Action of Antimicrobial Peptides against Bacterial Biofilms
Abstract
1. Biofilms
1.1. Biofilm Formation
1.2. Characteristics of Biofilms
1.3. Biofilm-Associated Infections
2. Antimicrobial Peptides
2.1. Mechanism of Action of AMPs against Planktonic Bacteria
2.2. Mechanism of Action against Biofilms
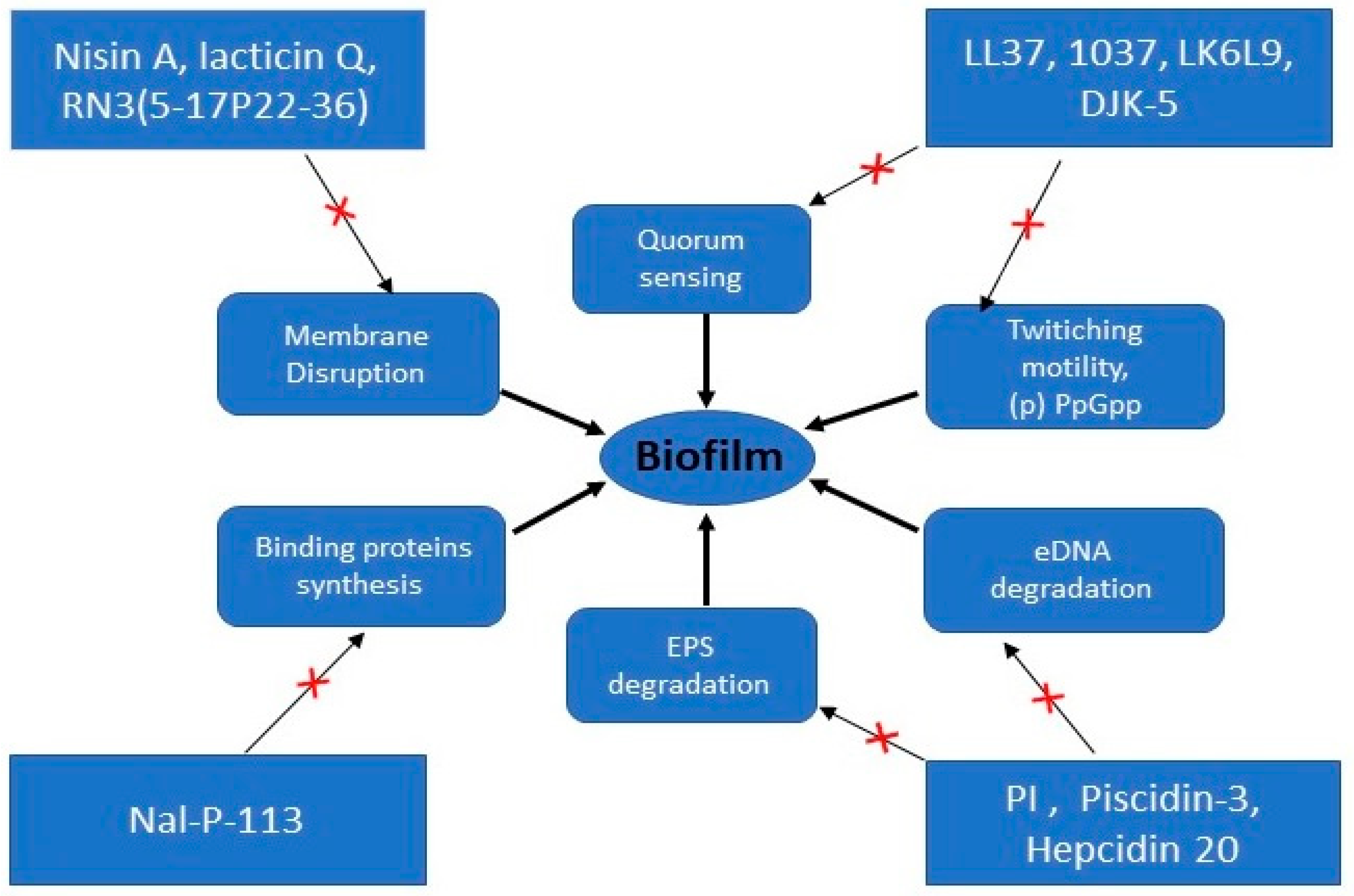
3. Biofilm Resistance to AMPs
3.1. Interaction with EPS
3.2. Adaptive Resistance Mechanism
3.3. Heterogeneity
3.4. Synergy of Anti-Biofilm AMPs with Antibiotics
4. Future Considerations
Funding
Acknowledgments
Conflicts of Interest
References
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Stoodley, P. Evolving concepts in biofilm infections. Cell. Microbiol. 2009, 11, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Stoodley, P.; Kathju, S.; Høiby, N.; Moser, C.; William Costerton, J.; Moter, A.; Bjarnsholt, T. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immun. Med. Microbiol. 2012, 65, 127–145. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Cheng, K.J.; Geesey, G.G.; Ladd, T.I.; Nickel, J.C.; Dasgupta, M.; Marrie, T.J. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 1987, 41, 435–464. [Google Scholar] [CrossRef] [PubMed]
- Anwar, H.; Dasgupta, M.K.; Costerton, J.W. Testing the susceptibility of bacteria in biofilms to antibacterial agents. Antimicrob. Agents Chemother. 1990, 34, 2043–2046. [Google Scholar] [CrossRef] [PubMed]
- Matz, C.; Bergfeld, T.; Rice, S.A.; Kjelleberg, S. Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 2004, 6, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Shirtliff, M.E.; Peters, B.M.; Jabra-Rizk, M.A. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett 2009, 299, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Ann. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- De la Fuente-Núñez, C.; Reffuveille, F.; Fernández, L.; Hancock, R.E. Bacterial biofilm development as a multicellular adaptation: Antibiotic resistance and new therapeutic strategies. Curr. Opin. Microbiol. 2013, 16, 580–589. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95. [Google Scholar] [CrossRef] [PubMed]
- Porat, Y.; Marynka, K.; Tam, A.; Steinberg, D.; Mor, A. Acyl-substituted dermaseptin S4 derivatives with improved bactericidal properties, including on oral microflora. Antimicrob. Agents Chemother. 2006, 50, 4153–4160. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente-Nunez, C.; Reffuveille, F.; Mansour, S.C.; Reckseidler-Zenteno, S.L.; Hernandez, D.; Brackman, G.; Coenye, T.; Hancock, R.E. D-enantiomeric peptides that eradicate wild-type and multidrug-resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 2015, 22, 196–205. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente-Núñez, C.; Reffuveille, F.; Haney, E.F.; Straus, S.K.; Hancock, R.E. Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PloS. Pathog. 2014, 10, e1004152. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilms: Microbial Life on Surfaces. Emerg. Inf. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Stoica, P.; Chifiriuc, M.C.; Rapa, M.; Lazăr, V. Overview of biofilm-related problems in medical devices. In Biofilms and Implantable Medical Devices; Deng, Y., Lv, W., Eds.; Woodhead Publishing: Cambridge, UK, 2017; pp. 3–23. [Google Scholar]
- Dalton, H.M.; March, P.E. Molecular genetics of bacterial attachment and biofouling. Curr. Opin. Biotechnol. 1998, 9, 252–255. [Google Scholar] [CrossRef]
- Garrett, T.R.; Bhakoo, M.; Zhang, Z. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- Whittaker, C.J.; Klier, C.M.; Kolenbrander, P.E. Mechanisms of adhesion by oral bacteria. Ann. Rev. Microbiol. 1996, 50, 513–552. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W. Introduction to biofilm. Int. J. Antimicrob. Agents. 1999, 11, 217–221. [Google Scholar] [CrossRef]
