Abstract
Design requirements for different mechanical metamaterials, porous constructions and lattice structures, employed as tissue engineering scaffolds, lead to multi-objective optimizations, due to the complex mechanical features of the biological tissues and structures they should mimic. In some cases, the use of conventional design and simulation methods for designing such tissue engineering scaffolds cannot be applied because of geometrical complexity, manufacturing defects or large aspect ratios leading to numerical mismatches. Artificial intelligence (AI) in general, and machine learning (ML) methods in particular, are already finding applications in tissue engineering and they can prove transformative resources for supporting designers in the field of regenerative medicine. In this study, the use of 3D convolutional neural networks (3D CNNs), trained using digital tomographies obtained from the CAD models, is validated as a powerful resource for predicting the mechanical properties of innovative scaffolds. The presented AI-aided or ML-aided design strategy is believed as an innovative approach in area of tissue engineering scaffolds, and of mechanical metamaterials in general. This strategy may lead to several applications beyond the tissue engineering field, as we analyze in the discussion and future proposals sections of the research study.
1. Introduction
Artificial intelligence (AI) and machine learning (ML) methods are reshaping data management, product design, materials science and mechanical engineering, among other industrially and socially relevant fields. As regards AI-aided discovery of materials, the Materials Genome Initiative [1,2,3] stands out as one of the pioneering large-scale projects, which has inspired other technological breakthroughs in innovative materials for advanced industrial applications [4,5]. Usually, AI and ML have been applied to the prediction of final properties and performance of materials from the chemical composition of the bulk materials under study [6,7,8]. In some cases, mechanical properties have been also predicted with ML techniques [9,10]. More recently, our team has focused on the forecasting of tribological properties of hierarchical topographies, advancing in the AI-aided design of textured surfaces and materials [11]. Besides, the progressive application of AI to the prediction, design and control of mechanical properties is already making an impact in the growing family of mechanical metamaterials [12,13], whose application fields include transport, energy, space and health, to cite a few.
Authors consider similar holistic approaches, aimed at promoting accelerated materials development through an intensive use of AI, if further research focusing on materials microstructures and mechanical properties can prove transformative towards high performance materials and devices in several industries.
The biomedical area can greatly benefit from innovative hierarchical mechanical metamaterials, whose multi-scale porous and lattice structures are essential for mimicking the biomechanical properties of human tissues and organs. This recapitulation of the mechanical properties of the cell microenvironment, through implanted artificial materials, constructs or “scaffolds”, helps the cells of damaged tissues receive the adequate mechanical stimuli and is one of the essential strategies in tissue engineering and regenerative medicine [14,15].
The design of successful tissue engineering scaffolds is a challenging engineering task, due to the complexity of the original tissues they aim to repair and the intricate connections between designed geometries, bulk materials and manufacturing tools. FEM simulations are commonly applied to CAD models of tissue engineering scaffolds, so as to analyze their prospective mechanical properties, usually focusing on their stiffness, as a parameter to optimize for enhanced biomimetic behavior [16,17]. However, there are many occasions in which FEM simulations cannot be applied to scaffolding structures or do not adequately predict final performance, due to geometrical complexity, large aspect ratios, or presence of manufacturing defects. Therefore, the use of AI and ML can prove valuable for deciphering this complexity of scaffolding structures and thus supporting bioinspired design approaches.
To mention some examples, AI and ML have been applied to the tissue engineering field for different purposes, in general related with the enhanced design of scaffolds: the ML-guided 3D printing of scaffolding geometries for minimizing defects has been reported [18], to predict vascularization in repair strategies [19] and to correlate in vitro performance with physico-chemical properties [20]. Conventional artificial neural networks are the most used methods for these AI-ML strategies. Nevertheless, the prediction of scaffolds’ mechanical performance directly from the CAD models, especially for cases in which FEM cannot be applied, may require advanced ML tools capable of using more complete descriptions of these complex geometries as input, and not just representative parameters. Quite recently, 2D convolutional neural networks (CNNs), with images as input, have been effectively applied to predicting multiple properties of porous materials, which constitutes a fundamental advance [21].
In the authors’ view, the use of 3D CNNs can constitute a novel and alternative (or complementary) method for predicting the properties of complex porous materials and structures, especially the mechanical properties of tissue engineering scaffolds. Taking into account the success of deep CNNs or 3D CNNs using medical images (i.e., computed tomography-CT-) as input for diagnostic purposes [22,23,24], similar ML strategies and schemes may apply to materials science and engineering.
Our rationale is as follows: the computed tomography of physical objects generates layered 2D images representing the whole geometry of a real object, proving useful as input for 3D CNNs for diagnostic purposes. Then, the use of digital tomography to obtain layered 2D images of virtual CAD models, like the CAD models of tissue engineering scaffolds, may be used as input for successfully predicting, in silico, the properties of the designed geometries.
To demonstrate our driving hypothesis, in this study, we create and characterize a collection of tissue engineering scaffolds and employ digital tomography to obtain the layered 2D images defining the geometries of the scaffolds of the collection. The 3D CNNs are trained using the layered images as input and the characterized properties, namely Young’s modulus, shear modulus and porosity, as outputs. Once trained and validated, employing a varied set of training and validation strategies, the predicting potential of the obtained 3D CNNs is tested with a new set of designs of tissue engineering scaffolds. To rapidly generate the tomographies from the virtual models, we apply a 3D printing slicer software in an innovative way, using the digital masks as the actual layered images that recreate the 3D geometry of each construct. The performance achieved, both in terms of predicting ability, computational speed and global cost, is remarkable.
To the authors’ best knowledge this study represents the first application of 3D CNNs to the AI-aided design of tissue engineering scaffolds. It is also an innovative approach in the area of mechanical metamaterials and may lead to a wide set of applications beyond the tissue engineering realm, as we analyze in the discussion and future proposals sections of the study. The following section describes the materials and methods employed, before dealing with the presentation and discussion of research results. Finally, the more relevant pending challenges, as well as some proposed and expected research directions, are also presented.
2. Materials and Methods
2.1. Creating a Library of Tissue Engineering Scaffolds with Well-Known Properties
Computer-aided design and finite element simulations are performed with the support of Autodesk Inventor 2021 (academic license). A collection of 20 lattices or scaffolding geometries is created by means of solid- and matrix-based operations and using Boolean tools. Geometrical diversity is sought, for which some designs have periodic features and remarkable symmetry, while others are conceived as irregular interconnections of trusses to increase the desired diversity. Some lattices are obtained by subtraction of already designed scaffolds to a bulk cubic geometry. In any case, all designed geometries can be considered tissue engineering scaffolds for different tissue repairs, as they resemble the common geometries used for 3D printed scaffolds for regenerative medicine. The cell units of the 20 designed scaffolds are shown in Figure 1, which summarizes the collection of CAD geometries. All of them are inscribed in a cube of 5 × 5 × 5 mm3.

Figure 1.
CAD library of tissue engineering scaffolds or mechanical metamaterials. Cell units of 5 × 5 × 5 mm3 are shown. Some geometries are obtained by direct Boolean operation, subtracting an already designed lattice to a cube of material.
Once designed, different properties of the scaffolding units are obtained. Porosity, defined as % of void within the 5 × 5 × 5 mm3 working volume, is directly measured with the CAD software. Two main mechanical properties are obtained for each lattice, the compression modulus and the shear modulus. The compression modulus of an elastic material is defined as the ratio between applied stress and resulting strain when that material is under compression. The shear modulus is defined as the ratio between applied shear stress and resulting shear strain. Due to the varied mechanical stimuli that tissue engineering scaffolds suffer in service, both properties are interesting from a biomechanical point of view.
