Comprehensive Biological Evaluation of Biomaterials Used in Spinal and Orthopedic Surgery
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Materials
2.2. Chemicals and Disposables
2.3. Preparation of Biomaterial’s Samples
2.4. Cell Culture
2.5. Cytotoxicity Test
2.6. Bacterial Colonization Test
2.7. Blood Platelets Adhesion Test
2.8. RNA and Protein Isolation
2.9. Microarray Analysis
2.10. 2D Electrophoresis
2.11. Mass Spectrometry Analysis
2.12. Statistical Analysis
3. Results
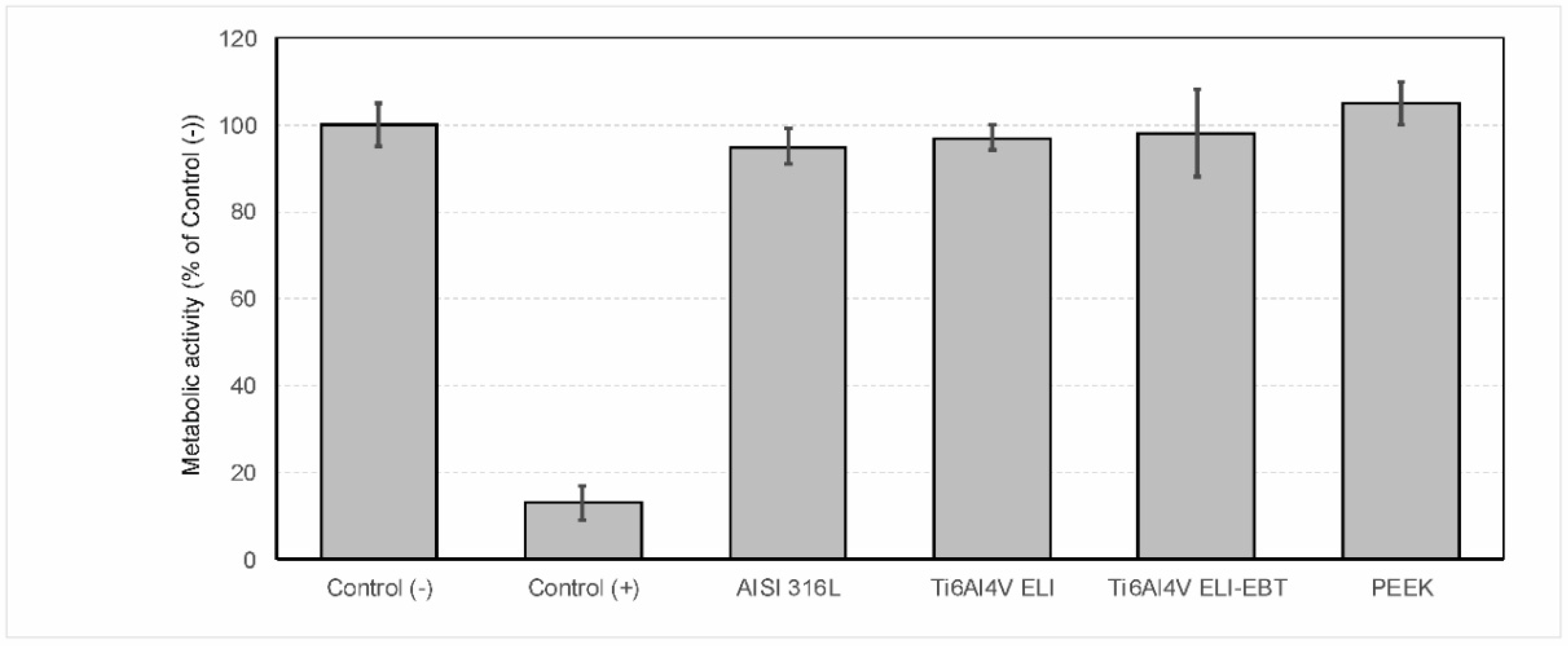
3.1. Cytotoxicity Test
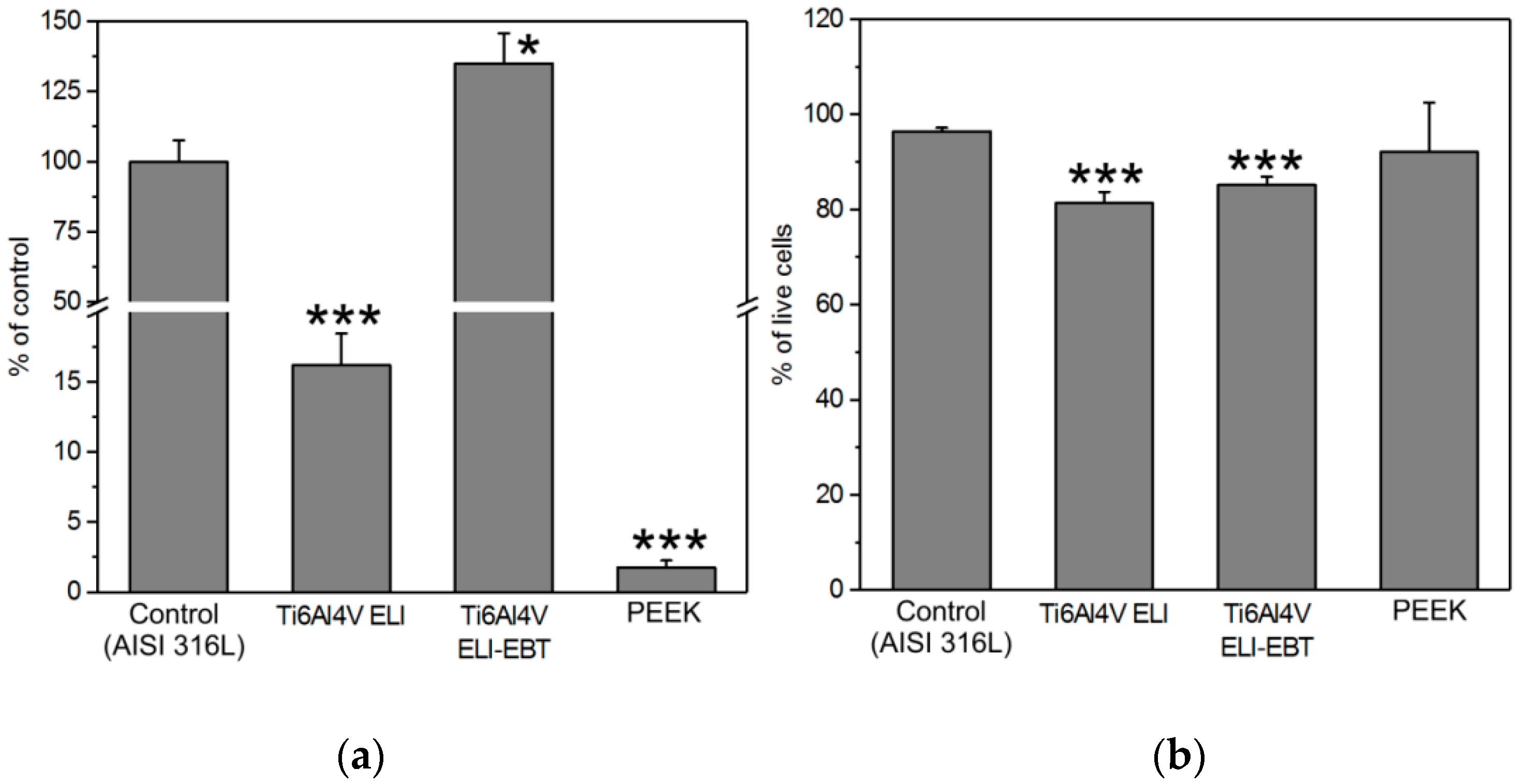
3.2. Bacterial Colonization Test
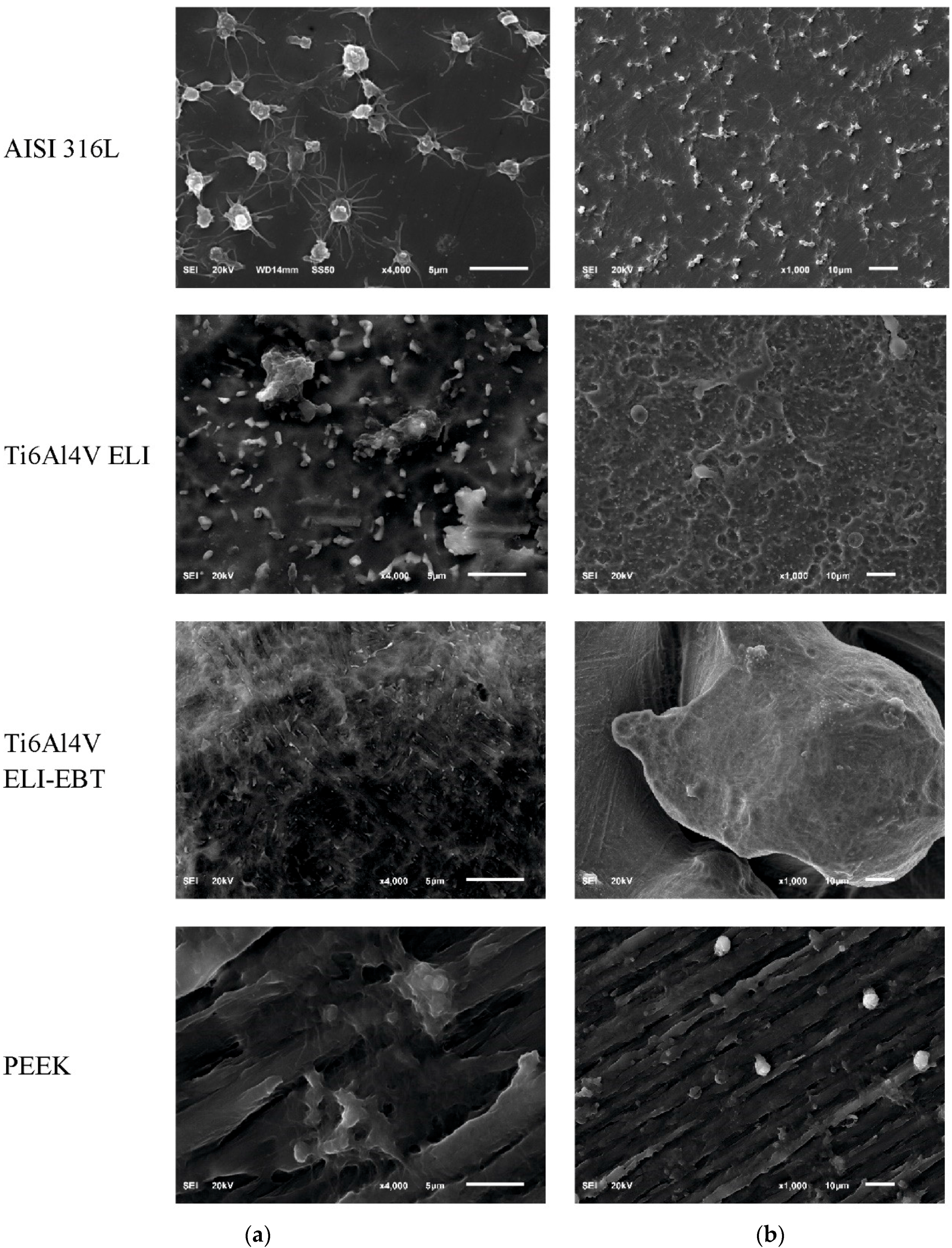
