A Novel Fluorescent Biosensor for Adenosine Triphosphate Detection Based on a Metal–Organic Framework Coating Polydopamine Layer
Abstract
:1. Introduction
2. Experimental
2.1. Materials
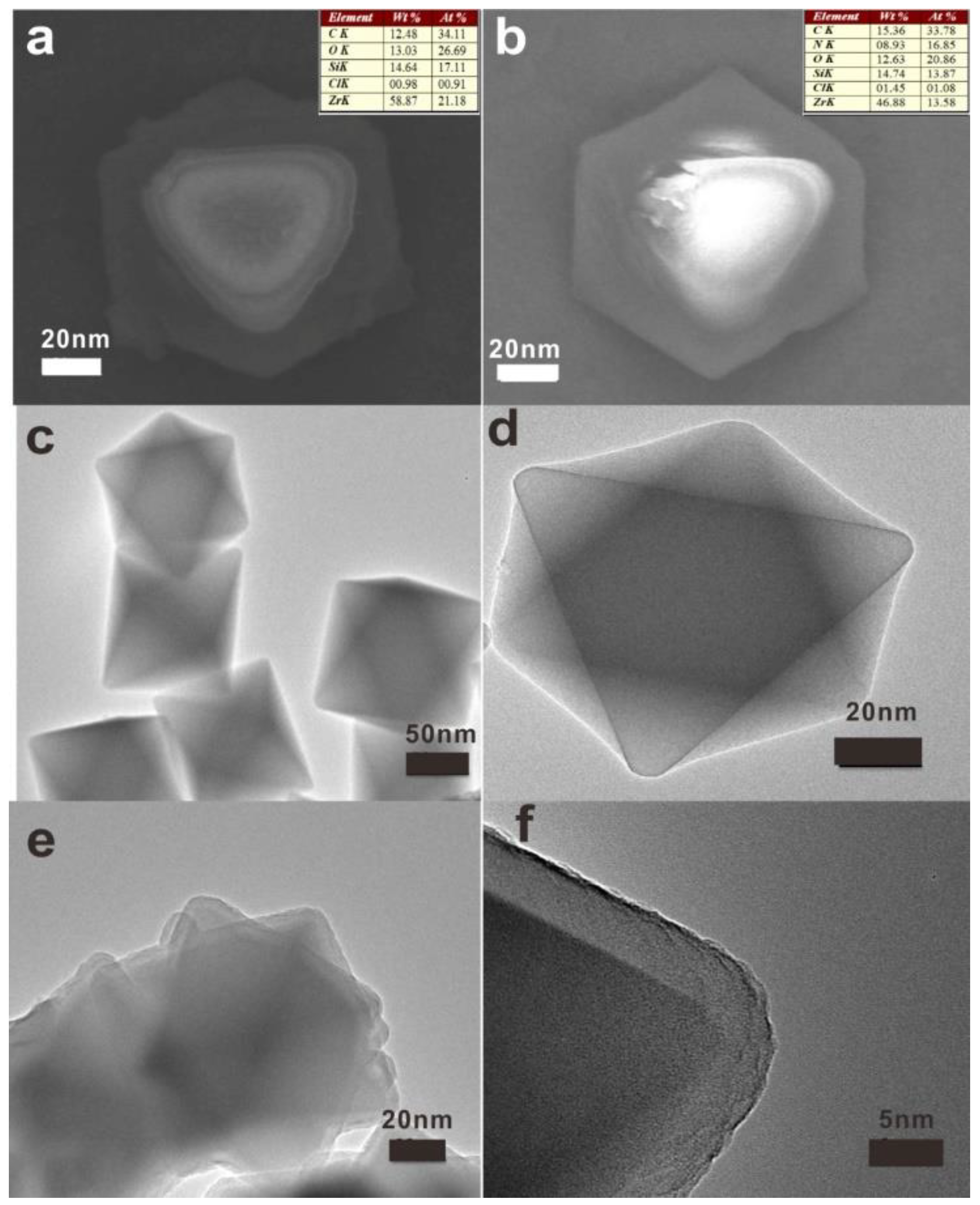
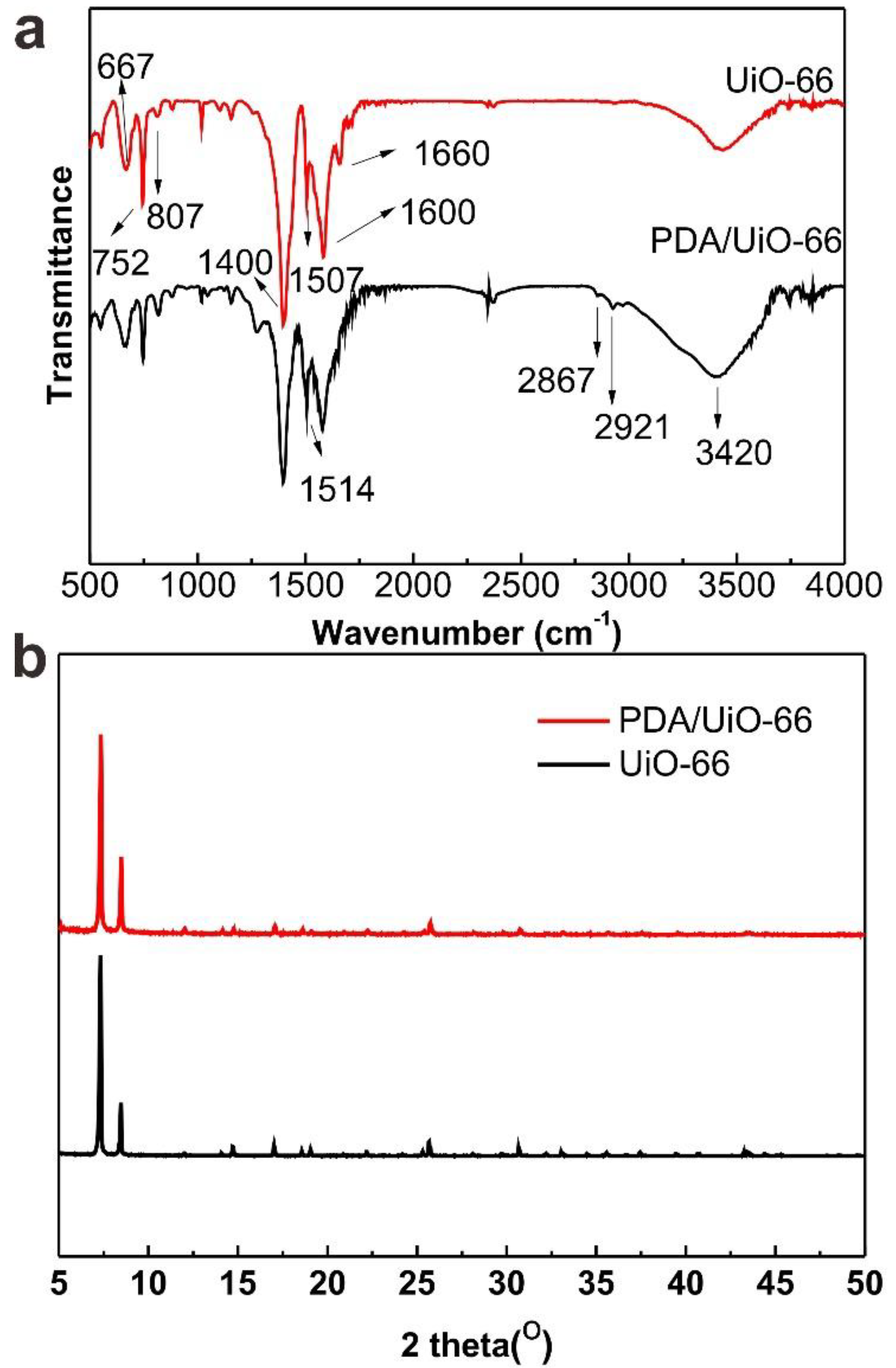
2.2. Preparation of UiO-66
2.3. Preparation of PDA/UiO-66
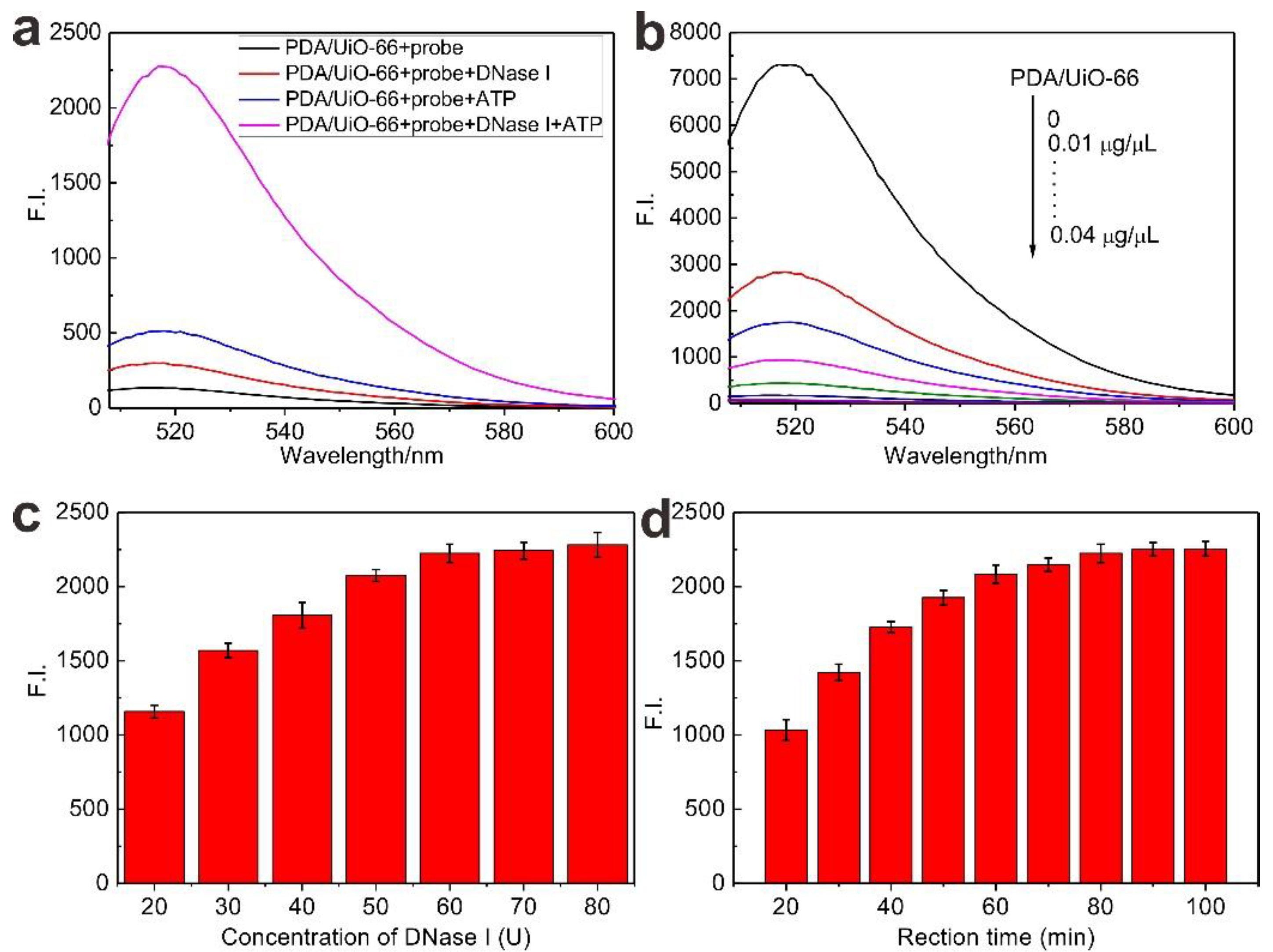
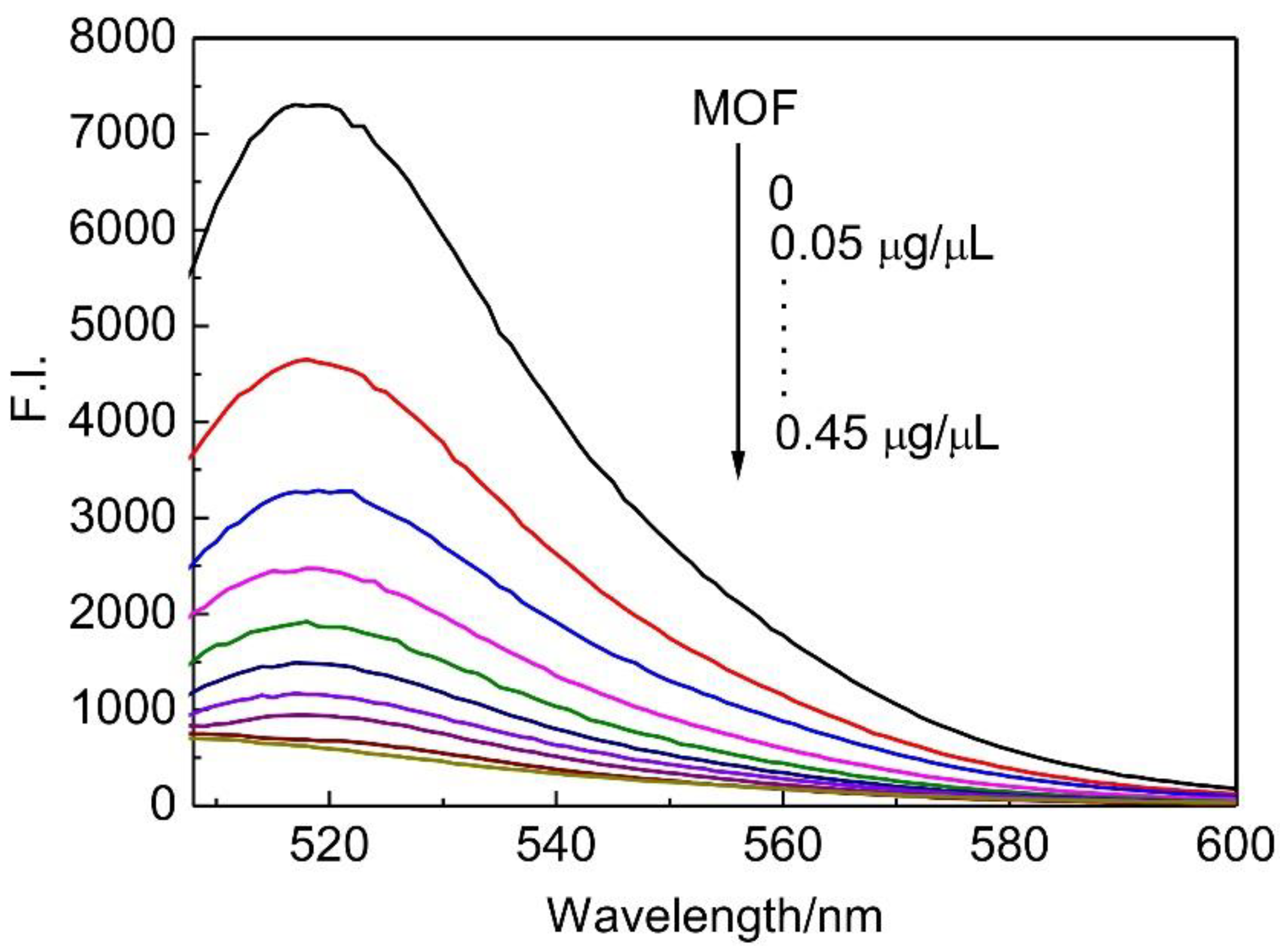
2.4. Fluorescence Measurements
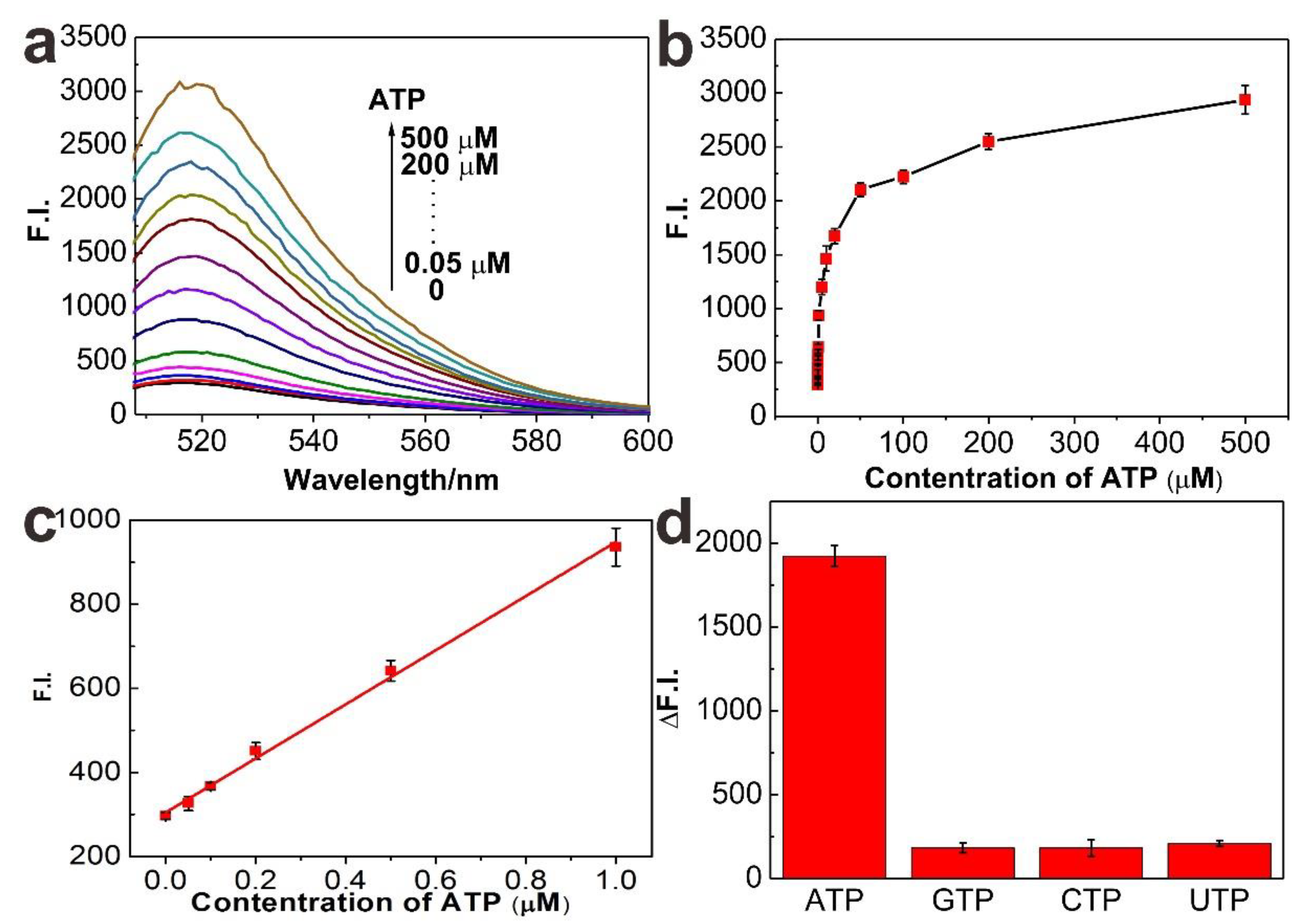
3. Results and Discussion
4. Conclusions
Author Contributions
Conflicts of Interest
Abbreviations/Nomenclature
| PET | photoinduced electron transfer |
| FRET | fluorescence resonance energy transfer |
| MOFs | metal-organic frameworks |
| ssDNA | single-stranded DNA |
| TNP | 2,4,6-trinitrophenol |
| ATP | adenosine triphosphate |
| PDA | polydopamine |
| LOD | detection limit |
| UTP | uridine triphosphate |
| CTP | cytosine triphosphate |
| GTP | guanidine triphosphate |
References
- Ke, Y.; Ong, L.L.; Shih, W.M.; Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 2012, 338, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xu, N.; Lei, J.; Ju, H. Regulative peroxidase activity of DNA-linked hemin by graphene oxide for fluorescence DNA sensing. Chem. Commun. 2014, 50, 6714–6717. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Li, Q.; Zhao, W.; Pang, Q.; Gao, H.; Xu, Z. In-situ construction of novel silver nanoparticle decorated polymeric spheres as highly active and stable catalysts for reduction of methylene blue dye. Appl. Catal. A 2018, 549, 102–111. [Google Scholar] [CrossRef]
