, C.; Weber, M.T. Quantification of Bacterial Colonization in Dental Hard Tissues Using Optimized Molecular Biological Methods. Front. Genet. 2020, 11, 599137. [Google Scholar] [CrossRef] [PubMed]
- Sedgley, C.M.; Nagel, A.C.; Shelburne, C.E.; Clewell, D.B.; Appelbe, O.; Molander, A. Quantitative real-time PCR detection of oral Enterococcus faecalis in humans. Arch. Oral Biol. 2005, 50, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Cherian, B.; Gehlot, P.M.; Manjunath, M.K. Comparison of the antimicrobial efficacy of octenidine dihydrochloride and chlorhexidine with and without passive ultrasonic irrigation-An invitro study. J. Clin. Diagn. Res. 2016, 10, ZC71–ZC77. [Google Scholar] [CrossRef]
- Bhuva, B.; Patel, S.; Wilson, R.; Niazi, S.; Beighton, D.; Mannocci, F. The effectiveness of passive ultrasonic irrigation on intraradicular Enterococcus faecalis biofilms in extracted single-rooted human teeth. Int. Endod. J. 2010, 43, 241–250. [Google Scholar] [CrossRef]
- Urban, K.; Donnermeyer, D.; Schäfer, E.; Bürklein, S. Canal cleanliness using different irrigation activation systems: A SEM evaluation. Clin. Oral Investig. 2017, 21, 2681–2687. [Google Scholar] [CrossRef]
- Conde, A.J.; Estevez, R.; Loroño, G.; Valencia de Pablo, O.; Rossi-Fedele, G.; Cisneros, R. Effect of sonic and ultrasonic activation on organic tissue dissolution from simulated grooves in root canals using sodium hypochlorite and EDTA. Int. Endod. J. 2017, 50, 976–982. [Google Scholar] [CrossRef]
- Nagendrababu, V.; Jayaraman, J.; Suresh, A.; Kalyanasundaram, S.; Neelakantan, P. Effectiveness of ultrasonically activated irrigation on root canal disinfection: A systematic review of in vitro studies. Clin. Oral Investig. 2018, 22, 655–670. [Google Scholar] [CrossRef]
- Căpută, P.E.; Retsas, A.; Kuijk, L.; Chávez de Paz, L.E.; Boutsioukis, C. Ultrasonic Irrigant Activation during Root Canal Treatment: A Systematic Review. J. Endod. 2019, 45, 31–44.e13. [Google Scholar] [CrossRef] [PubMed]
- Moreira, R.N.; Pinto, E.B.; Galo, R.; Falci, S.G.M.; Mesquita, A.T. Passive ultrasonic irrigation in root canal: Systematic review and meta-analysis. Acta Odontol. Scand. 2019, 77, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Chen, Z.; Jiang, Y.; Xue, F.; Li, B. Advances and challenges in viability detection of foodborne pathogens. Front. Microbiol. 2016, 7, 1833. [Google Scholar] [CrossRef] [Green Version]
- Codony, F.; Dinh-Thanh, M.; Agustí, G. Key Factors for Removing Bias in Viability PCR-Based Methods: A Review. Curr. Microbiol. 2020, 77, 682–687. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weber, M.-T.; Alkhafaji, Y.; Pioch, A.; Trips, E.; Basche, S.; Dannemann, M.; Kilistoff, A.; Hannig, C.; Sterzenbach, T. Quantification of Bacterial DNA from Infected Human Root Canals Using qPCR and DAPI after Disinfection with Established and Novel Irrigation Protocols. Materials 2022, 15, 1911. https://doi.org/10.3390/ma15051911
Weber M-T, Alkhafaji Y, Pioch A, Trips E, Basche S, Dannemann M, Kilistoff A, Hannig C, Sterzenbach T. Quantification of Bacterial DNA from Infected Human Root Canals Using qPCR and DAPI after Disinfection with Established and Novel Irrigation Protocols. Materials. 2022; 15(5):1911. https://doi.org/10.3390/ma15051911
Chicago/Turabian StyleWeber, Marie-Theres, Yousef Alkhafaji, Anne Pioch, Evelyn Trips, Sabine Basche, Martin Dannemann, Alan Kilistoff, Christian Hannig, and Torsten Sterzenbach. 2022. "Quantification of Bacterial DNA from Infected Human Root Canals Using qPCR and DAPI after Disinfection with Established and Novel Irrigation Protocols" Materials 15, no. 5: 1911. https://doi.org/10.3390/ma15051911
APA StyleWeber, M.-T., Alkhafaji, Y., Pioch, A., Trips, E., Basche, S., Dannemann, M., Kilistoff, A., Hannig, C., & Sterzenbach, T. (2022). Quantification of Bacterial DNA from Infected Human Root Canals Using qPCR and DAPI after Disinfection with Established and Novel Irrigation Protocols. Materials, 15(5), 1911. https://doi.org/10.3390/ma15051911