3.3. Blood Platelets Adhesion Test
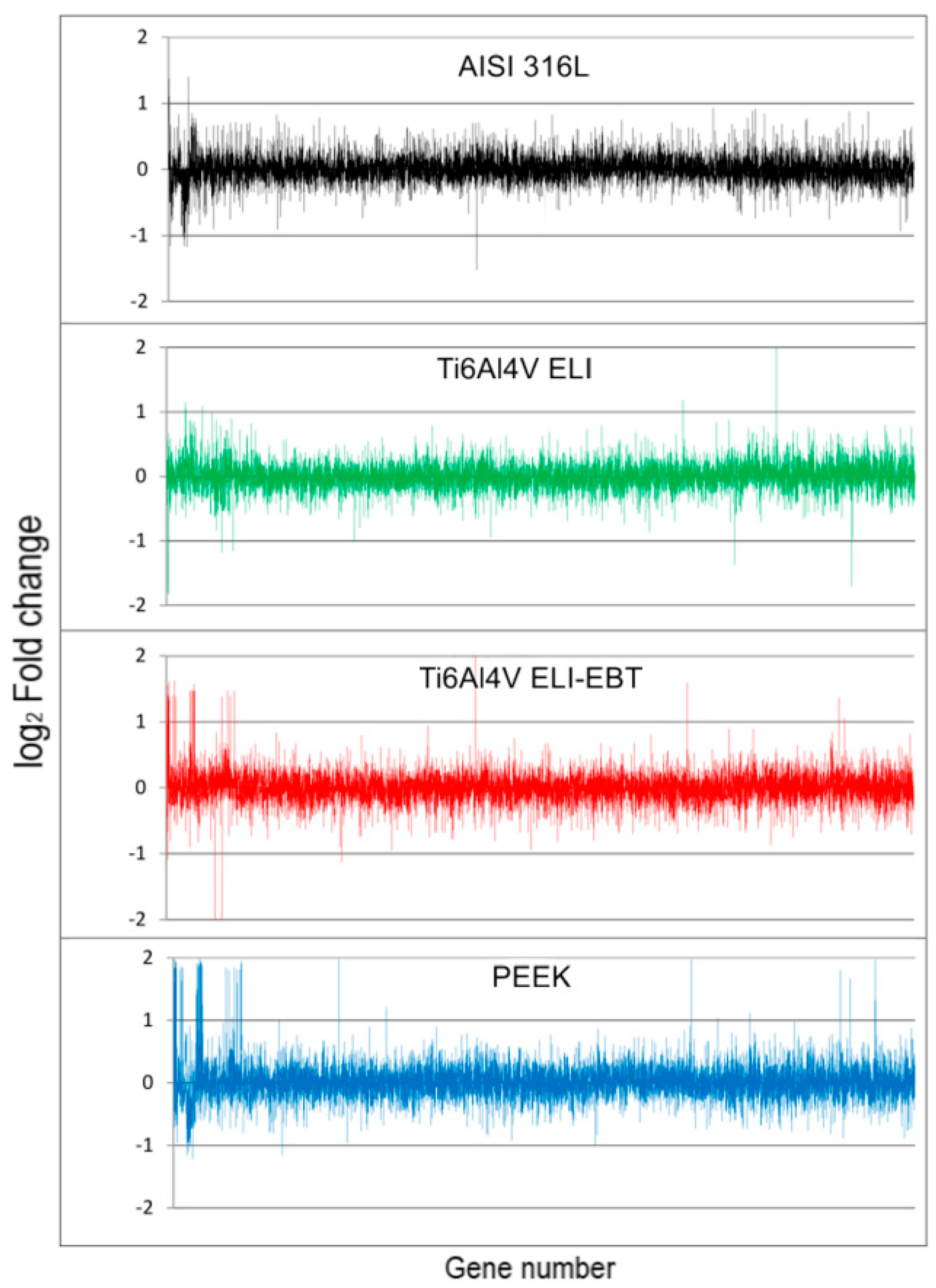
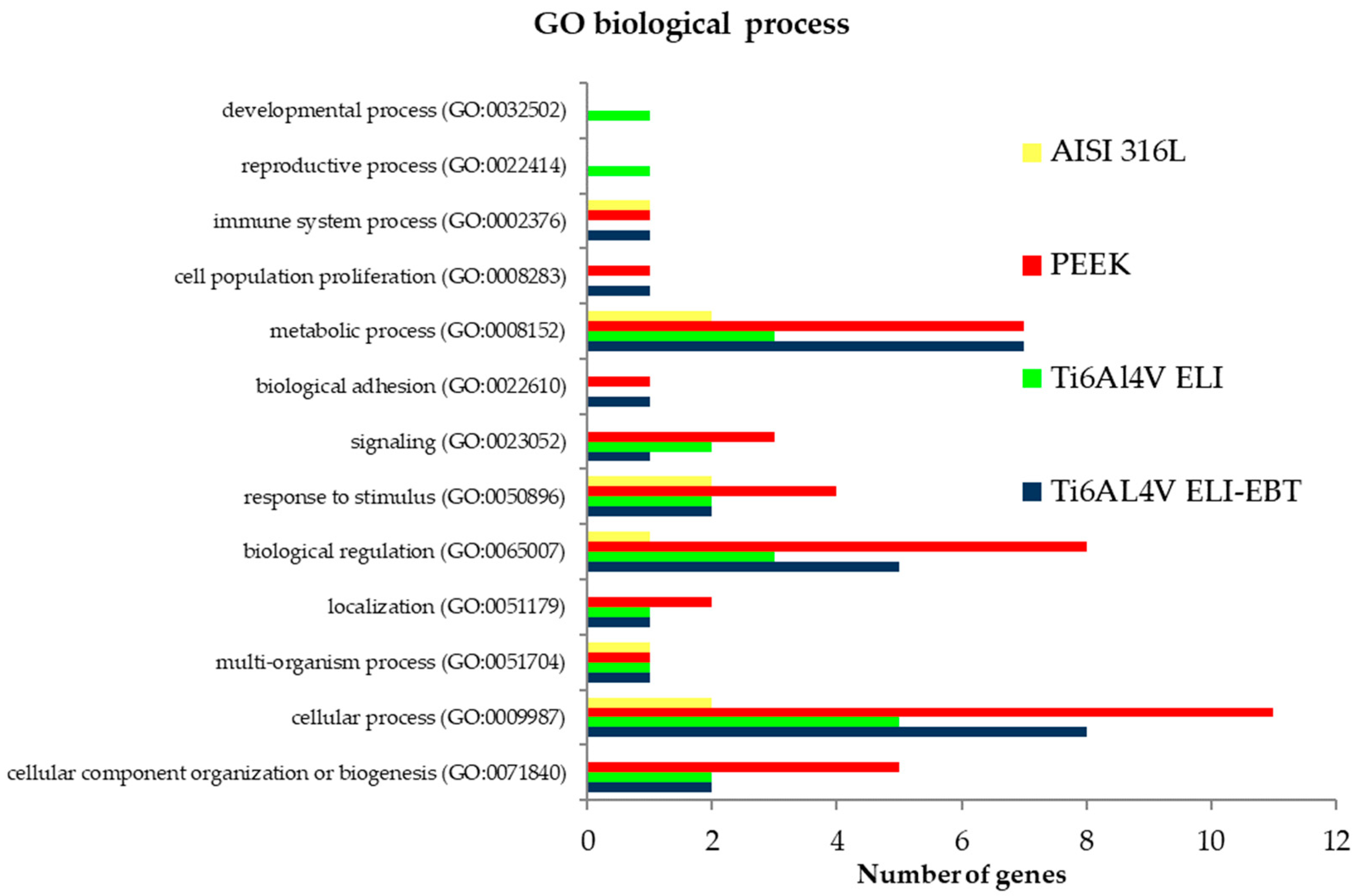
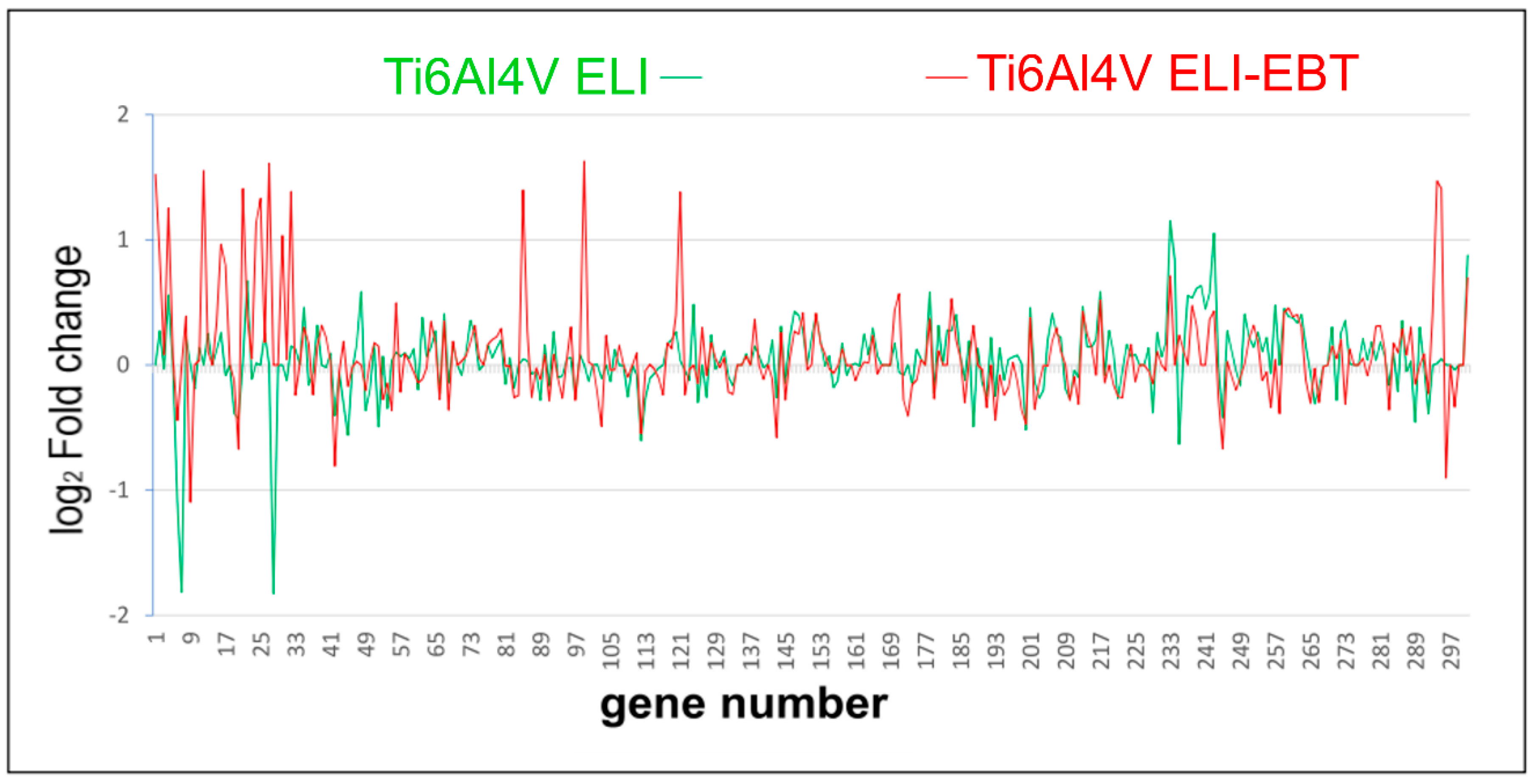
3.4. Microarray Analysis
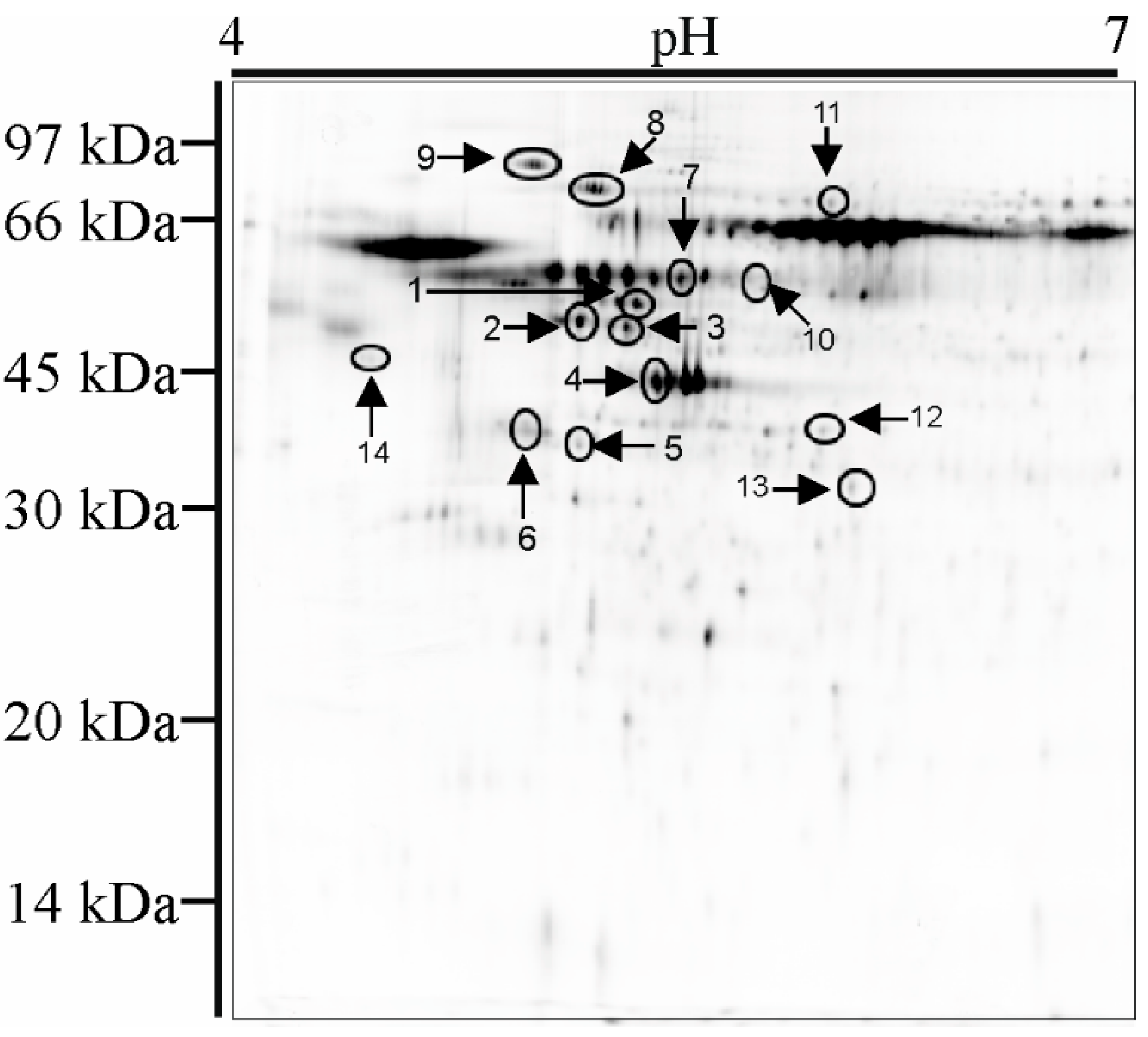
3.5. 2D Electrophoresis and Mass Spectrometry Analysis
4. Discussion
4.1. Cytotoxicity Test and Bacterial Colonization Test
4.2. Blood Platelets Adhesion Test
4.3. Microarray Analysis
4.4. 2D Electrophoresis and Mass Spectrometry Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Du, Z.; Zhu, Z.; Wang, Y. The degree of peri-implant osteolysis induced by PEEK, CoCrMo, and HXLPE wear particles: A study based on a porous Ti6Al4V implant in a rabbit model. J. Orthop. Surg. Res. 2018, 13, 1–9. [Google Scholar] [CrossRef]
