1. Introduction
Nanoparticles (NPs) have been explored for a wide range of applications, including biosensors and chemical sensors, bioimaging, catalysis, optics, electronics, drug delivery, and energy [
1]. The conventional chemical or physical methods for the production of nanoscale materials depend on the utilization of toxic stabilizing and capping agents, which pose risks to the environment and human health [
2]. Therefore, there is an increasing need to develop cost-effective, non-toxic, and environmentally green approaches that do not use toxic chemicals to produce NPs. The microbial synthesis of NPs, which takes advantage of microbial detoxification of metallic ions and precipitation of metals through reducing enzymes, intracellular or extracellular reducing reagents and metal-binding proteins, showed immense potential [
3,
4].
In particular, there is an increasing interest in the exploitation of the cysteine-rich metal-binding proteins, metallothioneins (MTs), in the development of biofactories for metal NP production. MTs are found in all eukaryotes, as well as some prokaryotes, and play an important role in intracellular metal distribution, accumulation, as well as protection of cells against heavy metal toxicity and oxidative stress [
5,
6]. The Cys-Xaa-Cys clusters of MTs can act as a reducing functional group for the reduction of metal ions, chelation, and accumulation of metal particles in the cells [
5,
6,
7]. Studies showed that MTs are involved in the detoxification of several metals, including zinc, mercury, copper and cadmium in microorganisms [
5,
6,
8]. Due to their important role in metal reduction and high metal binding affinity, studies have been seeking to integrate MTs in creating biological platforms for the production of metal NPs. Engineered bacteria expressing MT and/or phytochelatins have been shown to be able to produce diverse nanoparticles [
9,
10,
11].
A critical aspect in developing a microbial platform for NP synthesis is choosing a proper host microorganism. It is ideal that a biofactory can make NPs with consistent properties for their applications in various field of nanotechnology. Many types of microorganism have been shown to be able to synthesize various metal NPs, but with diverse properties [
12,
13,
14,
15,
16]. In addition, the conditions for cultivation and maintenance of microorganisms vary for different species [
14,
15,
16]. Among the microorganisms able to serve as potential biofactories for metal NP synthesis, the readily available bacteria,
E. coli, is of particular interest, due to the relatively easy and simple procedures to grow, maintain and manipulate them in the lab. The potential of engineered
E. coli as a common platform for synthesis of diverse NP has been explored; however, the efficiency of NP synthesis by the engineered bacteria and the potential to upgrade to large scale NP production were not examined [
9,
10,
11].
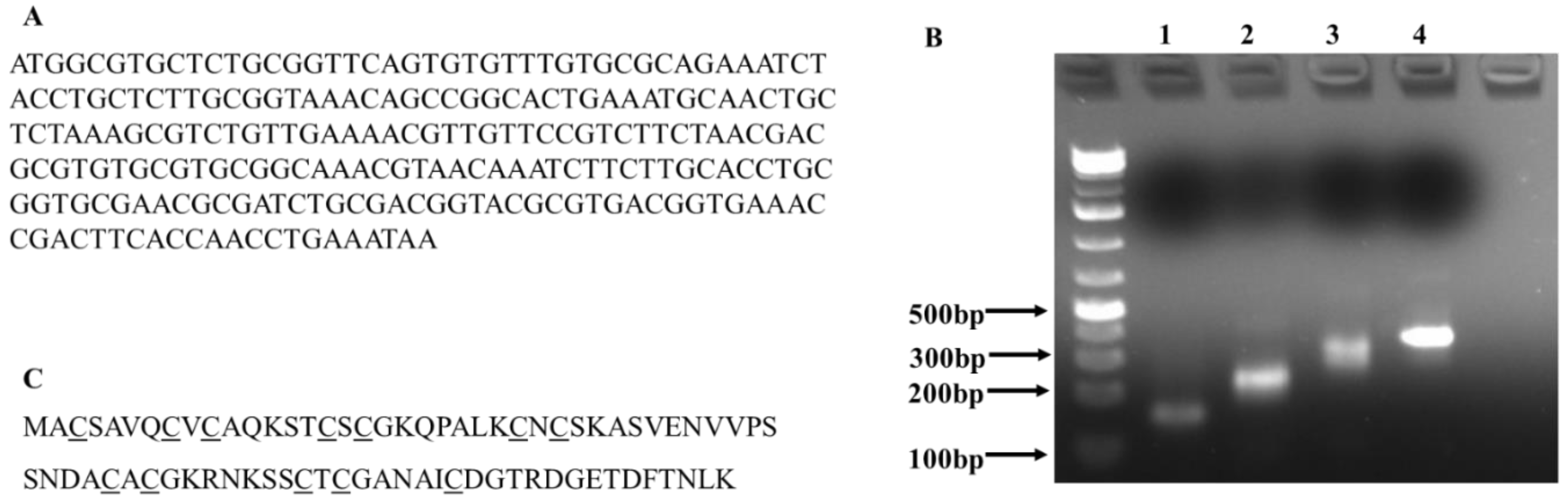
The goal of this study was to provide preliminary evidence for exploiting engineered E. coil as a platform for the large-scale production of metal NPs with controlled morphological properties. In this study, we chose C. albicans MT as a target protein and transferred a PCR-assembled C. albicans MT gene into E. coli DH5α cells to examine whether expression of C. albicans MT in E. coli would improve bacterial NP synthesis.