- de la Fuente-Núñez, C.; Cardoso, M.H.; de Souza Cândido, E.; Franco, O.L.; Hancock, R.E. Synthetic antibiofilm peptides. Biochimic. Biophys. Acta. Biomem. 2016, 1858, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Reffuveille, F.; de la Fuente-Núñez, C.; Mansour, S.; Hancock, R.E.W. A Broad-Spectrum Antibiofilm Peptide Enhances Antibiotic Action against Bacterial Biofilms. Antmicrob. Agents Chemother. 2014, 58, 5363–5371. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antmicrob. Agents Chemother. 1996, 40, 2517–2522. [Google Scholar] [CrossRef]
- Mah, T.-F.C.; O’toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends. Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Arciola, C.R.; Campoccia, D.; Speziale, P.; Montanaro, L.; Costerton, J.W. Biofilm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for biofilm-resistant materials. Biomaterials 2012, 33, 5967–5982. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Riddle of biofilm resistance. Antmicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Hoiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.J.; Moser, C.; Jensen, P.O.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Roe, F.; Rayner, J.; Elkins, J.G.; Lewandowski, Z.; Ochsner, U.A.; Hassett, D.J. Effect of catalase on hydrogen peroxide penetration into Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2000, 66, 836–838. [Google Scholar] [CrossRef] [PubMed]
- Fux, C.; Costerton, J.W.; Stewart, P.S.; Stoodley, P. Survival strategies of infectious biofilms. Trends Microbiol. 2005, 13, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Hoiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef]
- Mah, T.F. Biofilm-specific antibiotic resistance. Future Microbiol. 2012, 7, 1061–1072. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M. Biofilm formation: A clinically relevant microbiological process. Clin. Infec. Dis. 2001, 33, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Moser, C.; Bassi, G.; Coenye, T.; Donelli, G.; Hall-Stoodley, L.; Hola, V.; Imbert, C.; Kirketerp-Møller, K. ESCMID∗ guideline for the diagnosis and treatment of biofilm infections. Clin. Microbiol. Infect. 2015, 21, S1–S25. [Google Scholar] [CrossRef] [PubMed]
- Romling, U.; Kjelleberg, S.; Normark, S.; Nyman, L.; Uhlin, B.E.; Akerlund, B. Microbial biofilm formation: A need to act. J. Intern. Med. 2014, 276, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Romling, U.; Balsalobre, C. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 2012, 272, 541–561. [Google Scholar] [CrossRef] [PubMed]
- Bryers, J.D. Medical Biofilms. Biotechnol. Bioeng. 2008, 100, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Montanaro, L.; Arciola, C.R. Biofilm in implant infections: Its production and regulation. Int. J. Art. Organs 2005, 28, 1062–1068. [Google Scholar] [CrossRef]
- Costerton, W.; Veeh, R.; Shirtliff, M.; Pasmore, M.; Post, C.; Ehrlich, G. The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Investig. 2003, 112, 1466–1477. [Google Scholar] [CrossRef] [PubMed]
- Veerachamy, S.; Yarlagadda, T.; Manivasagam, G.; Yarlagadda, P.K. Bacterial adherence and biofilm formation on medical implants: A review. Proc. Inst. Mech. Eng. Part. H J. Eng. Med. 2014, 228, 1083–1099. [Google Scholar] [CrossRef] [PubMed]
- Bryers, J.D.; Ratner, B.D. Bioinspired implant materials befuddle bacteria. ASM News-Am. Soc. Microbiol. 2004, 70, 232. [Google Scholar]