- Zou, Y.; Jin, H.; Sun, F.; Dai, X.; Xu, Z.; Yang, S.; Liao, G. Design and synthesis of a lead sulfide based nanotheranostic agent for computer tomography/magnetic resonance dual-mode-bioimaging-guided photothermal therapy. ACS Appl. Nano Mater. 2018, 1, 2294–2305. [Google Scholar] [CrossRef]
- Xu, N.; Wang, Q.; Lei, J.; Liu, L.; Ju, H. Label-free triple-helix aptamer as sensing platform for “signal-on” fluorescent detection of thrombin. Talanta 2015, 132, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Chen, J.; Zeng, W.; Yu, C.; Yi, C.; Xu, Z. Facile preparation of uniform nanocomposite spheres with loading silver nanoparticles on polystyrene-methyl acrylic acid spheres for catalytic reduction of 4-nitrophenol. J. Phys. Chem. C 2016, 120, 25935–25944. [Google Scholar] [CrossRef]
- Verma, S.; Baig, R.B.N.; Nadagouda, M.N.; Varma, R.S. Titanium-based zeolitic imidazolate framework for chemical fixation of carbon dioxide. Green Chem. 2016, 18, 4855–4858. [Google Scholar] [CrossRef]
- Zhou, H.C.; Long, J.R.; Yaghi, O.M. Introduction to metal–organic frameworks. Chem. Rev. 2012, 112, 673–674. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Baig, R.B.N.; Nadagouda, M.N.; Varma, R.S. Fixation of carbon dioxide into dimethyl carbonate over titanium-based zeolitic thiophene-benzimidazolate framework. Sci. Rep. 2017, 7, 655. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Feng, D.; Liu, T.F.; Su, J.; Yuan, S.; Chen, Y.P.; Bosch, M.; Zou, X.; Zhou, H.C. A series of highly stable mesoporous metalloporphyrin Fe-MOFs. J. Am. Chem. Soc. 2014, 136, 13983–13986. [Google Scholar] [CrossRef] [PubMed]
- Horcajada, P.; Gref, R.; Baati, T.; Allan, P.K.; Maurin, G.; Couvreur, P.; Ferey, G.; Morris, R.E.; Serre, C. Metal–organic frameworks in biomedicine. Chem. Rev. 2012, 112, 1232–1268. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Deibert, B.J.; Li, J. Luminescent metal–organic frameworks for chemical sensing and explosive detection. Chem. Soc. Rev. 2014, 43, 5815–5840. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zheng, H.; Wei, X.; Lin, Z.; Guo, L.; Qiu, B.; Chen, G. Metal–organic framework (MOF): A novel sensing platform for biomolecules. Chem. Commun. 2013, 49, 1276–1278. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.L.; Wang, Z.; Zhao, S.N.; Meng, X.; Song, X.Z.; Feng, J.; Song, S.Y.; Zhang, H.J. A Metal–organic Framework/DNA Hybrid System as a Novel Fluorescent Biosensor for Mercury(II) Ion Detection. Chem. Eur. J. 2016, 22, 477–480. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.T.; Zhang, J.W.; Huang, G.; Du, Z.Y.; Jiang, H.L. An amine-functionalized metal–organic framework as a sensing platform for DNA detection. Chem. Commun. 2014, 50, 12069–12072. [Google Scholar] [CrossRef] [PubMed]