Compared to
E. coli MT, which has four cysteine out of 56 residues (7.1%), the
C. albicans MT has a relatively high content of cysteine residues (15.8%). Importantly, the
C. albicans has four repeats of Cys-Xaa-Cys presumably for binding metal ions, such as copper [
17,
18]. To examine whether the expression of this MT protein will improve NP synthesis, we used biosynthesis of silver NPs as an example to investigate the effects of
C. albicans MT on NP production. Our results showed that the
E. coli cells expressing
C. albicans MT produced silver NPs faster than
E. coli cells transformed with pUC19 empty vectors. Particularly, the silver NPs synthesized by engineered
E. coli are more uniform in size and shape.
2. Materials and Methods
2.1. Synthesis of C. Albicans MT Gene and Transformation of E. Coli DH5α Cells
To obtain the
C. albicans MT gene, first, the MT protein sequence of the
C. albicans was obtained from the NCBI protein database. Then the
C. albicans MT gene was codon-optimized for protein expression in
E. coli, and eight oligonucleotides for gene synthesis were designed using DNAworks. The oligonucleotides were synthesized by Eurofins Genomics (Louisville, KY, USA). The
C. albicans gene was synthesized in four consecutive PCR reactions, as described by D. Marsc [
19]. Assembled gene was inserted between HindIII and BamHI sites of plasmid pUC19 through in vivo homologous recombination in
E. coli DH5α cells. Plasmids were propagated in
E. coli DH5α cells, and clones carrying the error-free gene are confirmed by DNA sequencing (Eurofins Genomics, Louisville, KY, USA). To obtain genetically modified
E. coli cells for NP synthesis, a single colony of
E. coli DH5α cells transformed with the sequence-verified plasmid was inoculated in 5 mL of Luria-Bertani (LB) medium in the presence of 100 µg/mL of carbenicillin and cultured overnight at 37 °C with a continuous shake at 250 rpm. The overnight culture was centrifuged using Sorvall Legend X1R (Fisher Scientific, Pittsburgh, PA, USA) and the cell pellet was resuspended in 15 mL of fresh LB medium containing 100 µg/mL of carbenicillin. The cells were further cultured at 37 °C with a continuous shake for 30–45 min. Then glycerol was added to the cells to reach a final concentration of 15% and cells were stored at −80 °C for future use. All chemicals and reagents were purchased from Fisher Scientific (Pittsburgh, PA, USA) unless specified.
2.2. Monitoring Cell Growth
To examine whether the expression of
C. albicans MT in E. coli affects cell growth, we monitored cell growth along time of cultivation. 50 µL of the glycerol stock cells containing the
C. albicans MT gene inserted in pUC19 plasmid (pUC-Met) was inoculated in 10 mL of LB in the presence of 100 µg/mL carbenicillin and cultured overnight at 37 °C in a shaker incubator. Cells were spun down and resuspended in 100 mL of LB with 100 µg/mL carbenicillin. Optical density at 600 nm (OD
600nm) was monitored each hour after resuspension until OD
600nm reached between 0.6 and 0.8, then OD
600nm was measured at 24 h, 48 h and 72 h. It has been shown that MT expression was induced and upregulated when microorganisms are under heavy metal stress [
5,
6,
17,
18]. To further investigate whether the expression of
C. albicans MT in
E. coli impact the bacterial tolerance to heavy metal, we cultivated the bacteria in the presence of 0.2 mM CuSO
4. 0.2 mM of CuSO
4 was added to the cells after OD
600nm reached between 0.6 and 0.8. Cells were further cultivated at 37 °C for 17–19 h with a continuous shake. OD
600nm was measured again before cells were collected by centrifugation at 4000×
g for 10 min.
E. coli cells transformed with pUC19 empty vector (pUC19-Vec) was used as control.
The viability of the cells after they grew overnight in the presence of 0.2 mM CuSO4 was estimated by plating the cells from the overnight culture on agar plates. The overnight bacterial culture was diluted with LB medium at a ratio of 1:1000, 1:10,000, 1:100,000 and 1:1,000,000, and 100 µL of each dilution was plated on agar plates in the absence of CuSO4. Cells were allowed to grow overnight at 37 °C for 17 h to obtain colonies. Individual colonies were counted to estimate the viable cells after CuSO4 challenge.