- Castelli, P.; Caronno, R.; Ferrarese, S.; Mantovani, V.; Piffaretti, G.; Tozzi, M.; Lomazzi, C.; Rivolta, N.; Sala, A. New trends in prosthesis infection in cardiovascular surgery. Surg. Infect. 2006, 7 (Suppl. 2), S45–S47. [Google Scholar] [CrossRef]
- Carmen, J.C.; Roeder, B.L.; Nelson, J.L.; Ogilvie, R.L.R.; Robison, R.A.; Schaalje, G.B.; Pitt, W.G. Treatment of biofilm infections on implants with low-frequency ultrasound and antibiotics. Am. J. Infect. Cont. 2005, 33, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Paterson, I.K.; Hoyle, A.; Ochoa, G.; Baker-Austin, C.; Taylor, N.G. Optimising antibiotic usage to treat bacterial infections. Sci Rep 2016, 6, 37853. [Google Scholar] [CrossRef] [PubMed]
- Bahar, A.A.; Ren, D. Antimicrobial peptides. Pharmaceuticals 2013, 6, 1543–1575. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.L.; Hancock, R.E. Cationic host defense (antimicrobial) peptides. Curr. Opin. Immunol. 2006, 18, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Groenink, J.; Walgreen-Weterings, E.; van‘t Hof, W.; Veerman, E.I.; Nieuw Amerongen, A.V. Cationic amphipathic peptides, derived from bovine and human lactoferrins, with antimicrobial activity against oral pathogens. FEMS Microbiol. Lett. 1999, 179, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, J.P. Cationic antimicrobial peptides. BioDrugs 2003, 17, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-helical cationic antimicrobial peptides: Relationships of structure and function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef]
- Harris, F.; Dennison, S.R.; Phoenix, D.A. Anionic antimicrobial peptides from eukaryotic organisms. Curr. Prot. Pep. Sci. 2009, 10, 585–606. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta 2016, 1858, 1044–1060. [Google Scholar] [CrossRef]
- Segev-Zarko, L.; Saar-Dover, R.; Brumfeld, V.; Mangoni, M.L.; Shai, Y. Mechanisms of biofilm inhibition and degradation by antimicrobial peptides. Biochem. J. 2015, 468, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Bowdish, D.M.; Davidson, D.J.; Hancock, R.E. A re-evaluation of the role of host defence peptides in mammalian immunity. Curr. Protein Pept. Sci. 2005, 6, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Boman, H.G. Peptide antibiotics and their role in innate immunity. Ann. Rev. Immunol. 1995, 13, 61–92. [Google Scholar] [CrossRef] [PubMed]
- Tossi, A.; Sandri, L.; Giangaspero, A. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 2000, 55, 4–30. [Google Scholar] [CrossRef]
- Ebenhan, T.; Gheysens, O.; Kruger, H.G.; Zeevaart, J.R.; Sathekge, M.M. Antimicrobial peptides: Their role as infection-selective tracers for molecular imaging. BioMed. Res. Int. 2014, 2014, 867381. [Google Scholar] [CrossRef] [PubMed]
- Tossi, A.; Sandri, L. Molecular diversity in gene-encoded, cationic antimicrobial polypeptides. Curr. Pharm. Des. 2002, 8, 743–761. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef]
- Lehrer, R.I. Primate defensins. Nat. Rev. Microbiol. 2004, 2, 727–738. [Google Scholar] [CrossRef]
- Takahashi, D.; Shukla, S.K.; Prakash, O.; Zhang, G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie 2010, 92, 1236–1241. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389. [Google Scholar] [CrossRef]
- Selsted, M.E.; Harwig, S.S. Determination of the disulfide array in the human defensin HNP-2. A covalently cyclized peptide. J. Biol. Chem. 1989, 264, 4003–4007. [Google Scholar] [PubMed]
- Tang, Y.Q.; Selsted, M.E. Characterization of the disulfide motif in BNBD-12, an antimicrobial beta-defensin peptide from bovine neutrophils. J. Biol. Chem. 1993, 268, 6649–6653. [Google Scholar] [PubMed]