- Hallab, N.J.; Cunningham, B.W.; Jacobs, J.J. Spinal Implant Debris-Induced Osteolysis. Spine 2003, 28, S125–S138. [Google Scholar] [CrossRef] [PubMed]
- Hallab, N.J. A review of the biologic effects of spine implant debris: Fact from fiction. SAS J. 2009, 3, 143–160. [Google Scholar] [CrossRef]
- Dattani, R. Femoral osteolysis following total hip replacement. Postgrad. Med. J. 2007, 83, 312–316. [Google Scholar] [CrossRef]
- Buehler, M.J.; Cranford, S. Materiomics: Biological protein materials, from nano to macro. Nanotechnol. Sci. Appl. 2010, 3, 127–148. [Google Scholar] [CrossRef]
- Cranford, S.W.; De Boer, J.; Van Blitterswijk, C.A.; Buehler, M.J. Materiomics: An -omics Approach to Biomaterials Research. Adv. Mater. 2013, 25, 802–824. [Google Scholar] [CrossRef]
- Cranford, S.W.; Buehler, M.J. Biomateriomics; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar] [CrossRef]
- Groen, N.; Guvendiren, M.; Rabitz, H.; Welsh, W.J.; Kohn, J.; De Boer, J. Stepping into the omics era: Opportunities and challenges for biomaterials science and engineering. Acta Biomater. 2016, 34, 133–142. [Google Scholar] [CrossRef]