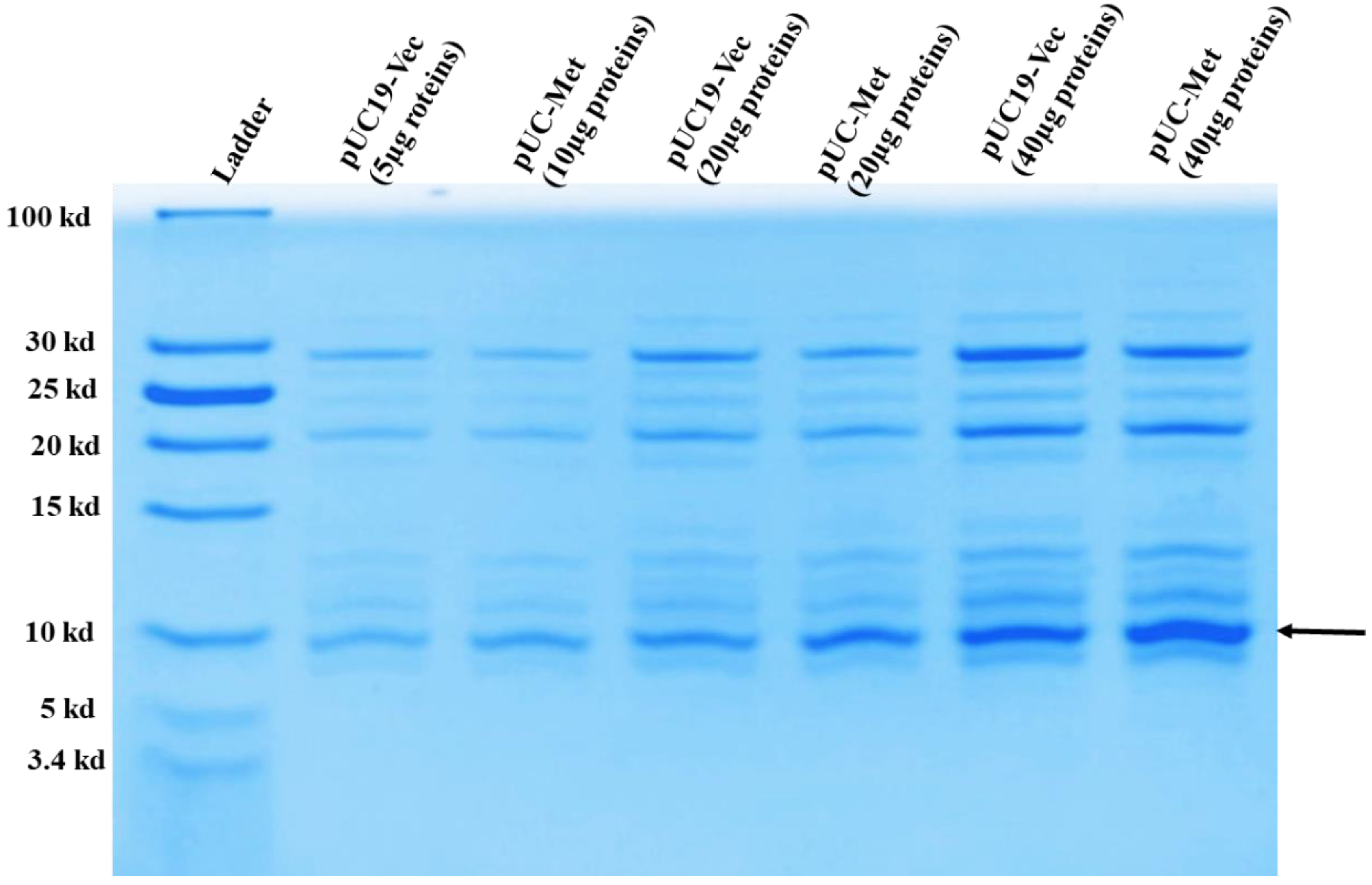
2.3. Determination of MT Protein Expression
C. albicans MT was identified as a copper binding protein, and its level increases under metal ion stresses [
5,
6,
17,
18]. To analyze the expression level of
C. albicans MT in
E. coli, the preparation of overnight culture for examining protein expression was carried out similarly as described in “Monitoring Cell Growth”. The overnight cell culture was centrifuged, and cells were resuspended in 100 mL of LB containing carbenicillin and grown at 37 °C. When OD
600nm reached between 0.6 and 0.8, bacteria were grown for another 8 h for MT protein expression analysis. Cell pellets were resuspended in lysis buffer containing 50 mM Tris-HCl, pH 8, 50 mM NaCl at a ratio of about 0.05 g wet cells/mL of buffer and sonicated to break apart the cells. The whole lysate was centrifuge again at 15,000×
g for 30 min to separate the soluble proteins from the cell membrane debris. Soluble proteins were passed through a GE Healthcare Vivaspin™ concentrator with a 30-kDa cutoff to remove the proteins larger than 30 kDa. Subsequently, the flow through was further concentrated with GE Healthcare Vivaspin™ concentrator with a 3-kDa cutoff. 10, 20, 40 µg of the concentrated proteins smaller than 30 kDa were run on 16% T, 4% C Tricine SDS-PAGE gel to separate the proteins. Proteins were stained in buffer containing 30% methanol, 10% glacial acetic acid, 0.1% Coomassie blue and 60% water for at least 2 h. Then the gel was destained in the same buffer without Coomassie blue, for overnight with continuous shake. Gel picture was taken using Gel Doc EZ Gel Documentation System (Biorad, Hercules, CA, USA).