The mechanical characterization of the different lattices, for obtaining the compression and shear moduli, is done in silico, using the FEM simulation capabilities of the used software. Lattices are meshed using tetrahedral elements of 0.05 mm. ABS, as conventional thermoplastic, is employed as bulk material. A normal or transversal distributed load of 25 N is applied, leading to an equivalent normal or shear stress considering that the lattices occupy a section of 5 × 5 mm2. As boundary condition, each lattice is fixed on the face opposite to the face where the force is applied. Once simulated, the equivalent compression or shear moduli are obtained. Dividing them by the actual compression or shear moduli of the bulk material used for the simulation (ABS) eliminates the influence of the raw material, leading to relative values only dependent on the actual lattice geometry, which are typically used for comparing mechanical metamaterials and in some materials selection strategies and in Ashby’s diagrams.
The results from in silico characterization for the different lattices are included in Table 1 and subsequently used for training the 3D CNNs. For training purposes, such values are normalized or scaled to the [0, 1] range, as this leads to better and faster 3D CNN convergence. For further processing, the CAD models of the designed lattices or unit cells are stored as binary .stl (standard tessellation language or stereolithography) files with a mesh precision of 0.05 mm, which proves adequate for employing Chitubox as slicing software for the digital tomographs, as explained further on.

Table 1.
Summary of lattices’ properties, characterized employing CAD measurement and FEM tools.
2.2. From 3D CAD Files to Digital Tomographies as Input for 3D CNNs
Chitubox v.1.8.1 basic (Chitubox, Zhongcheng Future Industrial Park, Hangcheng Avenue, Baoan District, Shenzhen, Guangdong, China 518128) is a free 3D printing software designed to edit and slice 3D CAD models. It also provides tools for CAD transformation including rotating, scaling, mirroring, repairing, hollowing, cloning, etc. With the help of Chitubox, the designed lattices are sliced, transforming their 3D geometry into a set of black and white images that resemble the layered images of CT-scans or MR imaging. A resolution of 1440 × 1440 pixels per slice is chosen and a distance of 0.25 mm between slices, along z-axis, is selected. Each slice is a cut of a scaffold, with a section of 5 mm2, and generates an image of 1440 × 1440 pixels. Thus, each pixel has a lateral size of 5 mm/1440. This leads to a set of 20 images per lattice capable of representing the 3D geometries with a remarkable level of detail, at least similar to the level of detail used when actually printing similar tissue engineering scaffolds.
Figure 2 provides examples of these digital tomographies, for different 3D CAD models of the designed lattices, achieved employing Chitubox as lithographic slicer for 3D printing. Results for lattices 1, 2 and 20 (from Figure 1) are presented in Figure 2 by means of example. Each lattice is transformed into 20 slices, many of which are coincident due to the periodic nature of these lattices. In these images, as happens in real CT-scans and in the Hounsfield scale utilized for the dicom (digital communications in medicine) standard, black represents empty spaces or voids and white corresponds to the actual scaffolding material. These sets of images are used as input for the 3D CNNs, for describing the geometries of the different lattices, while the in silico obtained properties (porosity, compression and shear moduli) are used as output, for training and validating purposes, as detailed in the following subsection.
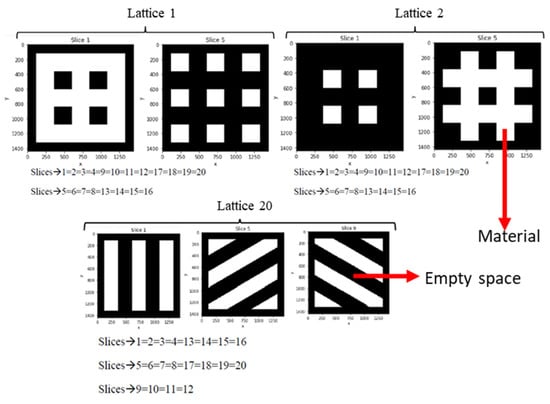
Figure 2.
Examples of digital tomographies, for different 3D CAD models of the designed lattices, achieved employing a lithographic slicer for 3D printing. Results for lattices 1, 2 and 20 (see Figure 1) are presented by means of example. Each lattice is transformed into 20 slices, many of which are coincident due to the periodic nature of the lattices.
The approach resembles pioneering experiences of 3D CNNs in medicine, but employing digital slices of CAD models, instead of real CT images.
2.3. Structuring and Training 3D CNNs for Predicting Mechanical Properties
For the development of the artificial intelligence/machine learning model, the Python programming language in version 3.8.5 (Phyton Software Foundation) is employed. This is the most used language in machine and deep learning since it is open source and provides the necessary tools to carry out this type of process effectively and relatively easily, thanks to available powerful dedicated libraries. Besides, Python interpreter allows to run programs written in Python language [25]. Different libraries for data analysis, data processing and deep learning are also employed, whose main features and application purposes are described below:
NumPy version 1.19.2: specialized in numerical calculation and data analysis for large volumes of data. This library incorporates matrices (arrays) that allow to represent data collections of the same type in several dimensions. It also incorporates very efficient functions for manipulating arrays [26]. This library is fundamental in study since a 3D CNN understands a 3D image as a 3D array.
Matplotlib version 3.3.4: develops quality 2D and 3D graphics with a few lines of code, uses static, animated and interactive figures, allows to take full control of line styles, axis properties, among other options [27]. In short, this library allows the visualization of data and results.
Scikit-learn version 0.24.1: main machine learning library in Python, providing different tools for predictive data analysis and calculation of metrics, such as mean square error [28], needed in supervised learning. It includes generalized linear regression models.
Scikit-image version 0.18.1: dedicated to image processing, it allows reading and displaying images from a file, binarizing, resizing, segmenting images, adjusting their contrast and color and other typical image processing operations [29].
TensorFlow version 2.3.0: compiles and trains artificial intelligence models with ease, using intuitive and high-level application programming interfaces (APIs), such as Keras, with immediate execution and allowing immediate model iteration and easy debugging [30].
Keras version 2.4.0: part of the TensorFlow library from TensorFlow version 2.0.0 and it has a more friendly code. It covers every step of the deep learning workflow, from data management, training, configuration and evaluation of the model to obtaining predictions or testing the artificial intelligence model. This makes it a widely used deep learning framework, whose guidelines are public [31].
For the installation of all the previous libraries, a Python distribution called Anaconda is used, which already includes the Python interpreter installed and the Numpy and Matplotlib libraries used. The other libraries are installed thanks to the Anaconda package manager called conda. In this case, the package manager is conda version 4.10.1.
Anaconda, in addition to the package manager, includes a desktop application called Anaconda Navigator, which allows to manage packages, as well as run applications such as the Jupyter Notebook development environment. In this case, Anaconda Navigator has Jupyter Notebook version 6.3.0 and it is the development environment used.
Once the software and libraries are installed, data are preprocessed and augmented, the structure for the 3D CNNs are defined. Finally, training, validation and testing strategies are designed and implemented.
Since each slice has very large dimensions (1440 × 1440 pixels), images are resized to 32 × 32 pixels using Scikit-image library again. Although the resolution is lower, the network is able to understand patterns and learns faster during training. Then, considering that 3D CNNs understand images as arrays, the 20 slices of each lattice are concatenated along a new axis, in this case the z axis. A total 20 3D arrays of 0 s and 1 s are obtained that represent the 20 CAD cellular scaffolds. The Numpy library is used for this operation.
