Core-Shell Imprinted Particles for Adenovirus Binding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Functionalized Core Particles
2.3. Monitoring of Functionalization
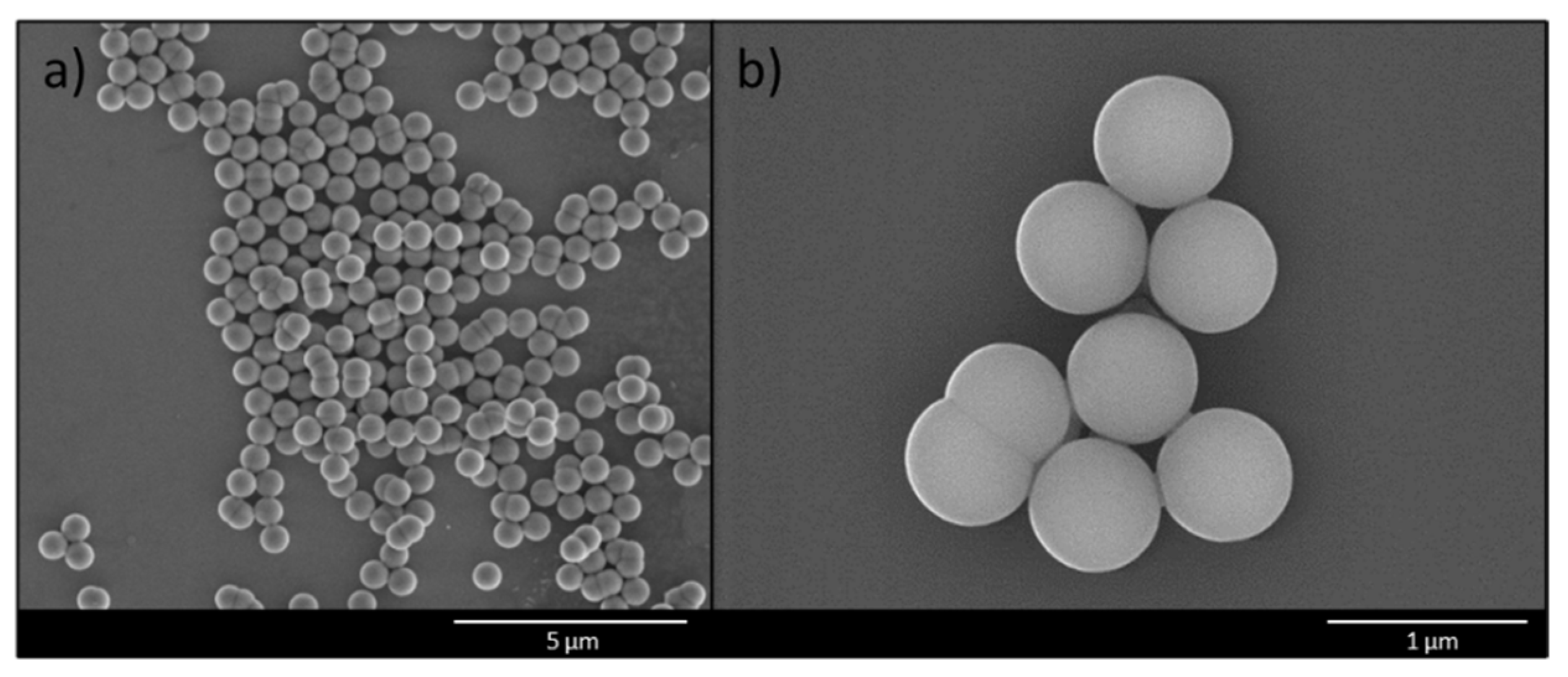
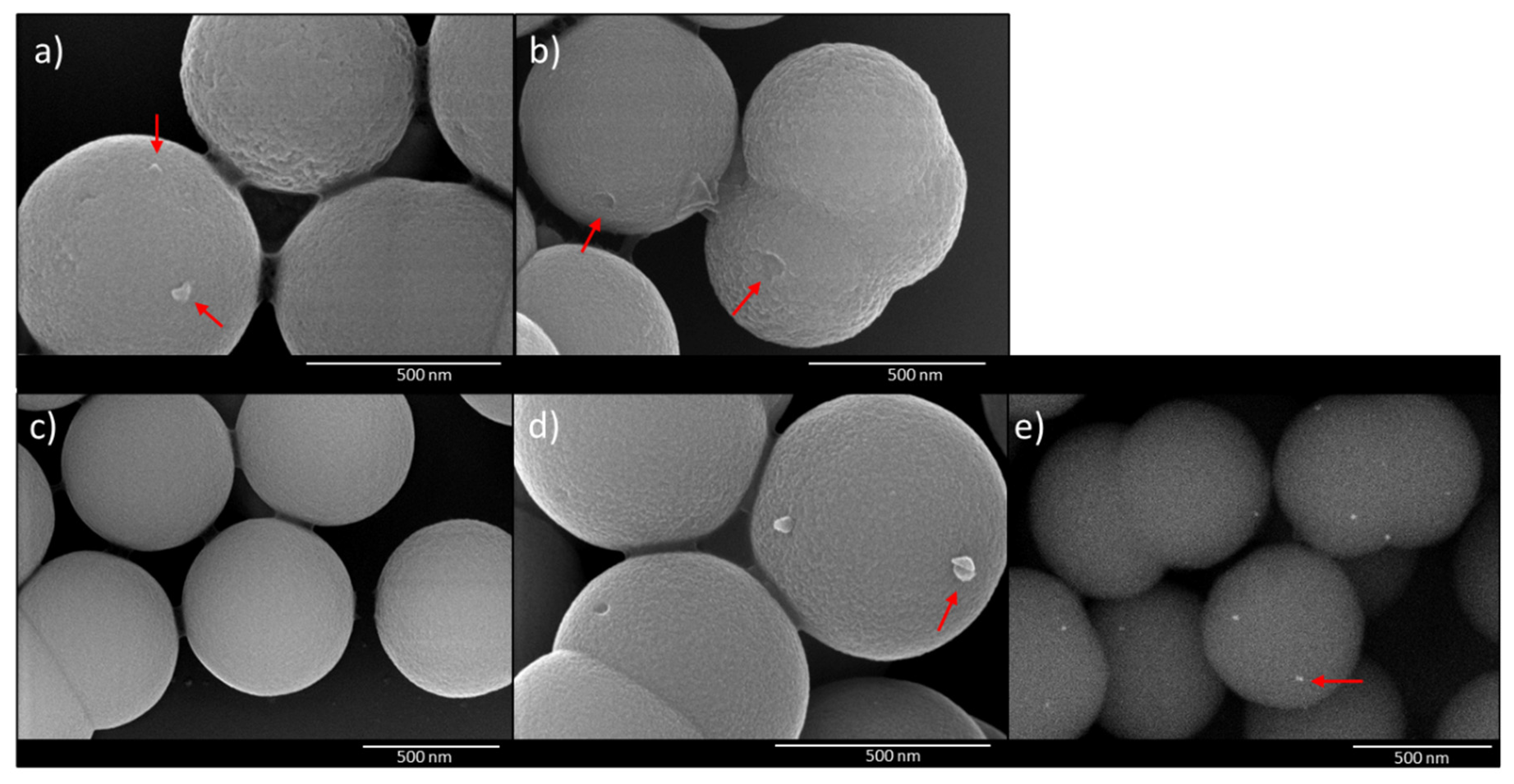
2.4. Scanning Electron Microscopy (SEM)
2.5. Imprinting and Rebinding
2.6. Binding Kinetics
2.7. AdV5 Detection in CCS
2.8. qPCR
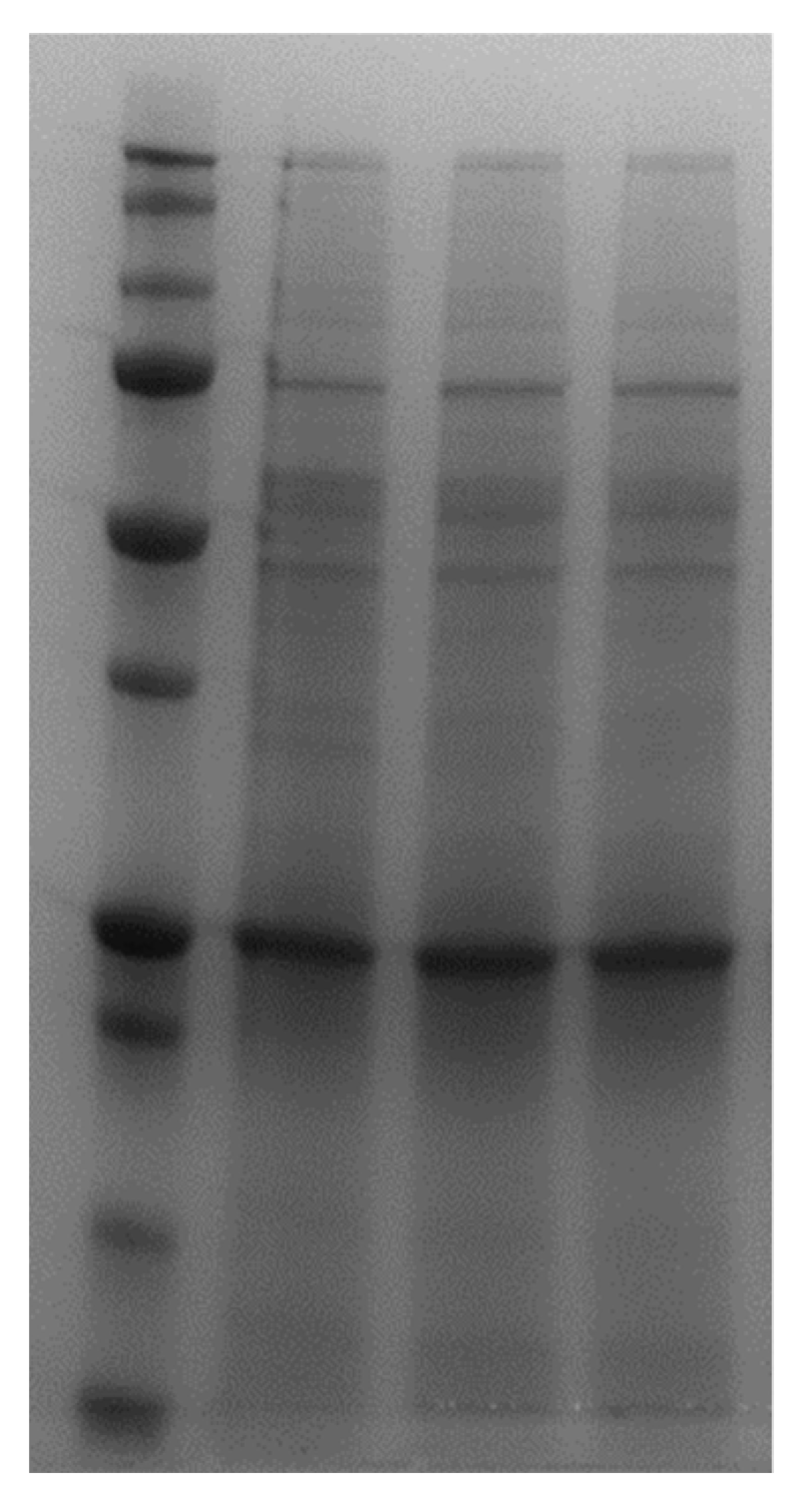
2.9. SDS-PAGE
3. Results and Discussion
3.1. Synthesis and Functionalization of Silica Core Particles
3.2. Binding Kinetics
3.3. Imprinting and Rebinding
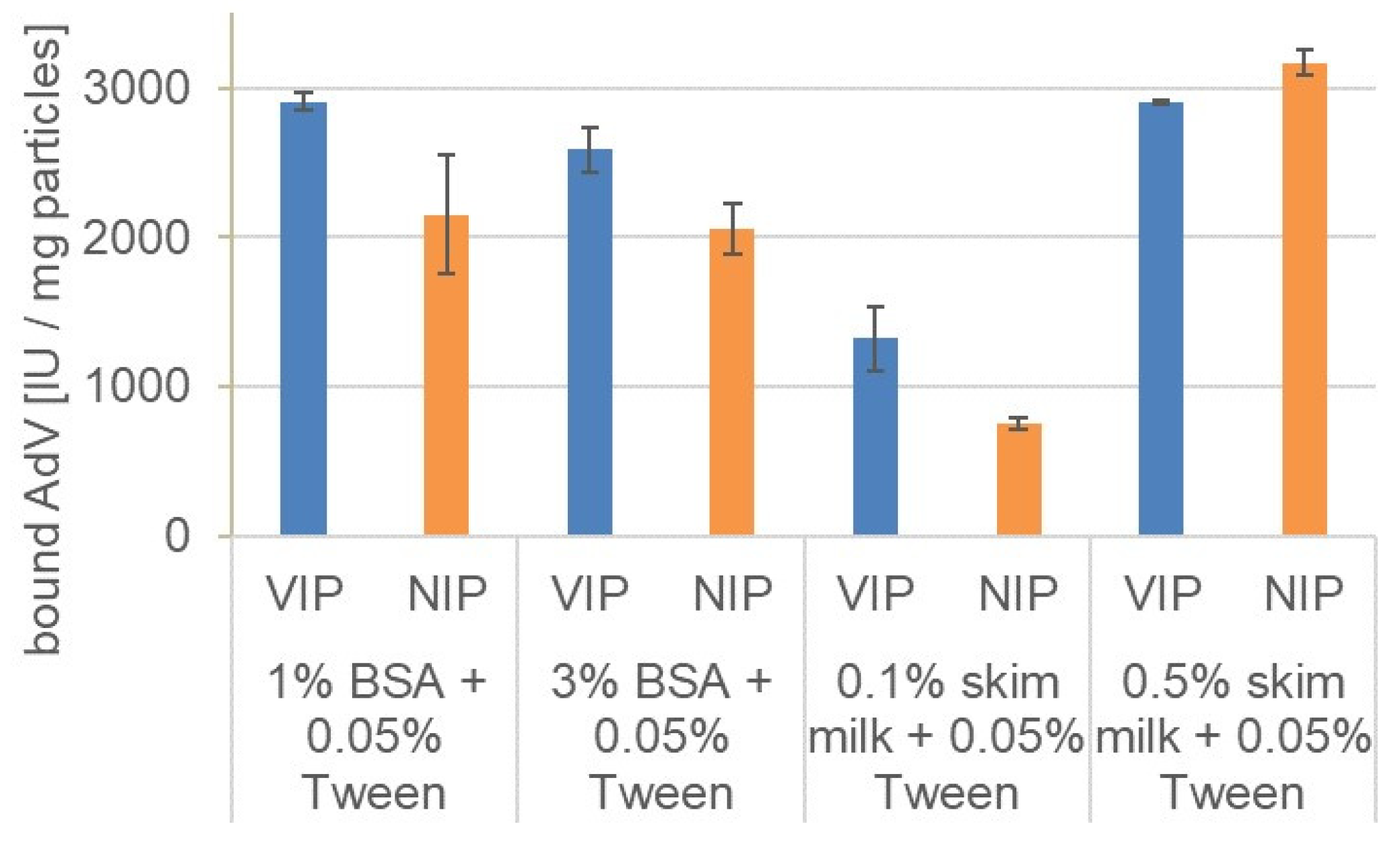
3.4. Rebinding in the Presence of Different Blocking Agents
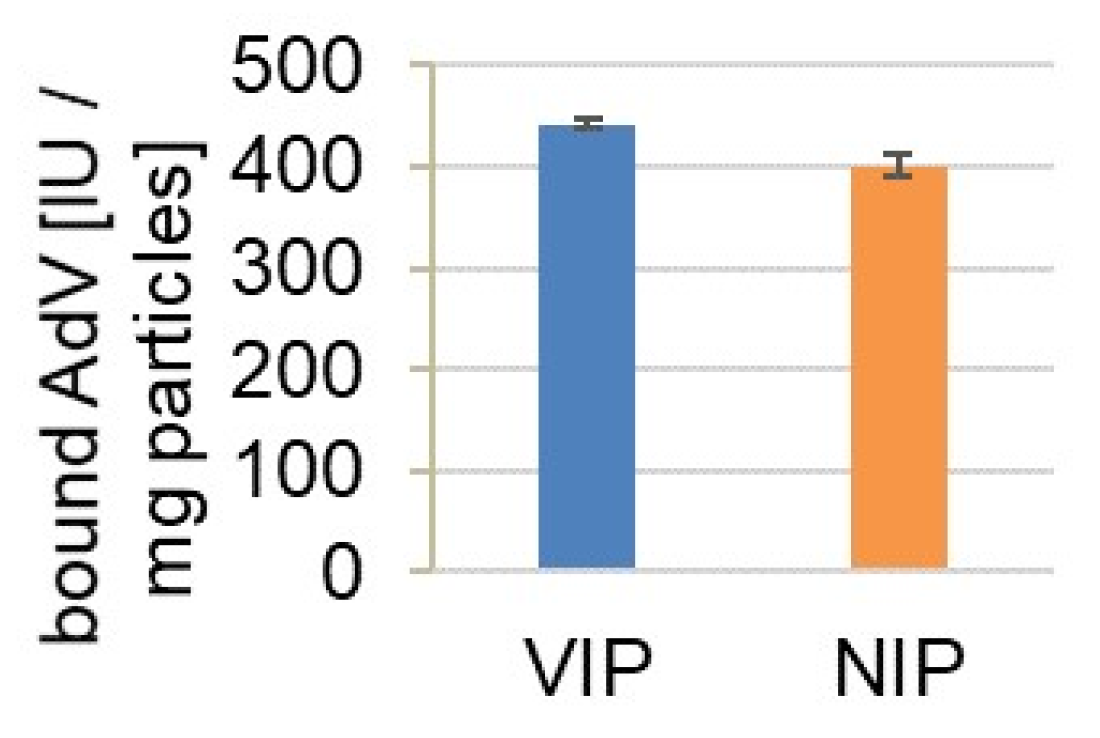
3.5. Binding of AdV5 from Cell Culture Supernatant
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haupt, K. Molecularly imprinted polymers in analytical chemistry. Analyst 2001, 126, 747–756. [Google Scholar] [CrossRef]
- Wei, S.; Mizaikoff, B. Recent advances on noncovalent molecular imprints for affinity separations. J. Sep. Sci. 2007, 30, 1794–1805. [Google Scholar] [CrossRef]
- Haupt, K.; Linares, A.V.; Bompart, M.; Bui, B.T.S. Molecularly Imprinted Polymers. In Molecular Imprinting; Haupt, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 325, pp. 1–28. [Google Scholar]