- Cruz, J.; Ortiz, C.; Guzman, F.; Fernandez-Lafuente, R.; Torres, R. Antimicrobial peptides: Promising compounds against pathogenic microorganisms. Curr. Med. Chem. 2014, 21, 2299–2321. [Google Scholar] [CrossRef] [PubMed]
- Su, L.Y.; Willner, D.L.; Segall, A.M. An antimicrobial peptide that targets DNA repair intermediates in vitro inhibits Salmonella growth within murine macrophages. Antimicrob. Agents Chemother. 2010, 54, 1888–1899. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Kragol, G.; Lovas, S.; Varadi, G.; Condie, B.A.; Hoffmann, R.; Otvos, L. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 2001, 40, 3016–3026. [Google Scholar] [CrossRef] [PubMed]
- Conti, S.; Radicioni, G.; Ciociola, T.; Longhi, R.; Polonelli, L.; Gatti, R.; Cabras, T.; Messana, I.; Castagnola, M.; Vitali, A. Structural and functional studies on a proline-rich peptide isolated from swine saliva endowed with antifungal activity towards Cryptococcus neoformans. Biochim. Biophys. Acta Biomem. 2013, 1828, 1066–1074. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochimic. Biophys. Acta 1999, 1462, 55–70. [Google Scholar] [CrossRef]
- Peschel, A.; Sahl, H.-G. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 2006, 4, 529. [Google Scholar] [CrossRef]
- Schuller, F.; Benz, R.; Sahl, H.G. The peptide antibiotic subtilin acts by formation of voltage-dependent multi-state pores in bacterial and artificial membranes. Eur. J. Biochem. 1989, 182, 181–186. [Google Scholar] [CrossRef]
- Tennessen, J.A. Molecular evolution of animal antimicrobial peptides: Widespread moderate positive selection. J. Evol. Biol. 2005, 18, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C.; Hristova, K. Antimicrobial Peptides: Successes, challenges and unanswered questions. J. Memb. Biol. 2011, 239, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Straus, S.K.; Hancock, R.E. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta 2006, 1758, 1215–1223. [Google Scholar] [CrossRef]
- Sang, Y.; Blecha, F. Antimicrobial peptides and bacteriocins: Alternatives to traditional antibiotics. Anim. Health Res. Rev. 2008, 9, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Haney, E.F.; Mansour, S.C.; Hancock, R.E. Antimicrobial Peptides: An Introduction. Meth. Mol. Biol. 2017, 1548, 3–22. [Google Scholar] [CrossRef]
- Sun, E.; Belanger, C.R.; Haney, E.F.; Hancock, R.E. Host defense (antimicrobial) peptides. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Elsevier: Amsterdam, the Netherlands, 2018; pp. 253–285. [Google Scholar]
- Okuda, K.; Zendo, T.; Sugimoto, S.; Iwase, T.; Tajima, A.; Yamada, S.; Sonomoto, K.; Mizunoe, Y. Effects of bacteriocins on methicillin-resistant Staphylococcus aureus biofilm. Antimicrob. Agents Chemother. 2013, 57, 5572–5579. [Google Scholar] [CrossRef]
- Pulido, D.; Prats-Ejarque, G.; Villalba, C.; Albacar, M.; González-López, J.J.; Torrent, M.; Moussaoui, M.; Boix, E. A novel RNase 3/ECP peptide for Pseudomonas aeruginosa biofilm eradication. Combining antimicrobial, lipopolysaccharide binding and cell agglutinating activities. Antimicrob. Agents Chemother. 2016, 60, 6313–6325. [Google Scholar] [CrossRef]
- Venge, P. Eosinophil cationic protein (ECP): Molecular and biological properties and the use of ECP as a marker of eosinophil activation in disease. Clin. Exp. Allergy 1999, 29, 1172–1186. [Google Scholar] [CrossRef]
- Acharya, K.R.; Ackerman, S.J. Eosinophil granule proteins: Form and function. J. Biol. Chem. 2014. [Google Scholar] [CrossRef]
- Luca, V.; Stringaro, A.; Colone, M.; Pini, A.; Mangoni, M.L. Esculentin(1-21), an amphibian skin membrane-active peptide with potent activity on both planktonic and biofilm cells of the bacterial pathogen Pseudomonas aeruginosa. Cell. Mol. Life. Sci. 2013, 70, 2773–2786. [Google Scholar] [CrossRef]