- Joarder, B.; Desai, A.V.; Samanta, P.; Mukherjee, S.; Ghosh, S.K. Selective and sensitive aqueous-phase detection of 2,4,6-trinitrophenol (TNP) by an amine-functionalized metal–organic framework. Chem. Eur. J. 2015, 21, 965–969. [Google Scholar] [CrossRef] [PubMed]
- Cavka, J.H.; Jakobsen, S.; Olsbye, U.; Guillou, N.; Lamberti, C.; Bordiga, S.; Lillerud, K.P. A new zirconium inorganic building brick forming metal organic frameworks with exceptional stability. J. Am. Chem. Soc. 2008, 130, 13850–13851. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Wang, H.; Ge, J.; Zhang, L.; Li, J.; Bai, D.; Chen, J.; Li, Z. Label-free and rapid detection of ATP based on structure switching of aptamers. Anal. Biochem. 2017, 526, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Nie, Z.; Xu, X.; Shen, Q.; Deng, C.; Chen, J.; Yao, S. A sensitive, label free electrochemical aptasensor for ATP detection. Talanta 2009, 78, 954–958. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, Y.; Ji, X.; He, Z. Target-protecting dumbbell molecular probe against exonucleases digestion for sensitive detection of ATP and streptavidin. Biosens. Bioelectron. 