- Gao, R.; Kong, X.; Wang, X.; He, X.; Chen, L.; Zhang, Y. Preparation and characterization of uniformly sized molecularly imprinted polymers functionalized with core–shell magnetic nanoparticles for the recognition and enrichment of protein. J. Mater. Chem. 2011, 21, 17863–17871. [Google Scholar] [CrossRef]
- Dinc, M.; Esen, C.; Mizaikoff, B. Recent advances on core–shell magnetic molecularly imprinted polymers for biomacromolecules. TrAC Trends Anal. Chem. 2019, 114, 202–217. [Google Scholar] [CrossRef]
- Matsunaga, T.; Hishiya, T.; Takeuchi, T. Surface plasmon resonance sensor for lysozyme based on molecularly imprinted thin films. Anal. Chim. Acta 2007, 591, 63–67. [Google Scholar] [CrossRef]
- Li, S.; Cao, S.; Whitcombe, M.; Piletsky, S. Size matters: Challenges in imprinting macromolecules. Prog. Polym. Sci. 2014, 39, 145–163. [Google Scholar] [CrossRef]
- Boysen, R.I.; Schwarz, L.J.; Nicolau, D.V.; Hearn, M.T.W. Molecularly imprinted polymer membranes and thin films for the separation and sensing of biomacromolecules: Other Techniques. J. Sep. Sci. 2017, 40, 314–335. [Google Scholar] [CrossRef] [PubMed]