- Nagant, C.; Pitts, B.; Stewart, P.S.; Feng, Y.; Savage, P.B.; Dehaye, J.P. Study of the effect of antimicrobial peptide mimic, CSA-13, on an established biofilm formed by P seudomonas aeruginosa. Microbiologyopen 2013, 2, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Overhage, J.; Campisano, A.; Bains, M.; Torfs, E.C.; Rehm, B.H.; Hancock, R.E. Human host defense peptide LL-37 prevents bacterial biofilm formation. Infect. Immun. 2008, 76, 4176–4182. [Google Scholar] [CrossRef] [PubMed]
- de la Fuente-Núñez, C.; Korolik, V.; Bains, M.; Nguyen, U.; Breidenstein, E.B.M.; Horsman, S.; Lewenza, S.; Burrows, L.; Hancock, R.E.W. Inhibition of Bacterial Biofilm Formation and Swarming Motility by a Small Synthetic Cationic Peptide. Antimicrob. Agents Chemother. 2012, 56, 2696–2704. [Google Scholar] [CrossRef] [PubMed]
- Jorge, P.; Lourenco, A.; Pereira, M.O. New trends in peptide-based anti-biofilm strategies: A review of recent achievements and bioinformatic approaches. Biofouling 2012, 28, 1033–1061. [Google Scholar] [CrossRef] [PubMed]
- Ansari, J.M.; Abraham, N.M.; Massaro, J.; Murphy, K.; Smith-Carpenter, J.; Fikrig, E. Anti-biofilm activity of a self-aggregating peptide against Streptococcus mutans. Front. Microbiol. 2017, 8, 488. [Google Scholar] [CrossRef] [PubMed]
- Gordya, N.; Yakovlev, A.; Kruglikova, A.; Tulin, D.; Potolitsina, E.; Suborova, T.; Bordo, D.; Rosano, C.; Chernysh, S. Natural antimicrobial peptide complexes in the fighting of antibiotic resistant biofilms: Calliphora vicina medicinal maggots. PLoS ONE 2017, 12, e0173559. [Google Scholar] [CrossRef] [PubMed]
- Brancatisano, F.L.; Maisetta, G.; Di Luca, M.; Esin, S.; Bottai, D.; Bizzarri, R.; Campa, M.; Batoni, G. Inhibitory effect of the human liver-derived antimicrobial peptide hepcidin 20 on biofilms of polysaccharide intercellular adhesin (PIA)-positive and PIA-negative strains of Staphylococcus epidermidis. Biofouling 2014, 30, 435–446. [Google Scholar] [CrossRef]
- Quiles, F.; Saadi, S.; Francius, G.; Bacharouche, J.; Humbert, F. In situ and real time investigation of the evolution of a Pseudomonas fluorescens nascent biofilm in the presence of an antimicrobial peptide. Biochimic. Biophys. Acta 2016, 1858, 75–84. [Google Scholar] [CrossRef]
- Libardo, M.D.J.; Bahar, A.A.; Ma, B.; Fu, R.; McCormick, L.E.; Zhao, J.; McCallum, S.A.; Nussinov, R.; Ren, D.; Angeles-Boza, A.M.; et al. Nuclease activity gives an edge to host-defense peptide piscidin 3 over piscidin 1, rendering it more effective against persisters and biofilms. FEBS J. 2017, 284, 3662–3683. [Google Scholar] [CrossRef]
- Pletzer, D.; Coleman, S.R.; Hancock, R.E. Anti-biofilm peptides as a new weapon in antimicrobial warfare. Curr. Opin. Microbiol. 2016, 33, 35–40. [Google Scholar] [CrossRef]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still magical? Ann. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Braeken, K.; Moris, M.; Daniels, R.; Vanderleyden, J.; Michiels, J. New horizons for (p) ppGpp in bacterial and plant physiology. Trends Microbiol. 2006, 14, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Åberg, A.; Shingler, V.; Balsalobre, C. (p) ppGpp regulates type 1 fimbriation of Escherichia coli by modulating the expression of the site-specific recombinase FimB. Mol. Microbiol. 2006, 60, 1520–1533. [Google Scholar] [CrossRef] [PubMed]