Regarding the architecture for the 3D CNNs, we opt for a structure involving input images (representative of the 3D geometries), convolutional and max pooling layers and fully connected dense layers leading to the outputs (porosity and mechanical properties), as schematically presented in Figure 3.
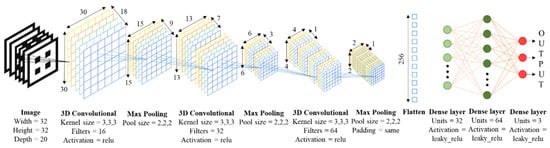
Figure 3.
Proposed structure for the 3D CNNs: from 3Dslices models to useful performance properties. Block diagram, structure and details of convolutional neural network.
Summarizing, the design of 3D CNNs’ architectures and their training and validation is carried out thanks to Keras library. To analyze which is the best model of all those designed, the following strategy is carried out. A total 70% of the 3D structures without any transformation are used as training data and 30% of them as validation. Regarding the loss function and the metric, the selected model is the one whose mean square error in training and validation is closer to 0 and the lowest. Moreover, if it is possible the validation MSE should be slightly lower than that of training. This means that the convolutional neural network “learns” from the training data and can generalize to data outside of that set. Therefore, the cross-validation method is used to estimate the precision of the different models. Data segregation, from the total training data set into training and validation data, is done using Scikit-learn library. The selected model consists of a 3D convolutional layer with 16 filters each with dimensions 3 × 3 × 3 pixels. A Relu activation function is used, and filters weights are applied randomly.
Subsequently, a 3D max pooling layer with a filter dimension of 2 × 2 × 2 pixels is placed. After this layer, a batch normalization layer is used and then a dropout layer with an index equal to 0.3. The batch normalization layer normalizes and scales its inputs by applying a transformation that keeps the mean output close to 0 and the output standard deviation close to 1 [32]. Standardizing the activations of the previous layer means that the subsequent layer makes about the propagation and distribution of the inputs during the weight update will not change dramatically. This has the effect of stabilizing and accelerating the deep neural network training process [33].
The dropout layer randomly sets the input units to 0 with a rate of speed at each step during the training time, which helps prevent overfitting. The inputs that are not set to 0 are scaled by 1/(1-rate) so that the sum of all the inputs does not change [34]. A dropout layer supposes the unlearning of the neural network, which prevents the network from learning in excess from the training data, “memorizing them”, and not being able to predict any new input data (problem well-known as overfitting). This succession of layers is then repeated, but this time the convolutional layer uses 32 random filters. Then it is repeated again, but the convolutional layer uses 64 random filters and the max pooling layer maintains the last dimension of the data it receives to avoid obtaining a negative dimension. Otherwise, there would be no dimensions to apply the max pooling layer. The combination of the three package of layers described adopts the shape of bottleneck, because the greater the depth of the convolutional neural network, the greater the abstraction. To extract all the feature information the deeper is the layer, smaller are the filters used per convolution layer and it is often used with a larger amount of filters. Finally, a flatten layer is placed, a succession of dense layers with 32 and 64 neurons respectively, and another dense layer with 3 neurons because the network predicts three variables (provides 3 outputs) from each input (lattice geometry as slices). All dense layers use a Leaky Relu activation function. Main features of CNN are seen in Figure 3, where the output shape of the layers is shown around the different blocks of convolution and the flatten layer.
Taking into account that a library of 20 scaffolding lattices may be limited as training and validation set for AI/ML strategies, data augmentation is performed. Such data expansion is achieved through rotations around z-axis, zooms and resizing, vertical flips, rotations around x- and y-axes, to cite some options used. Different examples of these data augmentation strategies are shown in Figure 4 by means of example and summarized in Table 2.
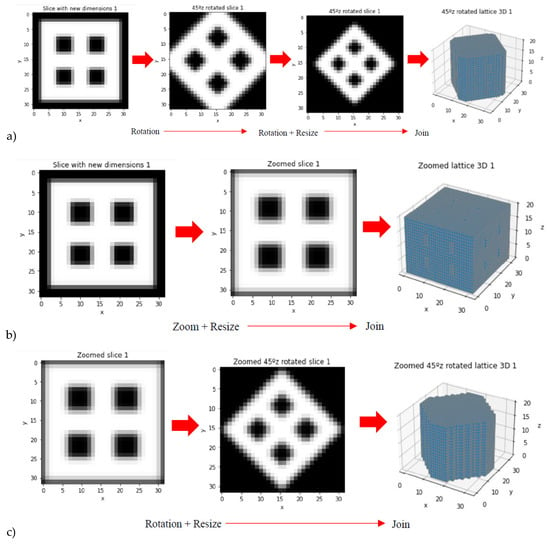
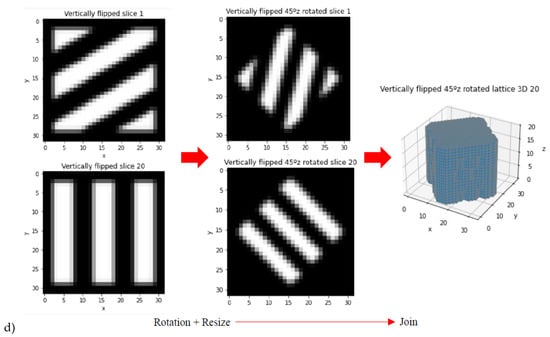
Figure 4.
Examples from data augmentation strategies: (a) Rotation around z axis and resizing. (b) Zoom. (c) Zoom, rotation around z axis and resizing. (d) Vertical flip and rotations around z axis.
The CNN shown above is designed based on two main examples [35,36]. The structure is adapted to solve the question dealt in this paper, using an iterative process to find a good performance. With the selected model, six training and validation strategies are carried out. The six strategies consist of expanding the training data and modifying the validation data randomly as summarized in Table 2 below.

Table 2.
Summary of the six training and validation strategies employed.
Table 2.
Summary of the six training and validation strategies employed.
| Strategy | Number of Lattices Used | Number of Lattices Used for Training | Number of Lattices Used for Validation | Data Augmentation Strategy |
|---|---|---|---|---|
| 1st strategy | 20 | 14 random ones | 6 random ones | - |
| 2nd strategy | 120 | 114 random ones | 6 random ones | Rotations around z-axis (15°, 30°, 45°, 60°, 75°) |
| 3rd strategy | 240 | 234 random ones | 6 random ones | Previous rotations plus zoomed-in lattices |
| 4th strategy | 360 | 354 random ones | 6 random ones | Previous rotations and zooms (240 lattices) plus vertical flips without zoom (120 lattices more) |
| 5th strategy | 480 | 474 random ones | 6 random ones | Addition of horizontal flips (120 lattices more) |
| 6th strategy | 680 | 674 random ones | 6 random ones | Addition of rotated initial lattices around x- and y-axes (15°, 30°, 45°, 60°, 75°, 200 lattices more) |
2.4. Testing and Validation of the Global Strategy
Once trained, the 3D CNNs are tested, and their predicting ability validated, in a real use case scenario. For this purpose, a new library of scaffolding geometries with 8 lattices (see Figure 5a), completely different from those included in the initial collection of 20, is designed. These lattices are sliced (see examples from Figure 5b), following the procedure described in Section 2.2 and the different 3D CNNs are employed to predict their properties. In parallel, the methods from Section 2.1 are applied to in silico characterize these new lattices. The obtained properties (Table 3) are compared to the predicted ones. The precision of the different 3D CNNs, obtained as detailed in Section 2.3 employing varied training strategies, is analyzed. Results are presented and discussed in the following section.