2016, 83, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Han, Y.; Guo, Y.; Dong, C. Porphyrin functionalized graphene nanosheets-based electrochemical aptasensor for label-free ATP detection. J. Mater. Chem. 2012, 22, 23900–23905. [Google Scholar] [CrossRef]
- Li, W.; Dong, Y.; Wang, X.; Li, H.; Xu, D. PolyA-tailed and fluorophore-labeled aptamer-gold nanoparticle conjugate for fluorescence turn-on bioassay using iodide-induced ligand displacement. Biosens. Bioelectron. 2015, 66, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; McGuirk, C.M.; Ross, M.B.; Wang, S.; Chen, P.; Xing, H.; Liu, Y.; Mirkin, C.A. General and direct method for preparing oligonucleotide-functionalized metal–organic framework Nanoparticles. J. Am. Chem. Soc. 2017, 139, 9827–9830. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liao, G.; Tian, J.; Xu, Z. Preparation of Novel fluorinated copolyimide/amine-functionalized sepia eumelanin nanocomposites with enhanced mechanical, thermal, and UV-shielding properties. Macromol. Mater. Eng. 2018, 303, 1700407. [Google Scholar] [CrossRef]
- Lin, L.S.; Cong, Z.X.; Cao, J.B.; Ke, K.M.; Peng, Q.L.; Gao, J.; Yang, H.H.; Liu, G.; Chen, X. Multifunctional Fe3O4@polydopamine core-shell nanocomposites for intracellular mRNA detection and imaging-guided photothermal therapy. ACS Nano 2014, 8, 3876–3883. [Google Scholar] [CrossRef] [PubMed]