- Hebels, D.G.; Carlier, A.; Coonen, M.L.; Theunissen, D.H.; De Boer, J. cBiT: A transcriptomics database for innovative biomaterial engineering. Biomaterials 2017, 149, 88–97. [Google Scholar] [CrossRef]
- ISO. Implants for Surgery—Metallic Materials—Part 3: Wrought Titanium 6-Aluminium 4-Vanadium Alloy; 5832-3: 2016; International Organization for Standardization: Geneva, Switzerland, 2016. [Google Scholar]
- ASTM. Standard Specification for Polyetheretherketone (PEEK) Polymers for Surgical Implant Applications; F2026-17; ASTM International: West Conshohocken, PA, USA, 2017. [Google Scholar] [CrossRef]
- Ciupik, L.; Kierzkowska, A. Technology-biomechanical evaluation of metal biomaterials derived by layer technology. Eng. Biomater. 2010, 14–18. [Google Scholar] [CrossRef]
- Walkowiak-Przybyło, M.; Klimek, L.; Okroj, W.; Jakubowski, W.; Chwiłka, M.; Czajka, A.; Walkowiak, B. Adhesion, activation, and aggregation of blood platelets and biofilm formation on the surfaces of titanium alloys Ti6Al4V and Ti6Al7Nb. J. Biomed. Mater. Res. Part A 2012, 100, 768–775. [Google Scholar] [CrossRef]
- Jakubowski, W.; Slosarczyk, A.; Paszkiewicz, Z.; Szymanski, W.; Walkowiak, B. Bacterial colonisation of bioceramic surfaces. Adv. Appl. Ceram. 2008, 107, 217–221. [Google Scholar] [CrossRef]
- Image Processing and Analysis in Java. Available online: https://imagej.nih.gov/ij/ (accessed on 17 September 2020).