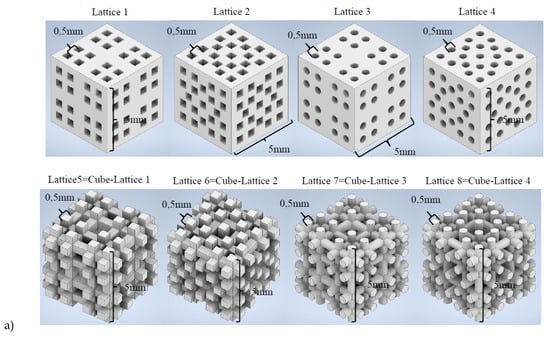
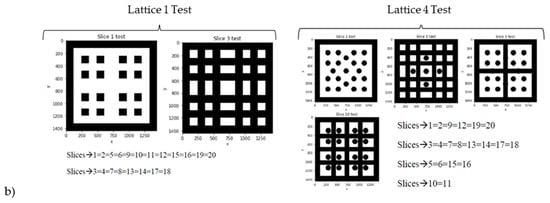
Figure 5.
Testing samples for validating the 3D CNNs performance. Eight new lattices are designed, and their properties are simulated, with the support of FEM simulations as reference, and predicted with the previously trained and validated 3D CNNs. (a) Computer-aided designs. (b) Digital tomographies as input to the 3D CNNs (examples for lattices 1 & 4).

Table 3.
Summary of new lattices’ properties, characterized employing CAD measurement and FEM tools.
3. Results and Discussion
3.1. CAD Models, Digital Tomographies, and Training and Validation of 3D CNNs
The initial CAD library of 20 scaffolding geometries, used for training and validating the 3D CNNs, is already an interesting result, which improves with the addition of the eight additional new scaffolds of Figure 5a, designed for testing the global AI/ML strategy. This designed collection of microstructured geometries is a starting point, aimed at creating the most comprehensive library of tissue engineering scaffolds with information about their biomechanical performance, which can be continuously updated. Such updates can be used for further training the 3D CNNs, once additional designs, simulation and testing results upon CAD files or physical prototypes are available. The scaffolds’ library already includes several CAD files in .ipt (Inventor parts) and .stl (standard tessellation language -or stereolithography-) formats, as well as their equivalent slices (or digital tomographies) stored in the form of arrays. The library, and its future additions, are openly available for researchers in the field, wishing to collaborate or test related approaches, linked to the AI- or ML-aided development of tissue engineering scaffolds and metamaterials.
Among other remarkable results of the study, authors would like to highlight the possibility of recapitulating the three-dimensional geometries of complex CAD objects by using a 3D printing slicer, Chitubox in this case, and employing the obtained slices as input for 3D CNNs. Examples of the application of the slicer are shown in Figure 2 for the initial training library and in Figure 5b for the new lattices used for testing the global AI-ML strategy. The use of digital tomographies is demonstrated useful for training, validating and testing deep convolutional neural networks, as the employment of images from CT scans had already proven highly useful for the progressive application of AI/ML methods in diagnostic medicine. Although recent studies have also combined machine learning and FEM simulations to predict the mechanical properties of biomaterials lattices and biomechanical structures [37,38], they have normally relied on conventional artificial neural networks (ANNs) with a few parameters as inputs/outputs that describe slight variations in thickness, size, length or density. Moreover, 3D CNNs, loaded with digital slices, receive the whole geometry as input and outperform simpler ANNs, especially when the geometrical complexity increases and when the diversity of geometrical inputs does not allow for a parametrization. It is also well-known that CNNs achieve better results in image processing than ANNs because CNNs preserve and analyze the sequence of data. These types of algorithms recognize the position and relationship between near pixels of the inputs.
Arguably, CT scans or magnetic resonance (MR) images carried out upon physical samples, materials, products and real patients, may synergize with the use of digital tomographies, like those obtained using 3D printing slicers upon CAD files, for fostering the application of AI/ML methods in a wide set of scientific-technological disciplines, from materials science and engineering to regenerative medicine. The training and validation work properly, the initial library of scaffolds and the errors decreasing with the addition of lattices to the training set, as seen in Figure 6a, thanks to the strategies for data augmentation described in Section 2.3.
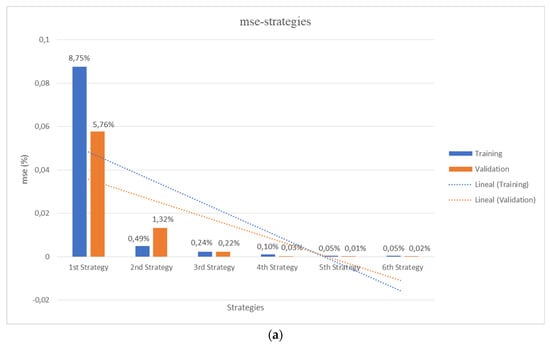
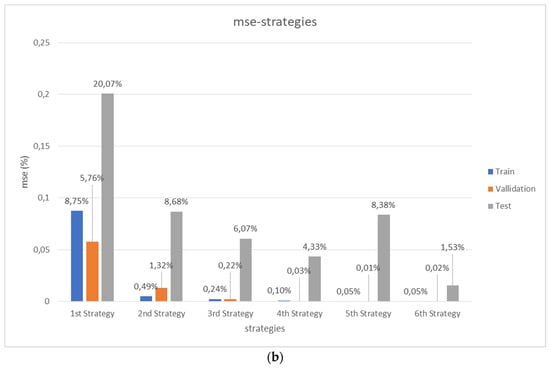
Figure 6.
Comparative performance of the different strategies. (a) Results from 3D CNNs training and validation according to the 6 detailed strategies with increasing number of input lattices (see Section 2.3). (b) Final performance, comparing the testing errors with those from previous training and validation processes for the different trained and validated 3D CNNs. Mean square errors (MSE) in % are presented.
The performance of the trained 3D CNNs in a real-life scenario, for predicting the mechanical properties of new lattices added to the library, is discussed in the following subsection.
3.2. Performance of the Structured and Trained 3D CNNs: Predictions vs. Real Performance
Once trained, the 3D CNNs are employed to predict the porosity and mechanical performance of the new design lattices of Figure 5, whose characteristic properties are shown in Table 3. The new lattices are sliced, following the same procedures applied to the original library of 20 samples, and evaluated with the available 3D CNNs that provide different porosities, compression and shear moduli as outputs. The predicted outputs are compared with values from Table 3. This corresponds to a real-life scenario, in which the trained networks face completely new geometries, never used previously for training or validation, and process them to predict their properties for classification or selection purposes.
Figure 6 schematically presents a comparative overview of the performance of the different strategies. On the one hand, Figure 6a shows the results from 3D CNNs training and validation according to the six detailed strategies with progressively increasing number of input lattices (as already described in Section 2.3). On the other hand, Figure 6b shows the final performance, comparing the testing errors with those from previous training and validation processes for the different trained and validated 3D CNNs. In all cases, the mean square errors (MSE, in %) are presented. Additional level of detail, for the different structures and strategies, is in the Appendix A, which presents a complete report of the characterized and predicted values for the different properties of the new lattice collection used for the final testing of the global strategy, according to the explanation provided in Section 2.4.