- Iskierko, Z.; Sharma, P.S.; Bartold, K.; Pietrzyk-Le, A.; Noworyta, K.; Kutner, W. Molecularly imprinted polymers for separating and sensing of macromolecular compounds and microorganisms. Biotechnol. Adv. 2016, 34, 30–46. [Google Scholar] [CrossRef] [PubMed]
- Gast, M.; Sobek, H.; Mizaikoff, B. Advances in imprinting strategies for selective virus recognition a review. TrAC Trends Anal. Chem. 2019, 114, 218–232. [Google Scholar] [CrossRef]
- Bolisay, L.D.; Culver, J.N.; Kofinas, P. Molecularly imprinted polymers for tobacco mosaic virus recognition. Biomaterials 2006, 27, 4165–4168. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Z.; Jain, V.; Yi, J.; Mueller, S.; Sokolov, J.; Liu, Z.; Levon, K.; Rigas, B.; Rafailovich, M.H. Potentiometric sensors based on surface molecular imprinting: Detection of cancer biomarkers and viruses. Sens. Actuators B Chem. 2010, 146, 381–387. [Google Scholar] [CrossRef]
- Wangchareansak, T.; Thitithanyanont, A.; Chuakheaw, D.; Gleeson, M.P.; Lieberzeit, P.A.; Sangma, C. Influenza A virus molecularly imprinted polymers and their application in virus sub-type classification. J. Mater. Chem. B 2013, 1, 2190–2197. [Google Scholar] [CrossRef]