- Keum, J.W.; Bermudez, H. Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem. Commun. 2009, 45, 7036–7038. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Lin, X.; Huang, Y.; Pan, R.; Zhu, Z.; Zhou, L.; Yang, C.J. Highly sensitive and selective detection of miRNA: DNase I-assisted target recycling using DNA probes protected by polydopamine nanospheres. Chem. Commun. 2015, 51, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Wu, W.; Liang, R.; Lin, R.; Wu, L. Highly dispersed palladium nanoparticles anchored on UiO-66(NH2) metal–organic framework as a reusable and dual functional visible-light-driven photocatalyst. Nanoscale 2013, 5, 9374–9382. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liao, G.; Zhang, S.; Pang, L.; Tong, H.; Zhao, W.; Xu, Z. Effect of adjustable molecular chain structure and pure silica zeolite nanoparticles on thermal, mechanical, dielectric, UV-shielding and hydrophobic properties of fluorinated copolyimide composites. Appl. Surf. Sci. 2018, 427, 437–450. [Google Scholar] [CrossRef]
- Xiao, L.; Deng, M.; Zeng, W.; Zhang, B.; Xu, Z.; Yi, C.; Liao, G. Novel robust superhydrophobic coating with self-cleaning properties in air and oil based on rare earth metal oxide. Ind. Eng. Chem. Res. 2017, 56, 12354–12361. [Google Scholar] [CrossRef]