- Pletzer, D.; Wolfmeier, H.; Bains, M.; Hancock, R.E.W. Synthetic Peptides to Target Stringent Response-Controlled Virulence in a Pseudomonas aeruginosa Murine Cutaneous Infection Model. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Rohde, H.; Frankenberger, S.; Zähringer, U.; Mack, D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur. J. Cell. Biol. 2010, 89, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Tan, H.; Cheng, T.; Shen, H.; Shao, J.; Guo, Y.; Shi, S.; Zhang, X. Human beta-defensin 3 inhibits antibiotic-resistant Staphylococcus biofilm formation. J. Surg. Res. 2013, 183, 204–213. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Lin, L.; Tan, L.-S.; Yu, H.-Y.; Cheng, J.-W.; Pan, Y.-P. Molecular pathways underlying inhibitory effect of antimicrobial peptide Nal-P-113 on bacteria biofilms formation of Porphyromonas gingivalis W83 by DNA microarray. BMC Microbiol. 2017, 17, 37. [Google Scholar] [CrossRef]
- Hinsa, S.M.; Espinosa-Urgel, M.; Ramos, J.L.; O’Toole, G.A. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003, 49, 905–918. [Google Scholar] [CrossRef]
- Andersen, R.N.; Ganeshkumar, N.; Kolenbrander, P.E. Cloning of the Streptococcus gordonii PK488 gene, encoding an adhesin which mediates coaggregation with Actinomyces naeslundii PK606. Infect. Immun. 1993, 61, 981–987. [Google Scholar]
- Otto, M. Bacterial evasion of antimicrobial peptides by biofilm formation. Curr. Top. Microbiol. Immunol. 2006, 306, 251–258. [Google Scholar]
- Vuong, C.; Voyich, J.M.; Fischer, E.R.; Braughton, K.R.; Whitney, A.R.; DeLeo, F.R.; Otto, M. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 2004, 6, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Gacesa, P. Bacterial alginate biosynthesis-recent progress and future prospects. Microbiology 1998, 144, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.R.; Linker, A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 1973, 116, 915–924. [Google Scholar] [PubMed]
- Chan, C.; Burrows, L.L.; Deber, C.M. Helix induction in antimicrobial peptides by alginate in biofilms. J. Biol. Chem. 2004, 279, 38749–38754. [Google Scholar] [CrossRef] [PubMed]
- Leid, J.G.; Willson, C.J.; Shirtliff, M.E.; Hassett, D.J.; Parsek, M.R.; Jeffers, A.K. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 2005, 175, 7512–7518. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.H.; Chan, C.; Burrows, L.L.; Deber, C.M. Hydrophobic interactions in complexes of antimicrobial peptides with bacterial polysaccharides. Chem. Biol. Drug. Des. 2007, 69, 405–412. [Google Scholar] [CrossRef]
- Doggett, R.G.; Harrison, G.M.; Stillwell, R.N.; Wallis, E.S. An atypical Pseudomonas aeruginosa associated with cystic fibrosis of the pancreas. J. Ped. 1966, 68, 215–221. [Google Scholar] [CrossRef]
- Stapper, A.P.; Narasimhan, G.; Ohman, D.E.; Barakat, J.; Hentzer, M.; Molin, S.; Kharazmi, A.; Hoiby, N.; Mathee, K. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J. Med. Microbiol. 2004, 53, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Colvin, K.M.; Irie, Y.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2012, 14, 1913–1928. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lai, Y.; Villaruz, A.E.; Cha, D.J.; Sturdevant, D.E.; Otto, M. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. USA 2007, 104, 9469–9474. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-J.; Bayer, A.S.; Mishra, N.N.; Meehl, M.; Ledala, N.; Yeaman, M.R.; Xiong, Y.Q.; Cheung, A.L. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect. Immun. 2012, 80, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Bacterial sensing of antimicrobial peptides. Cont. Microbiol. 2009, 16, 136–149. [Google Scholar] [CrossRef]