2.4. Biosynthesis of Silver NPs by Bacteria
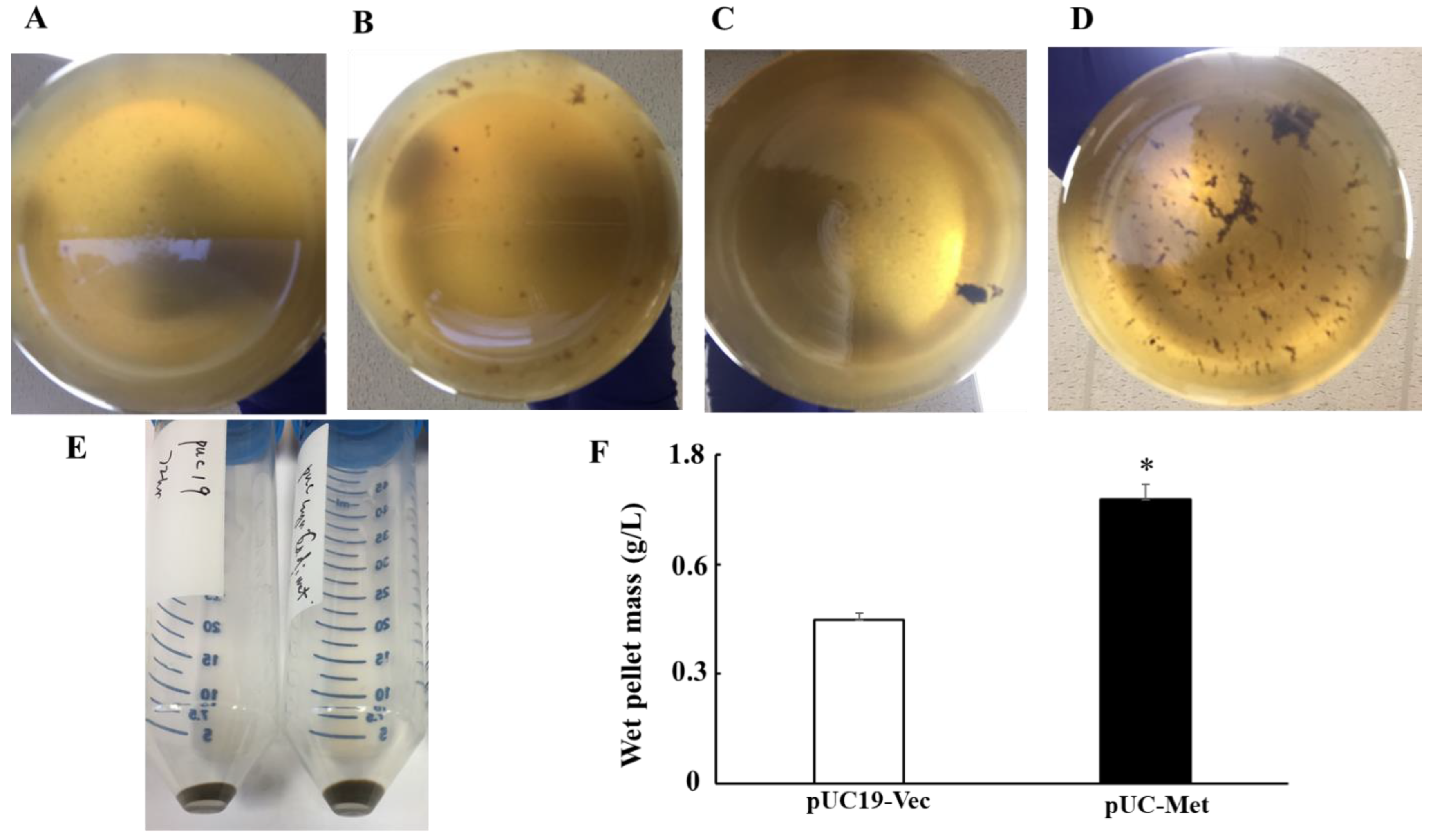
The preparation of overnight culture for silver NP synthesis was carried out similarly as described in “Monitoring Cell Growth”. The overnight cell culture was centrifuged, and cells were resuspended in 100 mL of LB in the presence of carbenicillin and grown at 37 °C. AgNO3 was added to the culture when OD600nm reached between 0.6–0.8. Cells were further cultured at 37 °C with continuous shaking for 72 h. Samples were taken to measure OD600nm at 24 h, 48 h and 72 h after addition of AgNO3. To estimate the yield of silver NPs in a larger scale, we carried out three experiments in which bacteria were grown in 1 L LB medium in the presence of 1 mM AgNO3 under the same conditions as for small scales. The wet weight of pellet collected after 72 h of cultivation was measured. After each OD600nm measurement, cultures sat for 30 min to allow particles to settle down on the bottom of the flask and take pictures of the precipitates. pUC19-Vec cells were used as a control in this experiment.