- Altintas, Z.; Gittens, M.; Guerreiro, A.; Thompson, K.-A.; Walker, J.A.; Piletsky, S.; Tothill, I.E. Detection of Waterborne Viruses Using High Affinity Molecularly Imprinted Polymers. Anal. Chem. 2015, 87, 6801–6807. [Google Scholar] [CrossRef]
- Dickert, F.L.; Hayden, O.; Bindeus, R.; Mann, K.-J.; Blaas, D.; Waigmann, E. Bioimprinted QCM sensors for virus detection?screening of plant sap. Anal. Bioanal. Chem. 2004, 378, 1929–1934. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, T.; Hu, L.; Gong, H.; Chen, C.; Chen, X.; Cai, C. Development of a thermosensitive molecularly imprinted polymer resonance light scattering sensor for rapid and highly selective detection of hepatitis A virus in vitro. Sens. Actuators B Chem. 2017, 253, 1188–1193. [Google Scholar] [CrossRef]
- Gast, M.; Kuehner, S.; Sobek, H.; Walther, P.; Mizaikoff, B. Enhanced Selectivity by Passivation: Molecular Imprints for Viruses with Exceptional Binding Properties. Anal. Chem. 2018, 90, 5576–5585. [Google Scholar] [CrossRef] [PubMed]
- Fabry, C.M.S.; Rosa-Calatrava, M.; Conway, J.F.; Zubieta, C.; Cusack, S.; Ruigrok, R.W.H.; Schoehn, G. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 2005, 24, 1645–1654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghebremedhin, B. Human adenovirus: Viral pathogen with increasing importance. Eur. J. Microbiol. Immunol. 2014, 4, 26–33. [Google Scholar] [CrossRef] [Green Version]