- Valenzano, L.; Civalleri, B.; Chavan, S.; Bordiga, S.; Nilsen, M.H.; Jakobsen, S.; Lillerud, K.P.; Lamberti, C. Disclosing the complex structure of UiO-66 metal organic framework: A synergic combination of experiment and theory. Chem. Mater. 2011, 23, 1700–1718. [Google Scholar] [CrossRef]
- He, S.; Song, B.; Li, D.; Zhu, C.; Qi, W.; Wen, Y.; Wang, L.; Song, S.; Fang, H.; Fan, C. A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv. Funct. Mater. 2010, 20, 453–459. [Google Scholar] [CrossRef]
- Zhu, C.; Zhao, Y.; Yan, M.; Huang, Y.; Yan, J.; Bai, W.; Chen, A. A sandwich dipstick assay for ATP detection based on split aptamer fragments. Anal. Bioanal. Chem. 2016, 408, 4151–4158. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Wang, R.; Sun, F.; Chen, H.; Wang, Z.; Na, N.; Ouyang, J. A nuclease-assisted label-free aptasensor for fluorescence turn-on detection of ATP based on the in situ formation of copper nanoparticles. Biosens. Bioelectron. 2017, 87, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, J.; Chen, J.; Shih, K. Noncovalent assembly of carbon nanoparticles and aptamer for sensitive detection of ATP. RSC Adv. 2014, 4, 38199–38205. [Google Scholar] [CrossRef]


| Probe | Detection Method | Lower Limit of Detection | Reference |
|---|---|---|---|
| FAM-labeled aptamer | Fluorescent detection using PDA/UiO-66 nanoparticles | 0.035 μM | This work |
| FAM-labeled aptamer | Fluorescent detection using UiO-66 nanoparticles | 0.2 μM | This work |