2.5. Collection of NPs
After 72 h of cultivation, cell culture was centrifuged at 4000× g for 10 min. The cell pellets were resuspended in 1.5 mL of 4 M urea, sonicated for three cycles of 45 s at an output of 20%, with an interval of 1 min between each cycle (Sonic Dismembrator, Model 50; Fisher Scientific, Pittsburgh, PA, USA). The lysate was then transferred to a 2-mL microcentrifuge tube and further spun at 10,000× g for 10 min (accuSpin™ Microcentrifuge, Fisher Scientific, Pittsburgh, PA, USA). The supernatant was discarded, and the pellet was resuspended in 1 mL of 4 M urea by 3–4 pulses of sonication, followed by centrifugation to remove the supernatant. The pellet was further washed twice with 1 mL of sterile deionized water. The final pellet was resuspended in sterile deionized water and stored in the dark at 4 °C for next step characterizations.
2.6. Fourier Transform Infrared (FTIR) Spectral Analysis
FTIR was used to reveal the functional groups that may be responsible for reducing and stabilizing the bioreduced silver NPs. The isolated NPs were diluted in deionized water, and The FTIR spectra were collected at a resolution of 2 cm−1 in transmission mode range between 4000 and 500 cm−1 using NICOLET IS10 FTIR (ThermoFisher, Waltham, MA, USA). Sterile deionized water was used as blank for this experiment.
2.7. Scanning Electron Microscopic and Scanning Transmission Electron Microscopic Characterization
We used scanning electron microscopy (SEM) and scanning transmission electron microscopy (STEM) to determine the morphological properties of the biologically synthesized silver NPs. The samples for SEM and TEM were prepared similarly. NPs washed with urea and deionized water were resuspended in sterile deionized water.
An FEI Tecnai F30 super-twin field-emission-gun transmission electron microscope (TEM) (manufactured by the FEI Company in Hillsboro, OR, USA) operating at 300 kV was used to acquire the TEM images, electron diffraction patterns and annular dark-field (ADF) scanning transmission electron microscopy (STEM) images. Chemical composition was explored using an X-ray energy dispersive spectrometer (EDS) attached to the TEM. A dual-beam focus ion beam (FIB) (FEI Helios 600 dual-beam FIB) was used to obtain the SEM images of the biosynthesized silver NPs. The size of NPs was analyzed using Gatan Digital Micrograph Software.
2.8. Statistical Analysis
All data are expressed as mean ±SD. Comparison between groups was evaluated with the Student t-test. Probability values of <0.05 were considered significant.
4. Discussion
Both wild type
E. coli and engineered
E. coli have been shown to be able to synthesize diverse NPs [
9,
10,
11]; however, it is not clear whether the engineered
E. coli cells perform better than non-engineered cells, and whether these cells have the potential to upgrade to large scale NP production. In this study, we explored the potential of application of
E. coli DH5α cells transformed with
C. albicans MT gene in the biosynthesis of more uniform silver NPs at lager scale. We used the
E. coli DH5α cells transformed with pUC19 empty plasmid vector as a control, since the
C. albicans MT gene was delivered into the cells by vector pUC19. Many studies have shown that MT is protective when cells are exposed to metal ions [
6,
17,
18]. MT proteins have high binding-affinity for heavy metal ions and serve as important reducing agents under heavy metal stress; thus, MT proteins are critical for proper accumulation, distribution and detoxification of the metal ions [
5,
17,
18]. Similarly, our results also showed that the
E. coli cells expressing
C. albicans MT (pUC-Met cells) tolerated CuSO
4 challenge better than the pUC19-Vec cells. During the prolonged cultivation in the absence of metal ion stress, the OD
600nm of pUC-Met cells was higher than that of pUC19-Vec cells at 24, 48 and 72 h after the OD
600nm reached 0.6–0.8 (
Figure 3A). Importantly, when the cells were subjected to copper ion stress for 17–19 h, the pUC-Met cells grew to a significantly higher OD
600nm than the pUC19-Vec cells (
Figure 3B). In particular, assessment of cell viability confirmed that pUC-Met cells had a higher survival rate than pUC19-Vec cells after bacteria were exposed to heavy metal stress (
Figure 3C). The better growth and higher survival of bacteria under heavy metal challenge suggested that the pUC-Met cells had increased tolerance to the metal ions, presumably due to the expression of