- Reddy, V.S.; Nemerow, G.R. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc. Natl. Acad. Sci. USA 2014, 111, 11715. [Google Scholar] [CrossRef] [Green Version]
- Flint, S.J.; Racaniello, V.R.; Rall, G.F.; Skalka, A.M.; Enquist, L.W. Principles of Virology; ASM Press: Washington, DC, USA, 2015. [Google Scholar]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral vector platforms within the gene therapy landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Stöber, W.; Fink, A.; Bohn, E. Controlled growth of monodisperse silica spheres in the micron size range. J. Colloid Interface Sci. 1968, 26, 62–69. [Google Scholar] [CrossRef]
- Dinc, M.; Basan, H.; Diemant, T.; Behm, R.J.; Lindén, M.; Mizaikoff, B. Inhibitor-assisted synthesis of silica-core microbeads with pepsin-imprinted nanoshells. J. Mater. Chem. B 2016, 4, 4462–4469. [Google Scholar] [CrossRef] [PubMed]
- Dinc, M.; Basan, H.; Hummel, T.; Müller, M.; Sobek, H.; Rapp, I.; Diemant, T.; Behm, R.J.; Lindén, M.; Mizaikoff, B. Selective Binding of Inhibitor-Assisted Surface-Imprinted Core/Shell Microbeads in Protein Mixtures. ChemistrySelect 2018, 3, 4277–4282. [Google Scholar] [CrossRef]
