Impact of the Partitioning Method on Multidimensional Adaptive-Chemistry Simulations
Abstract
1. Introduction
2. Theory
2.1. Adaptive-Chemistry Approach
- Dataset generation: A training dataset is generated from previously available multi-dimensional simulations of the system or, in alternative, canonical simulations (0D or 1D) carried out with detailed kinetic schemes.
- Partitioning of the thermochemical space: the high-dimensional space spanned by the simulations is partitioned by means of a clustering algorithm, and several groups of similar points (clusters) are found.
- Generation of reduced mechanisms: for each point of the dataset, a reduced mechanism is generated. In this work, Directed Relation Graph with Error Propagation (DRGEP) is used. Then, a mechanism for each cluster is created as the union of each individual mechanism of the points belonging to the cluster, generating a library of reduced mechanisms.
- Adaptive simulation: the CFD simulation of the multi-dimensional system of interest (2D or 3D) is carried out. At each time-step, the grid points are classified by means of an on-the-fly classifier evaluating their temperature and species mass fractions, and the most appropriate reduced mechanism among the ones contained in the library is locally adopted.
2.2. Self Organizing Maps
2.3. K-Means Clustering
2.4. Coupling SOM with K-Means
2.5. Local Principal Component Analysis
- Initialization: the clusters’ centroids are initialized;
- Partitioning: each observation from the training matrix is assigned to a cluster by means of Equation (3);
- Update: the clusters’ centroids position is updated according to the new partitioning;
- Local PCA: Principal Component Analysis is performed in each cluster.