C. albicans MT protein.
C. albicans MT has a relatively high content of cysteine residues that can be used to reduce silver ions, bind and accumulates silver particles in the cells, thus, protect the cells from the toxicity of silver ions. We proposed that the higher tolerance to metal ions would subsequently promote the cells to make more NPs if they are used as biofactories for metal NP synthesis. Indeed, when the cells were grown in the presence of 1 mM AgNO
3, the pUC-Met cells consistently produced more silver NPs at each examination time point (
Figure 4A–E), and the final yield of NPs from 1liter culture by pUC-Met cells was greater than that of the pUC19-Vec cells (
Figure 4E, 1 g/L vs. 0.74 g/L). The yield of a wet pellet of bacterial culture in the presence of 1 mM AgNO
3 for 72 h was not compromised by increasing culture medium volume from 0.1 L to 1 L, suggesting that upgrade to large scale NP production is very promising, although more experiments are needed to confirm the conclusion. Thus, our results demonstrated that expression of
C. albicans MT in
E. coli promoted cell growth and survival and enhanced biosynthesis of silver NPs in these cells.
Even if the mechanisms of bacterial synthesis of metallic NPs remain to be completely elucidated, it has been postulated that the presence of reducing groups that can donate electrons to metal ions, such as phenol, amide, thiols and other unsaturated bonds, of the organic matter in bacteria play an important role in reducing and binding metal ions [
12,
25]. To shed light into the mechanisms of bacterial NP synthesis, we analyzed the reducing functional groups using FTIR. FTIR identified compounds that probably contain phenol, amines, amide I and isothiocyanate functional groups in all cells (
Figure 5). We hypothesized that the oxidization of C–OH group in organic molecules to C=O (amide I) group probably was involved in the reduction of Ag
+ ions to Ag
0. In addition, the amide I groups may cap and stabilize the silver NPs once the Ag
+ ions were converted into Ag
0. The possible amines (N–H) of biomolecules, such as proteins, may as well act as binding and stabilizing agents of silver NPs. Furthermore, it is well accepted that the O–H stretching in alcohols and phenolic compounds are used to reduce the Ag
+ ions into Ag
0 in the biological synthesis of metal NPs [
22,
23,
24]. Therefore, the FTIR analysis suggested that the bacteria in our study probably used similar reducing agents to reduce Ag
+ ions to Ag
0 as reported [
12,
22,
23,
24,
25]. Interestingly, a possible isothiocyanate (N=C=S) was observed in the bacteria synthesized silver NPs. Isothiocyanates are sulfur-containing antioxidants that have antimicrobial activities [