- Gast, M.; Sobek, H.; Mizaikoff, B. Nanoparticle Tracking of Adenovirus by Light Scattering and Fluorescence Detection. Hum. Gene Ther. Methods 2019, 30, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Huyghe, B.G.; Liu, X.; Sutjipto, S.; Sugarman, B.J.; Horn, M.T.; Shepard, H.M.; Scandella, C.J.; Shabram, P. Purification of a Type 5 Recombinant Adenovirus Encoding Human p53 by Column Chromatography. Hum. Gene Ther. 1995, 6, 1403–1416. [Google Scholar] [CrossRef]
- Cumbo, A.; Lorber, B.; Corvini, P.F.-X.; Meier, W.; Shahgaldian, P. A synthetic nanomaterial for virus recognition produced by surface imprinting. Nat. Commun. 2013, 4, 1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbs, J. Effective Blocking Procedures; ELISA Technical Bulletin, Life Sciences No. 3; Corning Incorporated: Kennebunk, ME, USA, 2001. [Google Scholar]
- Lichtenberg, J.Y.; Ling, Y.; Kim, S. Non-Specific Adsorption Reduction Methods in Biosensing. Sensors 2019, 19, 2488. [Google Scholar] [CrossRef] [Green Version]


| Particle | Zeta-Potential [mV] |
| silica | −49.5 ± 3.5 |
| silica-APTES | 31.8 ± 5.1 |
| silica-APTES-glutaraldehyde | 10.8 ± 4.6 |