- Fabretti, F.; Theilacker, C.; Baldassarri, L.; Kaczynski, Z.; Kropec, A.; Holst, O.; Huebner, J. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 2006, 74, 4164–4171. [Google Scholar] [CrossRef] [PubMed]
- Skiada, A.; Markogiannakis, A.; Plachouras, D.; Daikos, G.L. Adaptive resistance to cationic compounds in Pseudomonas aeruginosa. Int. J. Antimicrob. Aents 2011, 37, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Bader, M.W.; Sanowar, S.; Daley, M.E.; Schneider, A.R.; Cho, U.; Xu, W.; Klevit, R.E.; Le Moual, H.; Miller, S.I. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 2005, 122, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, M.M.; Whiteley, M. Pseudomonas aeruginosa attachment and biofilm development in dynamic environments. Mol. Microbiol. 2004, 53, 1075–1087. [Google Scholar] [CrossRef]
- McPhee, J.B.; Lewenza, S.; Hancock, R.E. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 2003, 50, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Gunn, J.S.; Lim, K.B.; Krueger, J.; Kim, K.; Guo, L.; Hackett, M.; Miller, S.I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 1998, 27, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Gunn, J.S.; Miller, S.I. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 1996, 178, 6857–6864. [Google Scholar] [CrossRef]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Hall-Stoodley, L.; Stoodley, P. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef]
- Pamp, S.J.; Gjermansen, M.; Johansen, H.K.; Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 2008, 68, 223–240. [Google Scholar] [CrossRef] [PubMed]
- Folkesson, A.; Haagensen, J.A.; Zampaloni, C.; Sternberg, C.; Molin, S. Biofilm induced tolerance towards antimicrobial peptides. PLoS ONE 2008, 3, e1891. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.M.; Briers, Y.; Lamberigts, C.; Steenackers, H.; Robijns, S.; Landuyt, B.; Vanderleyden, J.; Schoofs, L.; Lavigne, R.; Luyten, W. Evaluation of the antibacterial and antibiofilm activities of novel CRAMP–vancomycin conjugates with diverse linkers. Org. Biomol. Chem. 2015, 13, 7477–7486. [Google Scholar] [CrossRef] [PubMed]
- Rudilla, H.; Fusté, E.; Cajal, Y.; Rabanal, F.; Vinuesa, T.; Viñas, M. Synergistic antipseudomonal effects of synthetic peptide AMP38 and carbapenems. Molecules 2016, 21, 1223. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, S.M.; de la Fuente-Núñez, C.; Baquir, B.; Faria-Junior, C.; Franco, O.L.; Hancock, R.E. Antibiofilm peptides increase the susceptibility of carbapenemase-producing Klebsiella pneumoniae clinical isolates to β-lactam antibiotics. Antimicrob. Agents Chemother. 2015, 59, 3906–3912. [Google Scholar] [CrossRef]
- Gopal, R.; Kim, Y.G.; Lee, J.H.; Lee, S.K.; Chae, J.D.; Son, B.K.; Seo, C.H.; Park, Y. Synergistic effects and antibiofilm properties of chimeric peptides against multidrug-resistant Acinetobacter baumannii strains. Antimicrob. Agents Chemother. 2014, 58, 1622–1629. [Google Scholar] [CrossRef] [PubMed]
- Grassi, L.; Maisetta, G.; Esin, S.; Batoni, G. Combination Strategies to Enhance the Efficacy of Antimicrobial Peptides against Bacterial Biofilms. Front. Microbiol. 2017, 8, 2409. [Google Scholar] [CrossRef] [PubMed]
- Jorge, P.; Grzywacz, D.; Kamysz, W.; Lourenço, A.; Pereira, M.O. Searching for new strategies against biofilm infections: Colistin-AMP combinations against Pseudomonas aeruginosa and Staphylococcus aureus single-and double-species biofilms. PLoS ONE 2017, 12, e0174654. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.Y.; Khanum, R. Antimicrobial peptides as potential anti-biofilm agents against multidrug-resistant bacteria. J. Microbiol. Immunol. Infect. 2017, 50, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Cools, T.L.; Struyfs, C.; Drijfhout, J.W.; Kucharikova, S.; Lobo Romero, C.; Van Dijck, P.; Ramada, M.H.S.; Bloch, C., Jr.; Cammue, B.P.A.; Thevissen, K. A Linear 19-Mer Plant Defensin-Derived Peptide Acts Synergistically with Caspofungin against Candida albicans Biofilms. Front. Microbiol. 2017, 8, 2051. [Google Scholar] [CrossRef] [PubMed]