26]. It is true that the strong reducing agent isothiocyanates can play a role in biosynthesis of silver NPs, it is yet to elucidate how and why the engineered bacteria produced these compounds that can kill themselves.
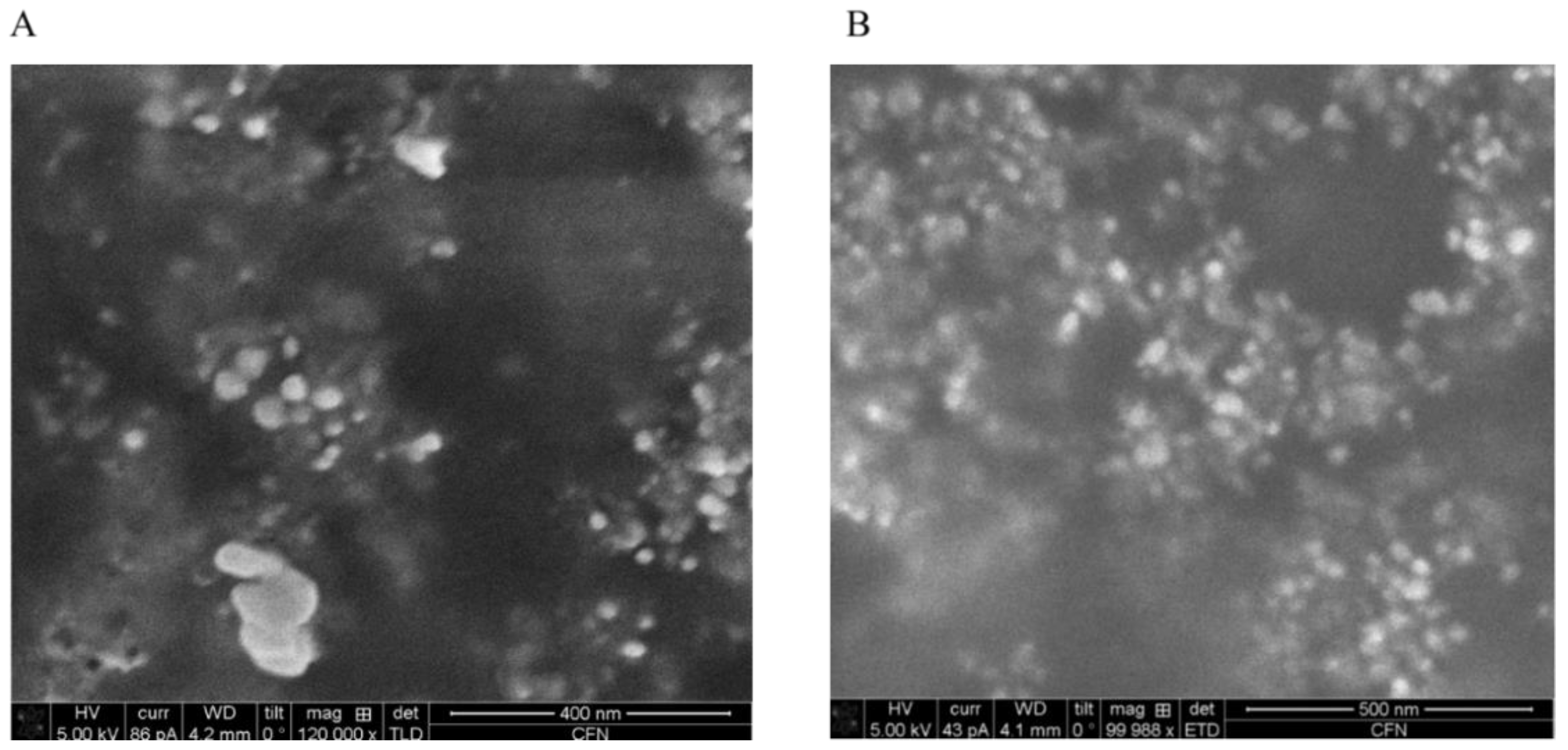
We expected that the FTIR could detect thiol groups, since the cysteine residues in the MTs contain thiol groups. However, FTIR could not catch the thiol groups associated with silver. We speculate that the thiol group, which usually is assigned to a weak peak around 2600 cm
−1, might be buried in the stretches of the strong peaks adjacent to 2600 cm
−1. To complement this drawback of FTIR, compositions of silver NPs were further assessed by STEM and SEM. Importantly, the compositional analysis confirmed that silver atoms were among the key elements in the NPs. Further, consistent with our prediction, sulfur atoms were indeed detected associated with these silver NPs, indicating that the thiol groups of proteins were involved in reducing silver ions and accumulating silver NPs (
Figure 6). Therefore, our results not only confirmed that the NPs were indeed silver NPs, but also supported that thiol groups were involved in reducing and binding silver NPs. Since
C. albicans MT has a higher level of cysteine residues that can generate a higher reducing power and metal-binding affinity in pUC-Met cells than the pUC-Met cells, we believe that the expression of
C. albicans MT in
E.
coli cells allowed the cells to convert more silver ions, and faster into silver NPs, and therefore, protected the cells from silver ion stress, as well as increased the bacterial NP yield.
Another finding of the SEM compositional assessment was the detection of chloride and copper associated with silver NPs. It has been shown that chloride is one of the key elements that
E. coli use to counter silver NP toxicity [
27]. The identification of chloride associated with silver NPs suggested that the
E. coli not only used thiol groups, but also chloride to reduce and accumulate silver under silver ion stress. It is not surprising that copper was also detected in the NPs, since
C. albicans MT was originally discovered as a copper-binding protein, and its expression was induced by copper ions [
17,
18].
FTIR analysis, SEM and STEM examinations obtained similar results in the NPs synthesized by pUC19-Vec and pUC-Met cells, suggesting the two groups of cells probably used similar mechanisms to reduce silver ions and produce NPs. However, the expression of C. albicans MT protein in E. coli conferred the engineered recombinant cells higher tolerance and better survival, thus, greater capacity to synthesize silver NPs when exposed to silver ions.