- Okroj, W.; Kaminska, M.; Klimek, L.; Szymanski, W.; Walkowiak, B. Blood platelets in contact with nanocrystalline diamond surfaces. Diam. Relat. Mater. 2006, 15, 1535–1539. [Google Scholar] [CrossRef]
- Okrój, W.; Walkowiak-Przybyło, M.; Rośniak-Bak, K.; Klimek, L.; Walkowiak, B. Comparison of microscopic methods for evaluating platelet adhesion to biomaterial surfaces. Acta Bioeng. Biomech. 2009, 11, 45–49. [Google Scholar]
- Kamińska, M.; Okrój, W.; Szymański, W.; Jakubowski, W.; Komorowski, P.; Nosal, A.; Szymanowski, H.; Gazicki-Lipman, M.; Jerczyńska, H.; Pawłowska, Z.; et al. Interaction of parylene C with biological objects. Acta Bioeng. Biomech. 2009, 11. [Google Scholar] [CrossRef]
- Geneontology; Panther Classification System. Available online: http://www.pantherdb.org/ (accessed on 12 October 2020).
- Pertea, M.; Salzberg, S.L. Between a chicken and a grape: Estimating the number of human genes. Genome Biol. 2010, 11, 206. [Google Scholar] [CrossRef]
- Pertea, M.; Shumate, A.; Pertea, G.; Varabyou, A.; Breitwieser, F.P.; Chang, Y.-C.; Madugundu, A.K.; Pandey, A.; Salzberg, S.L. CHESS: A new human gene catalog curated from thousands of large-scale RNA sequencing experiments reveals extensive transcriptional noise. Genome Biol. 2018, 19, 1–14. [Google Scholar] [CrossRef]
- Walkowiak, B.; Komorowski, P.; Walkowiak, M.-P. Means of Identification of Materials Engineering Products or Nanotechnologies. Polish Patent No. 223224, 11 July 2012. Available online: https://ewyszukiwarka.pue.uprp.gov.pl/search/pwp-details/P.399899 (accessed on 11 July 2012).
- Komorowski, P.; Walkowiak-Przybyło, M.; Walkowiak, B. Early cell response to contact with biomaterial’s surface. J. Biomed. Mater. Res. Part B Appl. Biomater. 2015, 104, 880–893. [Google Scholar] [CrossRef]
- Walkowiak-Przybyło, M.; Komorowski, P.; Walkowiak, B. Differences in the expression of cell cycle genes in osteoblasts and endothelial cells cultured on the surfaces of Ti6Al4V and Ti6Al7Nb alloys. J. Biomed. Mater. Res. Part A 2017, 105, 1607–1617. [Google Scholar] [CrossRef]
- Chatterjee, N.; Eom, H.-J.; Choi, J. A systems toxicology approach to the surface functionality control of graphene–cell interactions. Biomaterials 2014, 35, 1109–1127. [Google Scholar] [CrossRef]
- McNamara, L.E.; Burchmore, R.; Riehle, M.O.; Herzyk, P.; Biggs, M.J.; Wilkinson, C.D.; Curtis, A.S.; Dalby, M.J. The role of microtopography in cellular mechanotransduction. Biomaterials 2012, 33, 2835–2847. [Google Scholar] [CrossRef]
- Sagomonyants, K.B.; Jarman-Smith, M.L.; Devine, J.N.; Aronow, M.S.; Gronowicz, G.A. The in vitro response of human osteoblasts to polyetheretherketone (PEEK) substrates compared to commercially pure titanium. Biomaterials 2008, 29, 1563–1572. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; An, M.; Wang, Q.; Liu, X.; Lai, W.; Zhao, Y.; Wei, S.; Ji, J.-G. Quantitative proteomic analysis of human osteoblast-like MG-63 cells in response to bioinert implant material titanium and polyetheretherketone. J. Proteom. 2012, 75, 3560–3573. [Google Scholar] [CrossRef]
| Biomaterial | AISI 316L | PEEK | Ti6Al4V ELI | Ti6Al4V ELI-EBT |
|---|---|---|---|---|
| small aggregates (<10 platelets) | 34.8 ± 14.6 | 2.3 ± 1.1 | 1.7 ± 0.9 | 0.65 ± 1.0 |
| large aggregates (>10 platelets) | 3.7 ± 3.5 | 1.0 ± 1.3 | 1.4 ± 0.8 | 0.2 ± 0.5 |
| singular platelets | 28.8 ± 16.2 | 3.5 ± 3.7 | 0.0 ± 0.0 | 0.6 ± 0.7 |
| Biomaterial | AISI 316L | PEEK | Ti6Al4V ELI | Ti6Al4V ELI-EBT |
|---|---|---|---|---|
| Number of expressed genes | 8716 | 9160 | 8455 | 8975 |