| Graphene oxid-based multicolor fluorescent DNA nanoprobe | The biosensor based on the graphene oxide | 10 μM | [32] |
| Thiolated aptamer | A sandwich dipstick assay based on split aptamer fragments | 0.5 μM | [33] |
| Nuclease-assisted label-free aptamer | Fluorescent detection using copper nanoparticles | 0.093 μM | [34] |
| Noncovalent assembly of carbon nanoparticles and aptamer | Assay based on carbon nanoparticles | 0.1 μM | [35] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, P.; Liao, G. A Novel Fluorescent Biosensor for Adenosine Triphosphate Detection Based on a Metal–Organic Framework Coating Polydopamine Layer. Materials 2018, 11, 1616. https://doi.org/10.3390/ma11091616
Xu P, Liao G. A Novel Fluorescent Biosensor for Adenosine Triphosphate Detection Based on a Metal–Organic Framework Coating Polydopamine Layer. Materials. 2018; 11(9):1616. https://doi.org/10.3390/ma11091616
Chicago/Turabian StyleXu, Peipei, and Guangfu Liao. 2018. "A Novel Fluorescent Biosensor for Adenosine Triphosphate Detection Based on a Metal–Organic Framework Coating Polydopamine Layer" Materials 11, no. 9: 1616. https://doi.org/10.3390/ma11091616
APA StyleXu, P., & Liao, G. (2018). A Novel Fluorescent Biosensor for Adenosine Triphosphate Detection Based on a Metal–Organic Framework Coating Polydopamine Layer. Materials, 11(9), 1616. https://doi.org/10.3390/ma11091616