| Atomic Concentration [%] | ||||
|---|---|---|---|---|
| Particle | C 1s | N 1s | O 1s | Si 2p |
| silica | 12.76 | 0.14 | 57.12 | 29.97 |
| silica-APTES | 16.72 | 0.94 | 52.29 | 30.05 |
| silica-APTES-glutaraldehyde | 22.14 | 1.21 | 49.49 | 27.16 |
| Blocking Solution | Imprinting Factor |
| 1% BSA + 0.05% Tween 20 | 1.35 |
| 3% BSA + 0.05% Tween 20 | 1.26 |
| 0.1% skim milk + 0.05% Tween 20 | 1.76 |
| 0.5% skim milk + 0.05% Tween 20 | 0.92 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dietl, S.; Walther, P.; Sobek, H.; Mizaikoff, B. Core-Shell Imprinted Particles for Adenovirus Binding. Materials 2021, 14, 7692. https://doi.org/10.3390/ma14247692
Dietl S, Walther P, Sobek H, Mizaikoff B. Core-Shell Imprinted Particles for Adenovirus Binding. Materials. 2021; 14(24):7692. https://doi.org/10.3390/ma14247692
Chicago/Turabian StyleDietl, Sandra, Paul Walther, Harald Sobek, and Boris Mizaikoff. 2021. "Core-Shell Imprinted Particles for Adenovirus Binding" Materials 14, no. 24: 7692. https://doi.org/10.3390/ma14247692
APA StyleDietl, S., Walther, P., Sobek, H., & Mizaikoff, B. (2021). Core-Shell Imprinted Particles for Adenovirus Binding. Materials, 14(24), 7692. https://doi.org/10.3390/ma14247692