| Number of highly specifically overexpressed genes for one particular biomaterial at FC ≥ 2, and FC irrelevant for others. | 1 | 14 | 3 | 1 |
| Name of genes | VAC14 | PMCH | XLOC_l2_008203 | lnc-TNFRSF14-2 |
| GPR182 | SPAG9 | |||
| UQCRFS1 | PRR5-ARHGAP8 | |||
| SNX18 | ||||
| GFRA2 | ||||
| LOC102723429 | ||||
| MAP2K6 | ||||
| lnc-DYDC1-4 | ||||
| A_22_P00017766 | ||||
| lnc-ZC3H12D-2 | ||||
| A_22_P00022299 | ||||
| LOC101928894 | ||||
| DHRS4L1 | ||||
| PROM2 | ||||
| Number of highly specifically suppressed genes for one particular biomaterial at FC ≤ 0.5, and FC irrelevant for others. | 0 | 1 | 3 | 2 |
| Name of genes | - | TOMM20L | SCRG1 | A_22_P00001871 |
| USP14 | LOC100507053 | |||
| POMZP3 | ||||
| Number of specifically overexpressed genes for one particular biomaterial at FC ≥ 2, and FC also relevant for at least one but not all other. | 3 | 9 | 3 | 1 |
| Name of genes | HMOX1 | TMEM158 | SNORA16A | TMEM65 |
| G6PD | SMARCA4 | SNORD105B | ||
| RNU6ATAC | MMP14 | lnc-RP1-177G6.2.1-2 | ||
| MTRNR2L2 | ||||
| lnc-MINA-3 | ||||
| lnc-RNF13-2 | ||||
| HS3ST1 | ||||
| ADAMTS1 | ||||
| IL6ST | ||||
| Number of specifically suppressed genes for one particular biomaterial at FC ≤ 0.5, and FC also relevant for at least one but not all other. | 3 | 2 | 5 | 2 |
| Name of genes | IFIT1 | SNORD12C | TAF15 | CPA4 |
| SNORA48 | APH1A | FOXD3-AS1 | ANKRD1 | |
| SNORA2B | A_22_P00020320 | |||
| PPA2 | ||||
| NLE1 |
| AISI 316L | PEEK | Ti6Al4V ELI | Ti6Al4V ELI-EBT | |
|---|---|---|---|---|
| Number of detected spots | 739 | 755 | 656 | 700 |
| Upregulated proteins | ||||
| Number of proteins | 29 | 27 | 33 | 14 |
| Identified proteins gene name | ALBU | ALBU | ||
| Downregulated proteins | ||||
| Number of proteins | 131 | 114 | 190 | 119 |
| Identified proteins gene name | ACTB | ACTB | ACTB | ACTB |
| RLA0 | RLA0 | RLA0 | RLA0 | |
| RSSA | RSSA | LDHB | RSSA | |
| UCHL1 | UCHL1 | ENPL | ||
| LDHB | ATPB/PDIA6 | |||
| ENPL | CH60/HNRPK | |||
| HS90A | HNRPK | |||
| HS90B | TBB5 | |||
| TBA1B | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komorowski, P.; Siatkowska, M.; Kamińska, M.; Jakubowski, W.; Walczyńska, M.; Walkowiak-Przybyło, M.; Szymański, W.; Piersa, K.; Wielowski, P.; Sokołowska, P.; et al. Comprehensive Biological Evaluation of Biomaterials Used in Spinal and Orthopedic Surgery. Materials 2020, 13, 4769. https://doi.org/10.3390/ma13214769
Komorowski P, Siatkowska M, Kamińska M, Jakubowski W, Walczyńska M, Walkowiak-Przybyło M, Szymański W, Piersa K, Wielowski P, Sokołowska P, et al. Comprehensive Biological Evaluation of Biomaterials Used in Spinal and Orthopedic Surgery. Materials. 2020; 13(21):4769. https://doi.org/10.3390/ma13214769
Chicago/Turabian StyleKomorowski, Piotr, Małgorzata Siatkowska, Marta Kamińska, Witold Jakubowski, Marta Walczyńska, Magdalena Walkowiak-Przybyło, Witold Szymański, Katarzyna Piersa, Patryk Wielowski, Paulina Sokołowska, and et al. 2020. "Comprehensive Biological Evaluation of Biomaterials Used in Spinal and Orthopedic Surgery" Materials 13, no. 21: 4769. https://doi.org/10.3390/ma13214769
APA StyleKomorowski, P., Siatkowska, M., Kamińska, M., Jakubowski, W., Walczyńska, M., Walkowiak-Przybyło, M., Szymański, W., Piersa, K., Wielowski, P., Sokołowska, P., Białkowska, K., Makowski, K., Elgalal, M., Kierzkowska, A., Ciupik, L., & Walkowiak, B. (2020). Comprehensive Biological Evaluation of Biomaterials Used in Spinal and Orthopedic Surgery. Materials, 13(21), 4769. https://doi.org/10.3390/ma13214769