In agreement with the initial expectations, the training and validation errors rapidly decay with the increase of input/output data. Besides, such decreasing trend is also seen for the testing experiment, although the fifth strategy gives an unexpectedly high error for the testing set, possibly due to over-fitting or over-learning during the training of such specific 3D CNNs with the fifth strategy. In any case, although the first 3D CNN trained and validated with a set of 20 samples leads to a high prediction error for the testing set of 20% (MSE), the 3D CNNs trained with increasing number of lattices lead to MSE values of c.a. 9%, 6%, 4% and 1.5% for strategies 2, 3, 4 and 6 respectively. It is important to remark that the MSEs values are calculated on scaled output.
These results are quite remarkable and are in the common range achievable by FEM simulations upon CAD files or by mechanical testing upon manufactured samples. When these results are compared with those from FEM simulations, it is important to note that when geometries are too complex or involve multi-scale features, FEM simulations are sometimes impractical or extremely demanding in terms of computational resources and simulation time. However, for the described 3D CNNs, the computational times required for training and validating are c.a. 0 s, 3 s, 6 s, 9 s, 13 s and 18 s for strategies 1 to 6. Once implemented, the actual testing or prediction of porosity and mechanical properties of new samples is almost immediate.
Regarding precision of slicing and number of images employed as input, for converting the 3D geometries into a collection of sliced images, to have a sort of “digital tomography”, we employ a 3D printing slicer for photopolymerization systems (Chitubox), as has been explained. These types of slicers normally cut the CAD models according to the z-axis resolution of printing machines, which typically print with vertical steps ranging from 50 to 300 microns. We considered that a digital tomography, using a separation between slices of 250 microns, which provides 20 images for each 5 × 5 × 5 mm3 scaffold, would be adequate for illustrating the methodology. It is a common value, both in the 3D printing field and in the materials science field when performing tomographies of porous materials, as well as in medicine when exploring patients. A higher precision in the z-axis would arguably lead to an increased precision, but it is also true that the geometric features of the CAD models are in many cases periodic, and their details are normally larger than 500 microns, which even with 20 images per geometry leads to many repeated slices in each sample. We consider that the selected resolution in the z axis is an adequate compromise between precision and processing speed.
In terms of compromise between precision and speed, due to the fact that each geometry was represented by 20 images of 1440 × 1440 pixels, which led to a large collection of high-quality images, resizing of slices to 32 × 32 pixels is applied. We understood that resolution would be lower, as previously mentioned, but for processing purposes this resizing is interesting, as it reduces the number of pixels by a factor above 2000. We believe that this is a common practice and we have followed examples from previous studies and a library that provides this possibility. In the end, results show that the network can detect the patterns, somehow understanding that a higher presence of white pixels is linked to more dense and rigid structures, although precision could probably be improved by using higher quality images for training and much larger processing times.
Furthermore, the results show an exciting point about the use of zoom for data augmentation. Before evaluating the results, the authors assumed that the zoom could probably adversely affect the performance of 3D CNN. Quite the opposite, the use of zoom improves the performance of the AI demonstrating the neural network appreciate the relation between empty and material space (black and white pixels) in each image, and their connections with density and stiffness (whiter structures tend to be denser and stiffer). The improvement using zoom is slight, but it can be appreciated when comparing the errors between 2th and 3th strategies, where the unique difference is the use of data augmentation by zoom-in. This technique allows the networks to learn from where an image is taken (closer or further away) that is, the networks learn different features due to the relationships between different pixels, their positioning, their colors and not by the fact of moving away or approaching.
Interestingly, although physical properties like Young’s modulus are not scale independent and although authors used the zoom as an option for data augmentation, but without initial high expectations about its potential benefits, its employment has shown some benefits. We explain this considering that the zoom may help the network to predict some phenomena like stress concentration, in which the details of the connections between trusses play a relevant role, even if further studies are still needed to analyze this possibility and the potential generalization problems that using a zoom and other data augmentation strategies might generate.
The metric mean absolute error (MAE) for the strategies is included in Table 4. MAE would provide a fast and easy outlook about the performance in the different scenarios. As it can be seen in Figure 6, the analysis of the MSE, MAE metrics revealed the increased power for predicting the output when 3D CNN is trained with more varied data, thanks to the use of data augmentation. The 6th strategy outperforms the others in all outputs with a global MAE of 7.057. The strategy global MAE is calculated as the average of the MAE for the three outputs and presented in more detail in the Appendix A.

Table 4.
Report of mean absolute error. Characterized vs predicted values for the different properties of the new lattices. Please see Appendix A for additional details, metrics and information about different strategies and lattices.
4. Challenges and Future Proposals
4.1. Potentials, Limitations, and Challenges of the Study
AI and ML techniques have intrinsic limits, including the need of large data for achieving desired results, the “black box” problem, issues with over fitting, interpolative nature, making them work adequately only for data with similar features to those used for training, among others, which we have tried to avoid along our research. However, such common AI and ML drawbacks may lead to the failure of similar learning strategies to those applied here, once translated to other problems in the tissue engineering field or in connection with the prediction of properties for mechanical metamaterials.
Although the data set used started with a library of just 20 geometries, the employment of varied data augmentation processes proved useful for minimizing errors and leading to acceptable results, despite the considerable overfitting to the validation sets perceived. Considering the preliminary nature of the study, which aims at validating an innovative strategy for the AI-aided design of tissue scaffolds, we believe that the study may be of interest for researchers in the field and for progressing towards the AI-aided design of other porous materials and metamaterials. In any case, for solving related problems by applying similar methods, it would be important to count with larger data sets or to expand them even more for enhancing results.
In spite of the intrinsic limitations of AI and ML techniques in general, and of the 3D CNNs employed here in particular, for some applications they may outperform other well established simulation methods. For example, FEM simulations may lead to unaffordable computational costs when evaluating highly complex geometries, especially when aspect ratios are high or when multi-scale “fractal-like” features are present, for which the use of AI methods may prove competitive indeed, if they were adequately trained and validated. Such multi-scale and fractal-like features are common in the tissue engineering field, as the scaffolds are normally designed to mimic the intricate geometries of nature. Apart from the applicability of these tools to biomechanical problems, other studies have also shown the benefits of resorting to deep neural networks when dealing with extremely complex simulations, as a way for achieving an attractive trade-off between cost and accuracy [39].
Ideally, the developed AI and ML tools will lead, not just to predicting porosity and mechanical properties, but also to a better understanding of the behavior of tissue engineering scaffolds and to more adequate AI-aided processes for the engineering of biomaterials for regenerative medicine.
4.2. Future Research Proposals
Thinking of future research directions, in the authors’ opinion, it would be interesting to augment the library of scaffolding materials to a larger extent, so as to obtain a truly universal tool for supporting designers of tissue engineering scaffolds (and of mechanical metamaterials in general). This augmentation can benefit from open-source approaches, sharing CAD models, evaluated data and developed and trained networks among the research community. In this way, the effort dedicated to collecting the data can be amortized over many groups collaborating. To progress in such direction, we provide the whole characterized and predicted data of the new lattices in the Appendix A, as a complement to the data shown in Table 1 and Table 3. The CAD models of our collection are also available by email request, with the hope of initiating fruitful collaboration with colleagues.