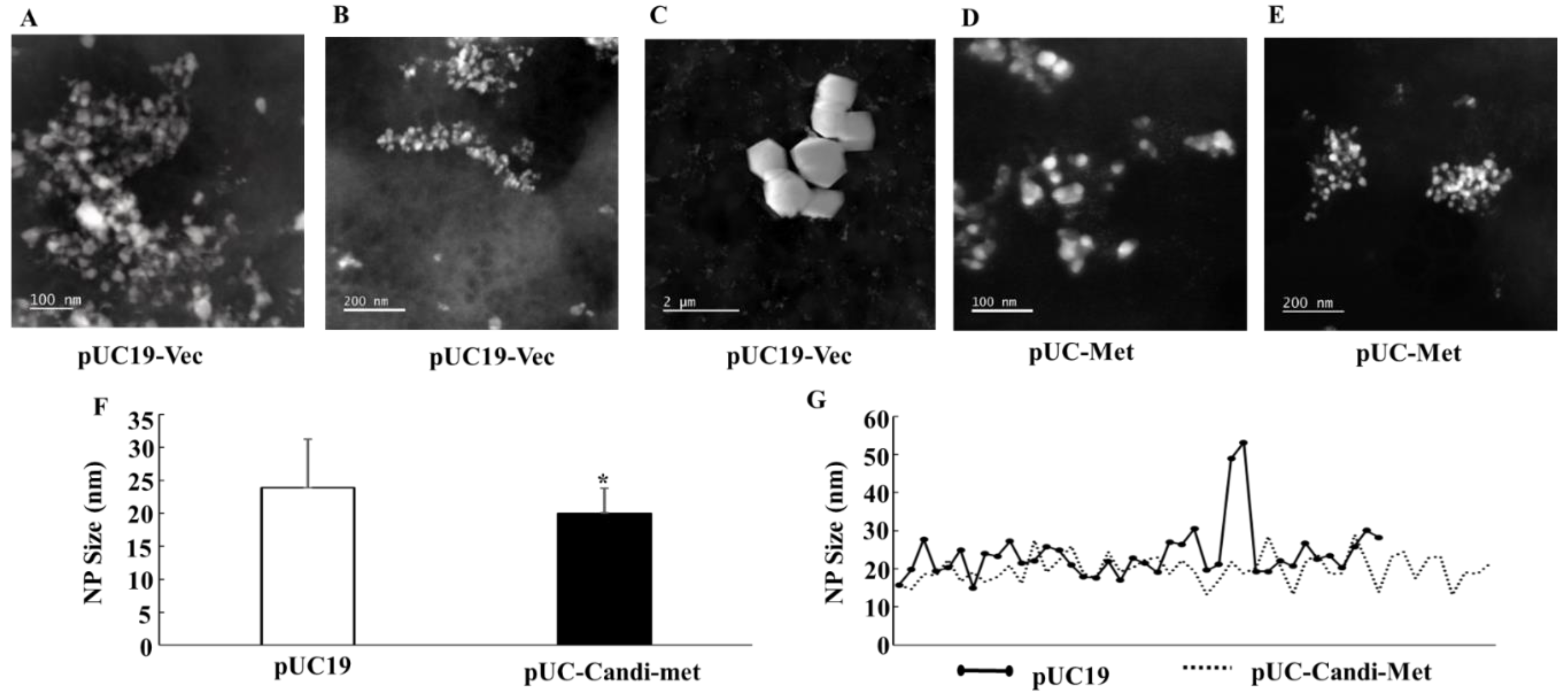
One of the problems associated with biologically synthesized NPs has been morphological heterogeneity of the NPs [
12,
13,
14]. A primary goal of this study was to investigate whether engineered
E. coli cells can improve the quality of NPs. Both SEM and STEM demonstrated that the two groups of
E. coli cells successfully produced silver particles in the nano-size range. The average size of NPs synthesized by pUC-Met cells is slightly, but significantly, smaller than the size of those produced by pUC19-Vec cells (
Figure 8F,G). However, there was a greater variation in the sizes of the NPs synthesized by pUC19-Vec cells (SD: 3.7 nm in pUC-Met vs. 7.3 nm in pUC19-Vec). Note that calculation of this SD did not include the large crystals mentioned below). Surprisingly, some micro-size silver crystals were formed by these cells (
Figure 8C). Overall, our data showed that NPs produced by pUC-Met cells not only had a more uniform size, but also appeared to have a more uniform shape. As shown in
Figure 7B,
Figure 8D,E, under SEM and STEM, it appeared that the NPs made by pUC-Met cells had a round shape, while the morphology of NPs from pUC19-Vec cells was more heterogeneous, which showed round, rectangular prism and hexagonal prism shapes (
Figure 7A and
Figure 8A,C). Thus, we demonstrated that both pUC19-Vec and pUC-Met bacterial groups were able to synthesize silver NPs. Nevertheless, the bacteria expressing
C. albicans MT had a better control in the dimension and morphology of silver NPs and a higher yield of NPs. The mechanisms of how the pUC-Met recombinant bacteria synthesized NPs with a more uniform size and shape remains to be elucidated. We speculate that more thiol groups in pUC-Met cells have a higher reducing power and a greater binding capacity for silver ions, which probably allows the cells to produce NP faster; meantime, more thiol groups can provide a larger silver binding capacity, which is more potent to stabilize the silver NPs once the silver ions are reduced. Faster reducing reactions along with a higher capacity to stabilize the NPs probably allowed the cells to synthesize NPs with a smaller size and higher uniformity.