- Mahlapuu, M.; Håkansson, J.; Ringstad, L.; Björn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef] [PubMed]
 activation
activation  inhibition.
inhibition.
 activation
activation  inhibition.
inhibition.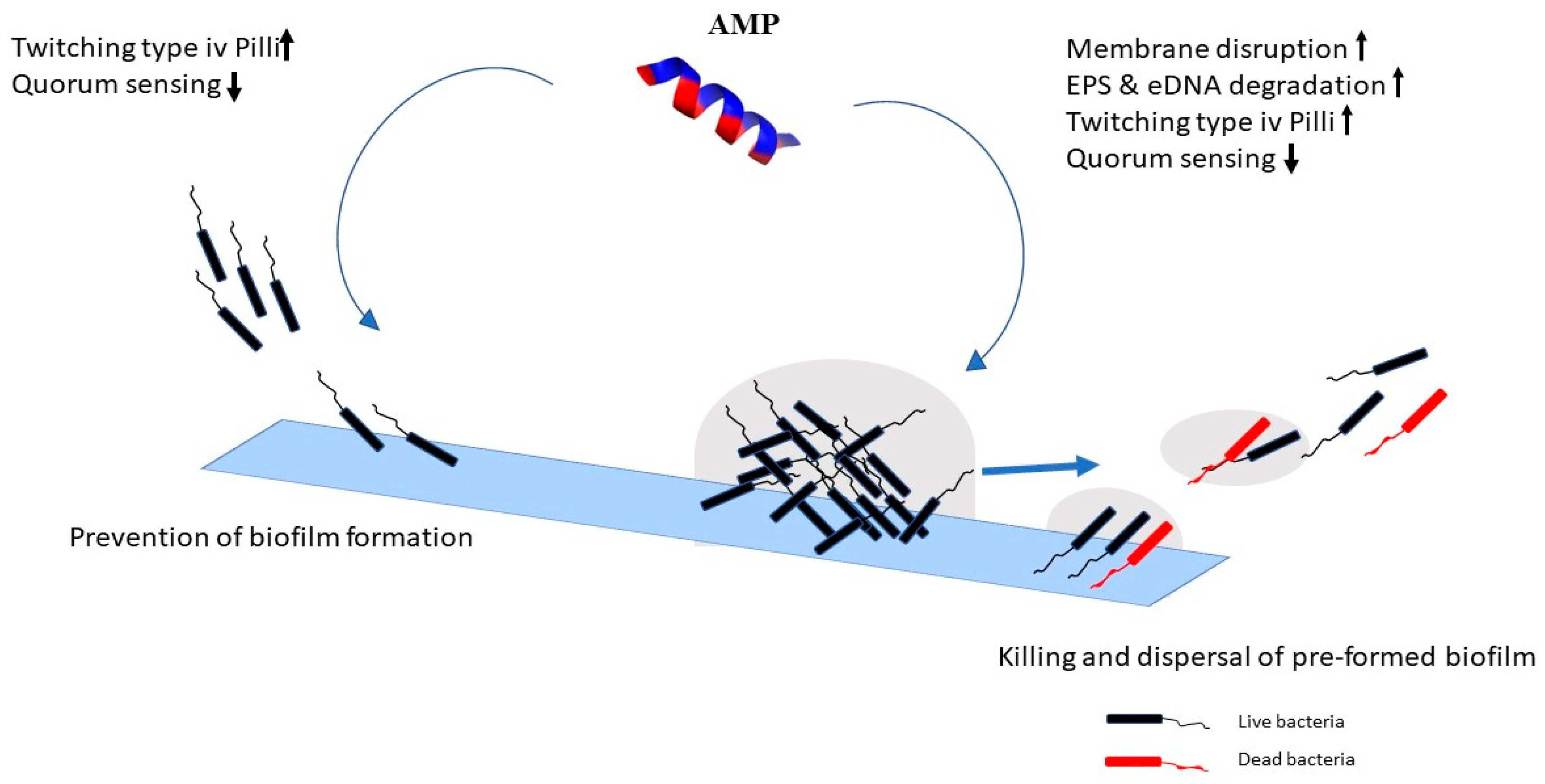
| AMPs | Sources | Amino Acids Sequence | Microorganisms | Proposed Mechanism of Action | Ref |
|---|---|---|---|---|---|
| LL-37 | Human | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | Pseudomonas aeruginosa | Reduces swimming and swarming motilities, promotes twitching motility, downregulates the genes required for biofilm formation and influences QS system | [84,85] |
| 1037 | Denovo | KRFRIRVRV | Pseudomonas aeruginosa | ||
| 1018 | Denovo | VRLIVAVRIWRR | Pseudomonas aeruginosa | Decrease intracellular (p) PpGpp | [14] |
| Esculentin-1a (1–21) | Denovo | GIFSKLAGKKIKNLLISGLKG | Pseudomonas aeruginosa | Disrupts cell membrane | [82] |
| Nisin A | Denovo | MSTKDFNLDLVSVSKKDSGASPR | Staphylococcus aureus | Depolarizes cell membrane | [78] |
| lacticin Q | Denovo | MAGFLKVVQLLAKYGSKAVQMAWANKGKILDWLNAGQAIDKVVSKIKQILGIK | Staphylococcus aureus | Depolarizes cell membrane | [78] |
| Nukacin ISK-1 | Denovo | KK-KSGVIPTVSHGCHMNSFQFVFTCC | Staphylococcus aureus | Depolarizes cell membrane | [78] |
| RN3(5-17P22-36) | Denovo | RPFTRAQWFAIQHISPRTIAMRAINNYRWR | Pseudomonas aeruginosa | Depolarizes and permeabilize cell membrane | [79] |
| S4 (1–16) | Denovo | ALWKTLLKKVLKAAAK | Pseudomonas aeruginosa | Disintegrates and release membrane lipids | [90] |
| P1 | Calliphora vicina | FVDRNRIPRSNNGPKIPIISNP | Escherichia coli, Staphylococcus aureus, Acinetobacter baumannii | Degrades biofilm matrix | [88] |
| L-K6L9 | Denovo | LKLLKKLLKKLLKLL | Pseudomonas aeruginosa | Degrades biofilms matrix | [52] |
| Piscidin-3 | Fish | FIHHIFRGIVHAGRSIGRFLTG | Pseudomonas aeruginosa | Degrades eDNA | [91] |
| PI | Tick | PARKARAATAATAATAATAAT | Streptococcus mutans | Interferes and degrade EPS matrix | [87] |
| Hepcidin 20 | Human | ICIFCCGCCHRSHCGMCCKT | Staphylococcus epidermidis | Acts on polysaccharide intercellular adhesin (PIA) | [88] |
| Nal-P-113 | Denovo | AKR-Nal-Nal-GYKRKF-Nal- | Porphyromonas gingivalis | Down regulates genes related to transport and binding proteins | [99] |
| Human β-defensin 3 (hBD-3) | Human | GIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK | Stahyloccocus epidermidis | Targets icaA, icaD and icaR genes | [98] |
| DJK-5 | Denovo | VQWRAIRVRVIR | Pseudomonas aeruginosa | Suppress spoT promoter activity | [96] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasir, M.; Willcox, M.D.P.; Dutta, D. Action of Antimicrobial Peptides against Bacterial Biofilms. Materials 2018, 11, 2468. https://doi.org/10.3390/ma11122468
Yasir M, Willcox MDP, Dutta D. Action of Antimicrobial Peptides against Bacterial Biofilms. Materials. 2018; 11(12):2468. https://doi.org/10.3390/ma11122468
Chicago/Turabian StyleYasir, Muhammad, Mark Duncan Perry Willcox, and Debarun Dutta. 2018. "Action of Antimicrobial Peptides against Bacterial Biofilms" Materials 11, no. 12: 2468. https://doi.org/10.3390/ma11122468
APA StyleYasir, M., Willcox, M. D. P., & Dutta, D. (2018). Action of Antimicrobial Peptides against Bacterial Biofilms. Materials, 11(12), 2468. https://doi.org/10.3390/ma11122468