In the tissue engineering field, many of the complex multi-scale geometries of porous tissue scaffolds are a consequence of more traditional manufacturing processes that do not follow an existing computer-aided design, as happens with 3D printing or additive manufacturing technologies. Such traditional scaffold fabrication processes usually obtain highly random porous structures, with biomimetic features, by phase-separation, solvent casting, gas-assisted injection molding or foaming. In those cases, digital tomographs of CAD files, like those used in this study, are not viable for obtaining the input images and expanding the library. As an alternative, the use of micro-CT or micro-MRI is proposed, measuring upon real manufactured samples and using them as input for increasing the versatility of the library and related 3D CNNs, constitutes another remarkable option.
Towards more holistic scaffold design strategies, apart from considering porosity and compression and shear moduli, other properties of the designed scaffolds may prove fundamental for a successful performance of the implantable construct. Characterizing the geometries of the library considering properties like diffusion coefficient, pressure drop of circulating fluid, natural frequencies of vibration, surface to volume ratio, to cite a few, and verifying the possibility of training the 3D CNNs for predicting them, may lead to a more complete and effective design tool. In addition, a higher degree of versatility is expectable if grayscale images, instead of black and white slices, are employed.
Once improved, automated design procedures may be implemented and the 3D CNNs may be used for selecting the best scaffolds according to a set of properties that should be optimized. This may help, in the future, to reach a tool capable of autonomously designing tissue engineering scaffolds and even mechanical metamaterials in their more general conception, if the presented strategies are applied to other application fields.
5. Conclusions
This study has dealt with the application of 3D convolutional neural networks to the prediction of different essential properties of tissue engineering scaffolds. The 3D CNNs have been trained using digital tomographies obtained from the CAD models and results from CAD measurements and FEM simulations. Their predictive performance has been analyzed by using the trained 3D CNNs to forecast the properties of a new set of tissue engineering scaffolds and the results using a collection of networks differently trained has also been discussed. Although the performance is not ideal and additional research efforts are needed, the results obtained validate an AI-based methodology for predicting the properties of complex structures, which may be applicable when the computational cost of other simulation methods results unaffordable. The study has dealt with the biomechanical performance of tissue engineering scaffolds, but similar strategies may be applied to a wide set of properties in the emergent area of metamaterials. Theoretically, these processes can be applied to the automated design or discovery of microstructures with desired mechanical properties, especially if the analyzed current limitations are answered and if multidisciplinary research approaches are promoted.
Author Contributions
Conceptualization, A.D.L. and F.F.-M.; methodology, software, validation, formal analysis, investigation, resources, writing, A.D.L., F.F.-M. and M.D.B.B.; data curation, M.D.B.B.; supervision, F.F.-M. and A.D.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data, source code of the developed software and developed 3D CNNs are available upon reasonable request.
Acknowledgments
The authors acknowledge reviewers for their comments and suggestions, which have helped to present the research results in a clearer way for the benefit of colleagues.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Porosity, Elastic Modulus and Shear Modulus Mean Absolute Error (MAE) estimation for each training strategy.
Table A1.
Porosity, Elastic Modulus and Shear Modulus Mean Absolute Error (MAE) estimation for each training strategy.
| Lattice nº | Simulated Relative Porosity (%) | Predicted Relative Porosity (%) | Relative Porosity AE | Relative Porosity MAE | Simulated Rel. Elastic Modulus E_Relative (%) | Predicted Rel. Elastic Modulus E_Relative (%) | Rel. Elastic Modulus E_Relative AE | Rel. Elastic Modulus E_Relative MAE | Simulated Rel. Shear Modulus G_Relative (%) | Predicted Rel. Shear Modulus G_relative (%) | Rel. Shear Modulus G_Relative AE | Rel. Shear Modulus G_Relative MAE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1st strategy | ||||||||||||
| Lattice 1 | 35.138 | −0.569 | 35.706 | 39.073 | 42.645 | 63.123 | 20.478 | 26.814 | 13.755 | 27.629 | 13.874 | 13.193 |
| Lattice 2 | 47.527 | −0.750 | 48.277 | 21.503 | 29.878 | 8.374 | 8.988 | 12.163 | 3.175 | |||
| Lattice 3 | 28.650 | −0.559 | 29.209 | 52.309 | 82.533 | 30.224 | 18.775 | 25.614 | 6.839 | |||
| Lattice 4 | 38.641 | −0.780 | 39.421 | 35.758 | 56.540 | 20.782 | 14.793 | 30.589 | 15.796 | |||
| Lattice 5 | 64.862 | 25.843 | 39.019 | 18.462 | −0.074 | 18.536 | 3.126 | 1.489 | 1.637 | |||
| Lattice 6 | 52.473 | −0.764 | 53.237 | 25.252 | 77.633 | 52.381 | 4.658 | 33.474 | 28.816 | |||
| Lattice 7 | 71.350 | 76.944 | 5.594 | 14.336 | −0.072 | 14.408 | 2.118 | 0.077 | 2.041 | |||
| Lattice 8 | 61.359 | −0.763 | 62.123 | 17.785 | 67.116 | 49.331 | 1.143 | 34.510 | 33.367 | |||
| 2nd strategy | ||||||||||||
| Lattice 1 | 35.138 | 12.707 | 22.431 | 18.476 | 42.645 | 62.542 | 19.897 | 18.926 | 13.755 | 24.392 | 10.638 | 9.263 |
| Lattice 2 | 47.527 | 57.088 | 9.561 | 21.503 | 4.106 | 17.397 | 8.988 | 4.646 | 4.342 | |||
| Lattice 3 | 28.650 | 29.877 | 1.226 | 52.309 | 77.738 | 25.429 | 18.775 | 29.877 | 11.102 | |||
| Lattice 4 | 38.641 | 38.019 | 0.622 | 35.758 | 24.999 | 10.759 | 14.793 | 13.045 | 1.749 | |||
| Lattice 5 | 64.862 | 49.150 | 15.713 | 18.462 | 19.016 | 0.554 | 3.126 | 7.370 | 4.243 | |||
| Lattice 6 | 52.473 | 7.153 | 45.320 | 25.252 | 71.796 | 46.544 | 4.658 | 27.718 | 23.060 | |||
| Lattice 7 | 71.350 | 53.194 | 18.155 | 14.336 | 17.750 | 3.414 | 2.118 | 6.110 | 3.992 | |||
| Lattice 8 | 61.359 | 26.582 | 34.777 | 17.785 | 45.202 | 27.417 | 1.143 | 16.120 | 14.977 | |||

Table A2.
Porosity, Elastic Modulus and Shear Modulus Mean Absolute Error (MAE) estimation for each training strategy.
Table A2.
Porosity, Elastic Modulus and Shear Modulus Mean Absolute Error (MAE) estimation for each training strategy.
| Lattice nº | Simulated Relative Porosity (%) | Predicted Relative Porosity (%) | Relative Porosity AE | Relative Porosity MAE | Simulated Rel. Elastic Modulus E_Relative (%) | Predicted Rel. Elastic Modulus E_Relative (%) | Rel. Elastic Modulus E_Relative AE | Rel. Elastic Modulus E_Relative MAE | Simulated Rel. Shear Modulus G_Relative (%) | Predicted Rel. Shear Modulus G_Relative (%) | Rel. Shear Modulus G_Relative AE | Rel. Shear Modulus G_Relative MAE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3rd strategy | ||||||||||||
| Lattice 1 | 35.138 | 41.831 | 6.694 | 17.646 | 42.645 | 18.270 | 24.375 | 20.207 | 13.755 | 12.084 | 1.670 | 5.634 |
| Lattice 2 | 47.527 | 91.267 | 43.740 | 21.503 | −0.201 | 21.704 | 8.988 | −0.187 | 9.175 | |||
| Lattice 3 | 28.650 | 32.668 | 4.018 | 52.309 | 26.848 | 25.461 | 18.775 | 14.455 | 4.320 | |||
| Lattice 4 | 38.641 | 89.796 | 51.155 | 35.758 | −0.239 | 35.997 | 14.793 | −0.160 | 14.954 | |||
| Lattice 5 | 64.862 | 60.900 | 3.963 | 18.462 | −0.137 | 18.599 | 3.126 | −0.101 | 3.227 | |||
| Lattice 6 | 52.473 | 32.554 | 19.919 | 25.252 | 22.132 | 3.120 | 4.658 | 12.956 | 8.298 | |||
| Lattice 7 | 71.350 | 66.871 | 4.479 | 14.336 | −0.185 | 14.521 | 2.118 | −0.143 | 2.261 | |||
| Lattice 8 | 61.359 | 68.560 | 7.201 | 17.785 | −0.098 | 17.883 | 1.143 | −0.025 | 1.167 | |||
| 4th strategy | ||||||||||||
| Lattice 1 | 35.138 | 27.745 | 7.393 | 14.577 | 42.645 | 45.714 | 3.069 | 12.108 | 13.755 | 18.809 | 5.054 | 6.823 |
| Lattice 2 | 47.527 | 86.467 | 38.940 | 21.503 | 0.292 | 21.211 | 8.988 | −0.003 | 8.991 | |||
| Lattice 3 | 28.650 | 17.930 | 10.720 | 52.309 | 60.546 | 8.237 | 18.775 | 24.280 | 5.505 | |||
| Lattice 4 | 38.641 | 65.936 | 27.296 | 35.758 | 16.456 | 19.303 | 14.793 | 6.952 | 7.841 | |||
| Lattice 5 | 64.862 | 61.508 | 3.354 | 18.462 | 22.591 | 4.130 | 3.126 | 7.012 | 3.886 | |||
| Lattice 6 | 52.473 | 26.997 | 25.476 | 25.252 | 55.402 | 30.150 | 4.658 | 20.239 | 15.581 | |||
| Lattice 7 | 71.350 | 68.602 | 2.748 | 14.336 | 21.723 | 7.387 | 2.118 | 4.622 | 2.503 | |||
| Lattice 8 | 61.359 | 62.052 | 0.693 | 17.785 | 21.164 | 3.378 | 1.143 | 6.365 | 5.222 | |||

Table A3.
Porosity, Elastic Modulus and Shear Modulus Mean Absolute Error (MAE) estimation for each training strategy.
Table A3.
Porosity, Elastic Modulus and Shear Modulus Mean Absolute Error (MAE) estimation for each training strategy.
| Lattice nº | Simulated Relative Porosity (%) | Predicted Relative Porosity (%) | Relative Porosity AE | Relative Porosity MAE | Simulated Rel. Elastic Modulus E_Relative (%) | Predicted Rel. Elastic Modulus E_Relative (%) | Rel. Elastic Modulus E_Relative AE | Rel. Elastic Modulus E_Relative MAE | Simulated Rel. Shear Modulus G_Relative (%) | Predicted Rel. Shear Modulus G_Relative (%) | Rel. Shear Modulus G_Relative AE | Rel. Shear Modulus G_Relative MAE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5th strategy | ||||||||||||
| Lattice 1 | 35.138 | 35.598 | 0.460 | 15.496 | 42.645 | 42.575 | 0.070 | 18.713 | 13.755 | 13.167 | 0.588 | 9.289 |
| Lattice 2 | 47.527 | 47.341 | 0.187 | 21.503 | 29.543 | 8.040 | 8.988 | 17.428 | 8.440 | |||
| Lattice 3 | 28.650 | 33.927 | 5.276 | 52.309 | 43.521 | 8.789 | 18.775 | 14.347 | 4.428 | |||
| Lattice 4 | 38.641 | 31.366 | 7.275 | 35.758 | 43.336 | 7.578 | 14.793 | 22.453 | 7.660 | |||
| Lattice 5 | 64.862 | 44.621 | 20.242 | 18.462 | 35.414 | 16.952 | 3.126 | 7.862 | 4.736 | |||
| Lattice 6 | 52.473 | 21.655 | 30.818 | 25.252 | 67.866 | 42.614 | 4.658 | 25.450 | 20.793 | |||
| Lattice 7 | 71.350 | 47.203 | 24.147 | 14.336 | 36.286 | 21.950 | 2.118 | 7.237 | 5.118 | |||
| Lattice 8 | 61.359 | 25.800 | 35.559 | 17.785 | 61.499 | 43.714 | 1.143 | 23.696 | 22.553 | |||
| 6th strategy | ||||||||||||
| Lattice 1 | 35.138 | 24.881 | 10.257 | 7.665 | 42.645 | 33.416 | 9.229 | 10.264 | 13.755 | 18.620 | 4.865 | 3.242 |
| Lattice 2 | 47.527 | 57.971 | 10.444 | 21.503 | 18.641 | 2.862 | 8.988 | 7.243 | 1.745 | |||
| Lattice 3 | 28.650 | 24.990 | 3.660 | 52.309 | 33.358 | 18.951 | 18.775 | 18.512 | 0.263 | |||
| Lattice 4 | 38.641 | 36.310 | 2.331 | 35.758 | 26.531 | 9.227 | 14.793 | 14.152 | 0.641 | |||
| Lattice 5 | 64.862 | 62.012 | 2.850 | 18.462 | 5.179 | 13.283 | 3.126 | 3.943 | 0.817 | |||
| Lattice 6 | 52.473 | 34.515 | 17.957 | 25.252 | 25.039 | 0.213 | 4.658 | 13.551 | 8.893 | |||
| Lattice 7 | 71.350 | 79.106 | 7.756 | 14.336 | 3.789 | 10.547 | 2.118 | 0.402 | 1.717 | |||
| Lattice 8 | 61.359 | 67.421 | 6.062 | 17.785 | −0.013 | 17.799 | 1.143 | 8.139 | 6.997 | |||
References
- Interagency Materials Genome Initiative Web Portal. Available online: https://www.mgi.gov (accessed on 23 June 2021).
- NIST Gateway to Materials Genome Information. Available online: https://mgi.nist.gov/ (accessed on 23 June 2021).
- Liu, Z.H. Perspective on Materials Genome®. Chin. Sci. Bull. 2014, 59, 1619–1623. [Google Scholar] [CrossRef]
- Qian, C.; Siler, T.; Ozin, G.A. Exploring the possibilities and limitations of a nanomaterials genome. Small 2015, 11, 64–69. [Google Scholar] [CrossRef]
- Raccuglia, P.; Elbert, K.C.; Adler, P.D.F.; Falk, C.; Wenny, M.B.; Mollo, A.; Zeller, M.; Friedler, S.A.; Schrier, J. Machine-learning-assisted materials discovery using failed experiments. Nature 2016, 533, 73–76. [Google Scholar] [CrossRef]
- Lu, W.; Xiao, R.; Yang, J.; Li, H.; Zhang, W. Data-mining aided materials discovery and optimization. J. Mater. 2017, 3, 191–201. [Google Scholar] [CrossRef]
- Winkler, D.A. Biomimetic molecular design tools that learn, evolve, and adapt. Beilstein J. Org. Chem. 2017, 13, 1288–1302. [Google Scholar] [CrossRef] [Green Version]
- Jose, R.; Ramakrishna, S. Materials 4.0: Materials big data enabled materials discovery. Appl. Mater. Today 2018, 10, 127–132. [Google Scholar] [CrossRef]
- Santos, I.; Nieves, J.; Penya, Y.K.; Bringas, P. Machine-learning-based mechanical properties prediction in foundry production. In Proceedings of the ICCAS-SICE 2009—ICROS-SICE International Joint Conference 2009, Fukuoka, Japan, 18–21 August 2009; pp. 4536–4541. [Google Scholar]
- Merayo, D.; Rodríguez-Prieto, Á.; Camacho, A. Prediction of physical and mechanical properties for metallic materials selection using big data and artificial neural networks. IEEE Access 2017, 20, 1–9. [Google Scholar] [CrossRef]
- Díaz Lantada, A.; Franco-Martínez, F.; Hengsbach, S.; Rupp, F.; Thelen, R.; Bade, K. Artificial intelligence aided design of microtextured surfaces: Application to controlling wettability. Nanomaterials 2020, 10, 2287. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Alavi, A.H. Artificial intelligence-enabled smart mechanical metamaterials: Advent and future trends. Int. Mater. Rev. 2021, 66, 365–393. [Google Scholar] [CrossRef]
- Bonfanti, S.; Guerra, R.; Font-Clos, F.; Rayneau-Kirkhope, D.; Zapperi, S. Automatic design of mechanical metamaterial actuators. Nat. Commun. 2020, 11, 4162. [Google Scholar] [CrossRef] [PubMed]
- Langer, R.; Vacanti, J.P. Tissue engineering. Science 1993, 260, 920–926. [Google Scholar] [CrossRef] [Green Version]
- Khademhosseini, A.; Vacanti, J.P.; Langer, R. Progress in tissue engineering. Sci. Am. 2009, 300, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Boccaccio, A.; Ballini, A.; Pappalettere, C.; Tullo, D.; Cantore, S.; Desiate, A. Finite element method (FEM), mechanobiology and biomimetic scaffolds in bone tissue engineering. Int. J. Biol. Sci. 2011, 7, 112–132. [Google Scholar] [CrossRef] [PubMed]
- Egan, P.F.; Gonella, V.C.; Engensperger, M.; Ferguson, S.J.; Shea, K. Computationally designed lattices with tuned properties for tissue engineering using 3D printing. PLoS ONE 2017, 12, e0182902. [Google Scholar]
- Conev, A.; Litsa, E.E.; Perez, M.R.; Diba, M.; Mikos, A.G.; Kavraki, L.E. Machine learning-guided three-dimensional printing of tissue engineering scaffolds. Tissue Eng. Part A 2020, 26, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ge, H.; Zhou, X.; Yan, J.; Chi, Q.; Zhang, Z. Prediction of vascular tissue engineering results with artificial neural networks. J. Biomed. Inform. 2005, 38, 417–421. [Google Scholar] [CrossRef] [Green Version]
- Sujeeun, L.Y.; Goonoo, N.; Ramphul, H.; Chummun, I.; Gimié, F.; Baichoo, S.; Bhaw-Luximon, A. Correlating in vitro performance with physico-chemical characteristics of nanofibrous scaffolds for skin tissue engineering using supervised machine learning algorithms. R. Soc. Open Sci. 2020, 7, 201293. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, A.; Babaei, M.; Shams, R.; Wang, Y.D.; Chung, T. DeePore: A deep learning workflow for rapid and comprehensive characterization of porous materials. Adv. Water Resour. 2020, 146, 103787. [Google Scholar] [CrossRef]
- Serj, M.F.; Lavi, B.; Hoff, G.; Valls, D.P. A deep convolutional neural network for lung cancer diagnostic. arXiv 2018, arXiv:1804.08170. [Google Scholar]
- Bassi, P.R.A.S.; Attux, R. A deep convolutional neural network for COVID-19 detection using chest X-rays. Res. Biomed. Eng. 2021, 1–10. [Google Scholar] [CrossRef]
- Ghaderzadeh, M.; Asadi, F.; Jafari, R.; Bashash, D.; Abolghasemi, H.; Aria, M. Deep convolutional neural network-based computer-aided detection system for COVID-19 using multiple lung scans: Design and Implementation Study. J. Med. Internet Res. 2021, 23, e27468. [Google Scholar] [CrossRef] [PubMed]
- García, I.E.G. Desarrollo de una red Neuronal Convolucional Volumétrica para la Síntesis de Imagen en Resonancia Magnética Nuclear Cerebral. Ph.D. Thesis, Universidad Politécnica de Valencia, Valencia, Spain, 2019. [Google Scholar]
- La librería Numpy. Aprende con Alf. Available online: https://aprendeconalf.es/docencia/python/manual/numpy/ (accessed on 23 June 2021).
- Matplotlib: Python Plotting—Matplotlib 3.4.2 Documentation. 2021. Available online: https://matplotlib.org/ (accessed on 23 June 2021).
- Scikit-Learn: Machine Learning in Python—Scikit-Learn 0.24.2 Documentation. Available online: https://scikit-learn.org/stable/ (accessed on 23 June 2021).
- Santos, P.G.D.L. Introducción a Scikit-Image, Procesamiento de Imágenes en Python. 2016. Available online: https://numython.github.io/posts/2016/01/introduccion-scikit-image-procesamiento/ (accessed on 23 June 2021).
- TensorFlow. Available online: https://www.tensorflow.org/?hl=es-419 (accessed on 23 June 2021).
- Keras: The Python Deep Learning API. Available online: https://keras.io/guides/ (accessed on 23 June 2021).
- Keras Batch Normalization Layer. Available online: https://keras.io/api/layers/normalization_layers/batch_normalization/ (accessed on 23 June 2021).
- Brownlee, J. A gentle introduction to batch normalization for deep neural networks. In Machine Learning Mastery; Machine Learning Mastery: San Juan, Puerto Rico, 2019; Available online: https://machinelearningmastery.com/batch-normalization-for-training-of-deep-neural-networks/ (accessed on 23 June 2021).
- Keras Dropout Layer. Available online: https://keras.io/api/layers/regularization_layers/dropout/ (accessed on 23 June 2021).
- Zunair, H. 3D Image Classification from CT Scans. Available online: https://keras.io/examples/vision/3D_image_classification/ (accessed on 23 June 2021).
- Chan, C.H.M. Step by Step Implementation: 3D Convolutional Neural Network in Keras. In Towards Data Science; 2020; Available online: https://towardsdatascience.com/about (accessed on 26 July 2021).
- Alwattar, T.A.; Mian, A. Development of an Elastic Material Model for BCC Lattice Cell Structures Using Finite Element Analysis and Neural Networks Approaches. J. Compos. Sci. 2019, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Jafari Chashmi, M.; Fathi, A.; Shirzad, M.; Jafari-Talookolaei, R.-A.; Bodaghi, M.; Rabiee, S.M. Design and Analysis of Porous Functionally Graded Femoral Prostheses with Improved Stress Shielding. Designs 2020, 4, 12. [Google Scholar] [CrossRef]
- Sinitskiy, A.V.; Pande, V. Deep Neural Network Computes Electron Densities and Energies of a Large Set of Organic Molecules Faster than Density Functional Theory (DFT). arXiv 2018, arXiv:1809.02723. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).