2.6. Directed Relation Graph with Error Propagation
3. Case Description
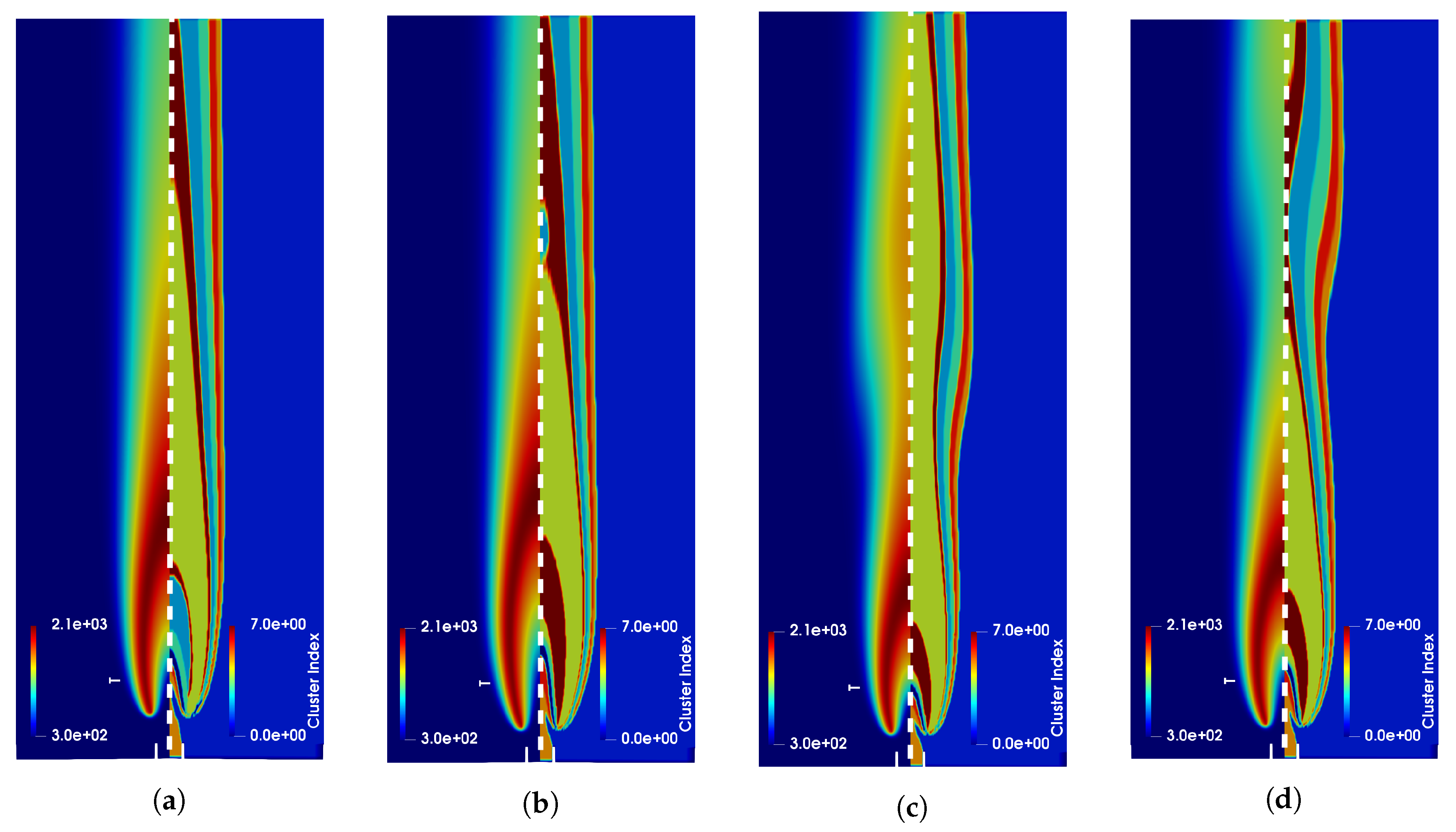
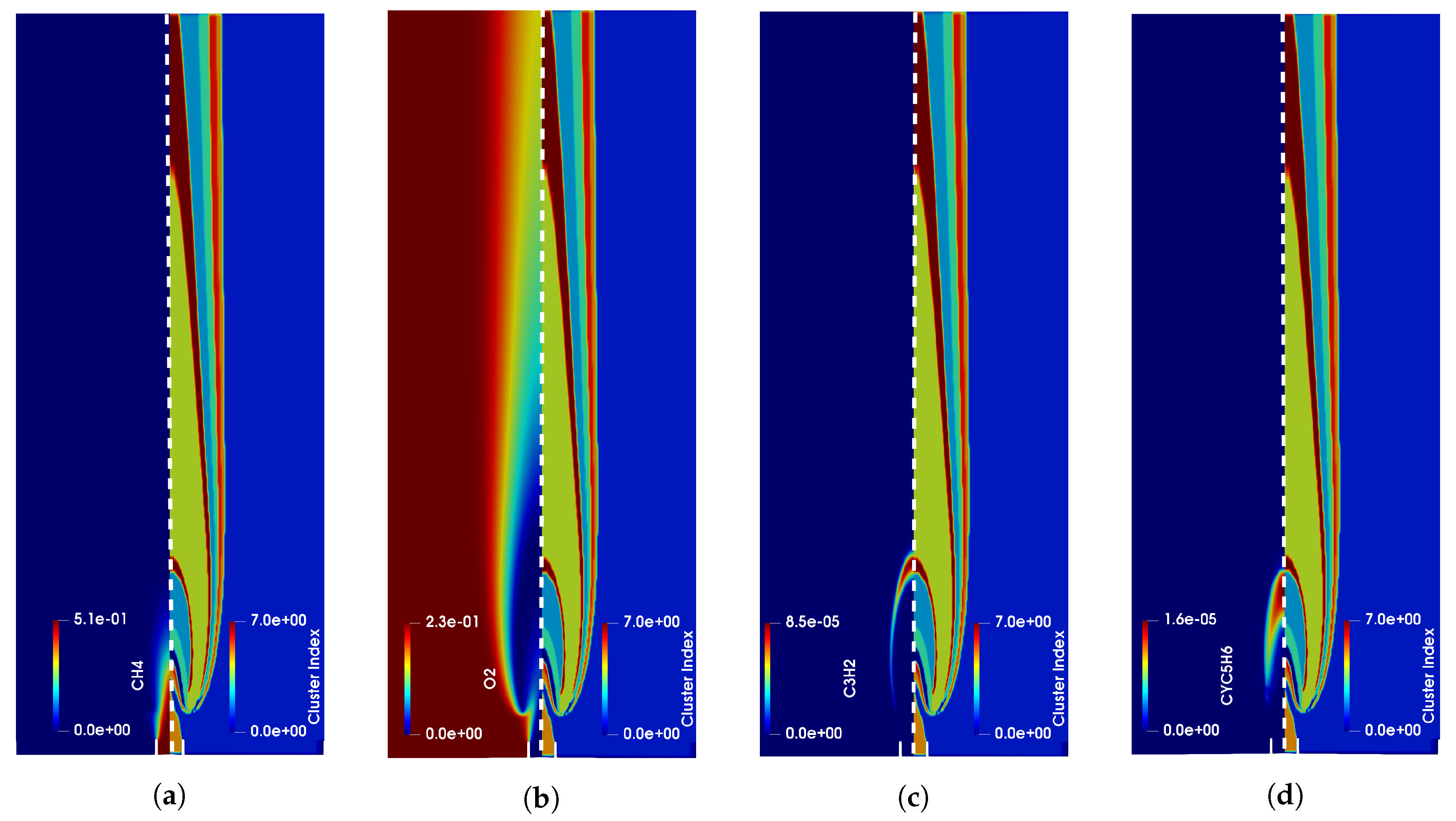
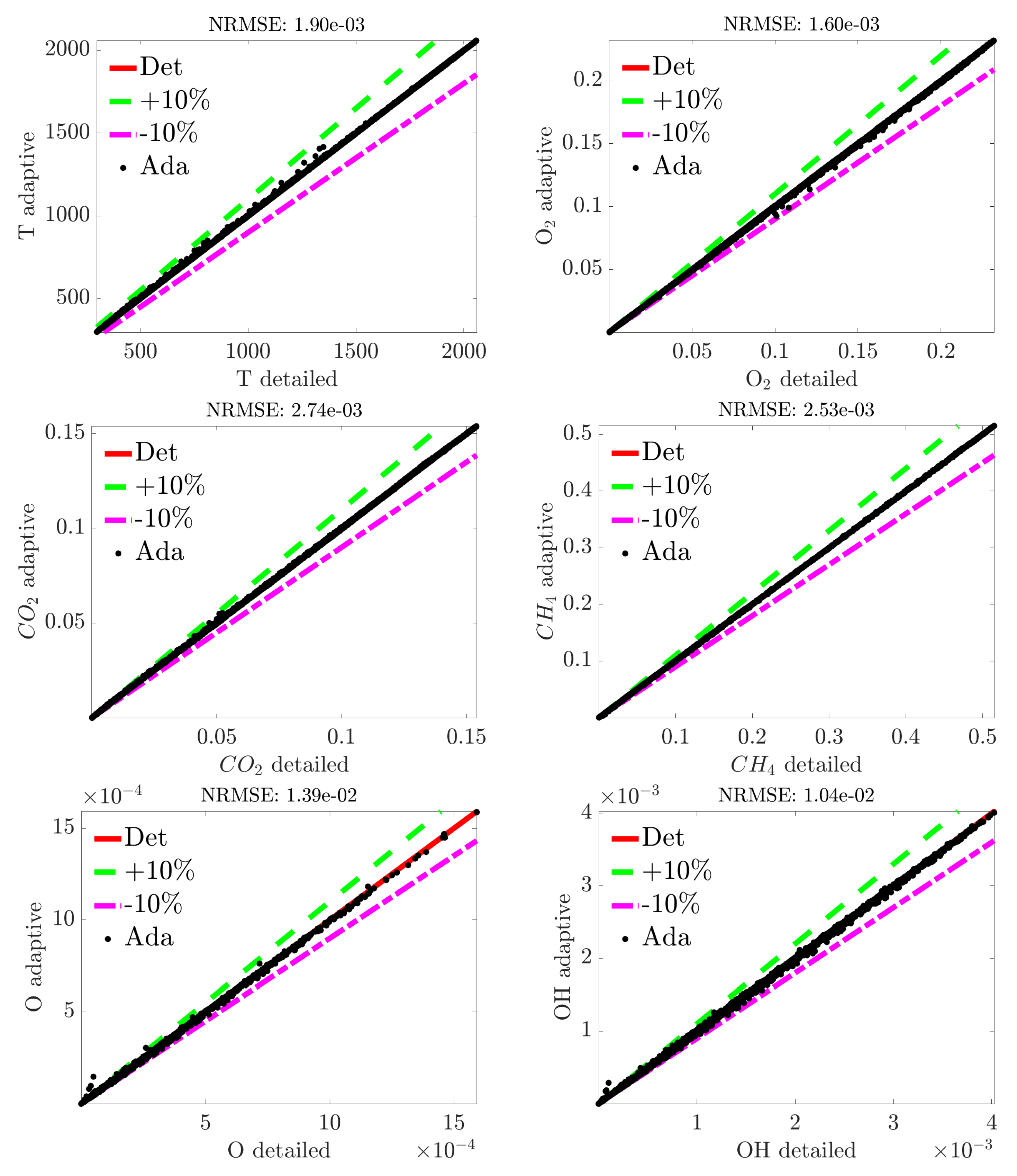
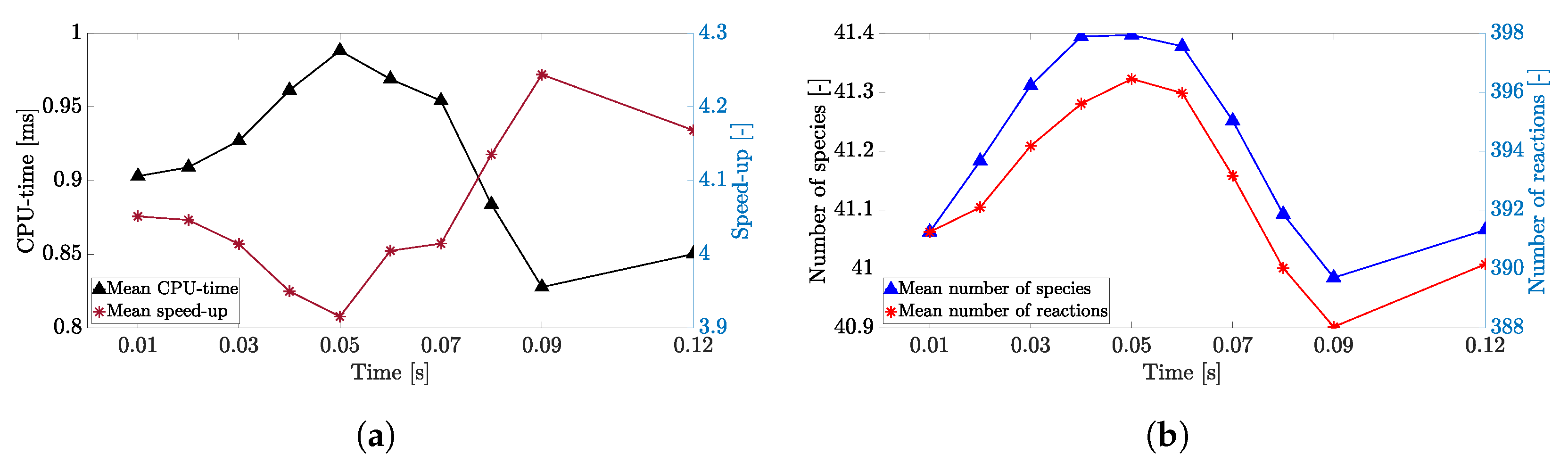
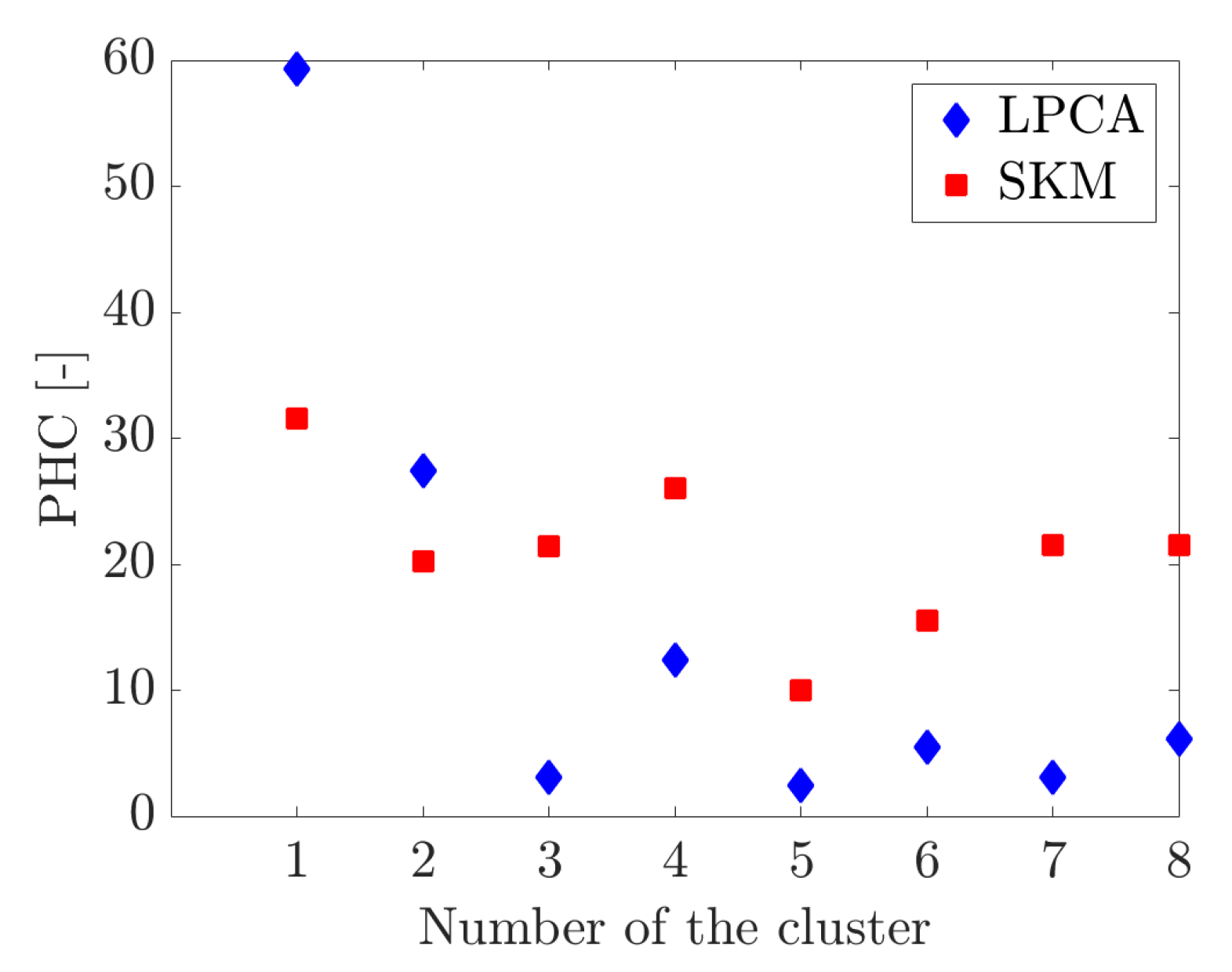
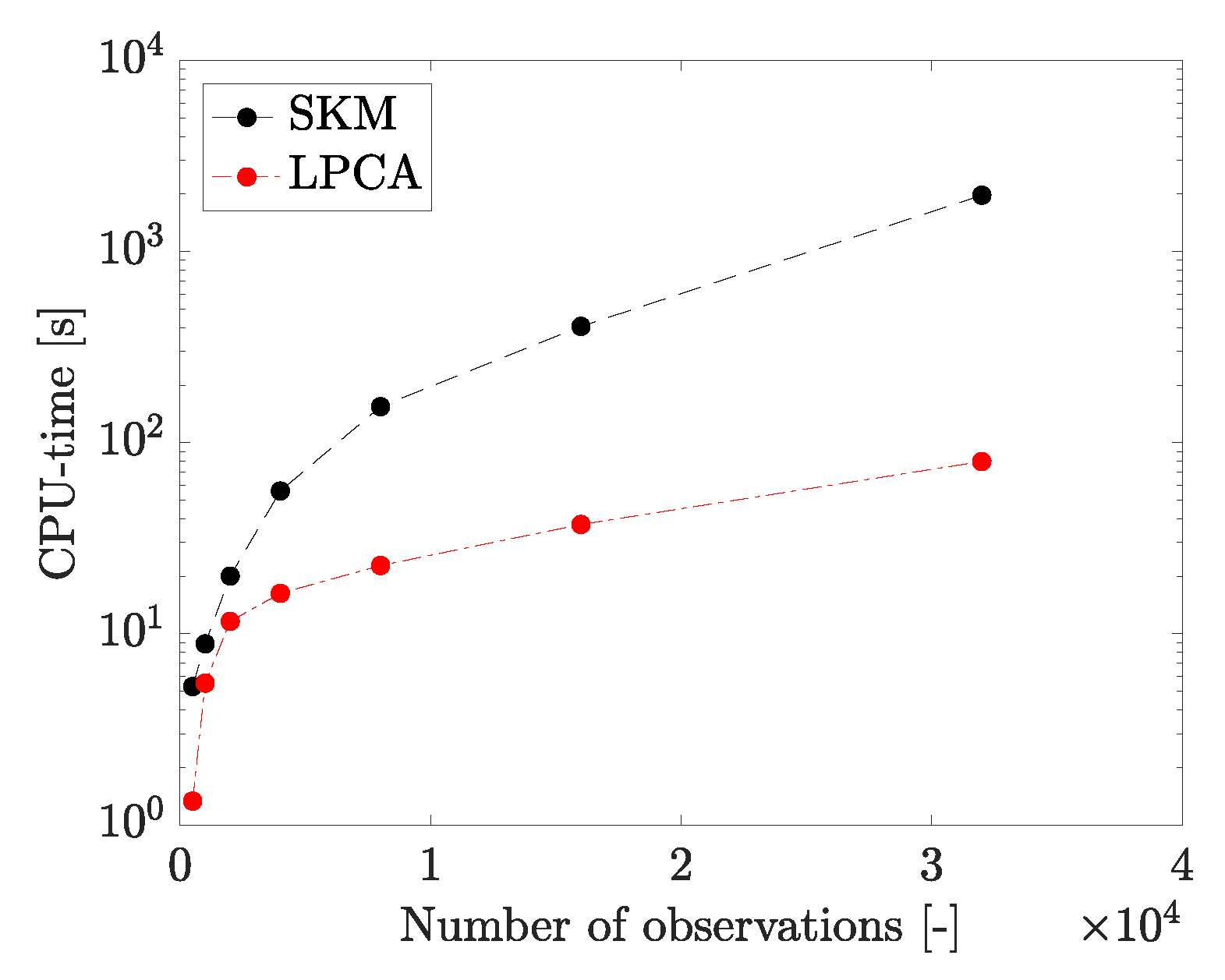
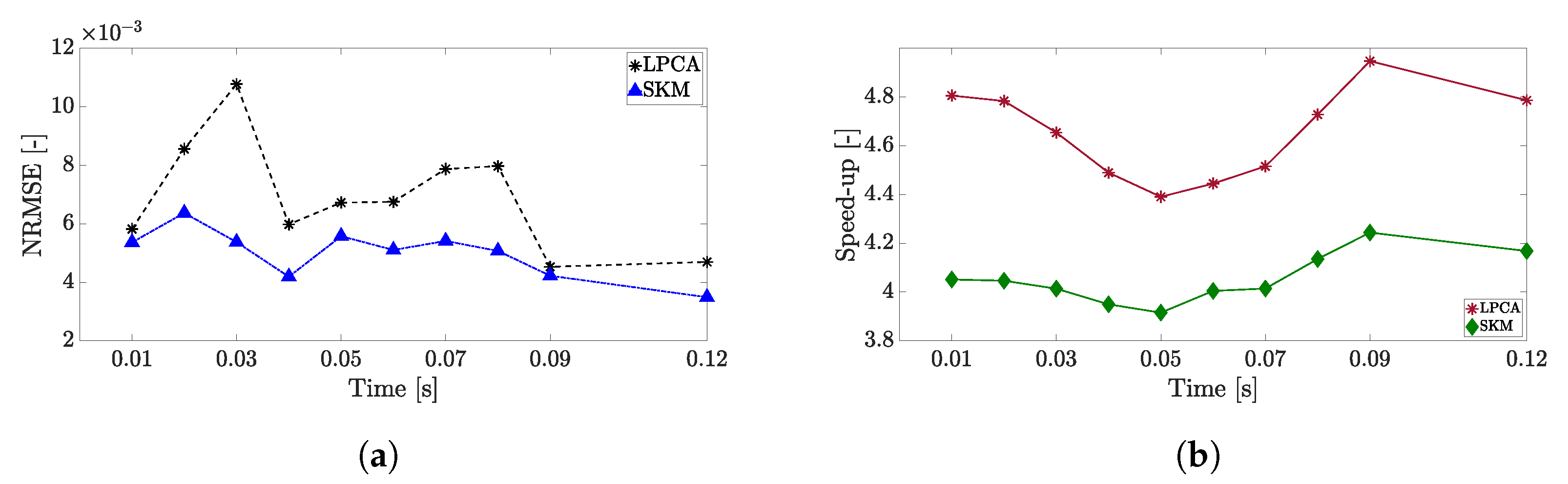
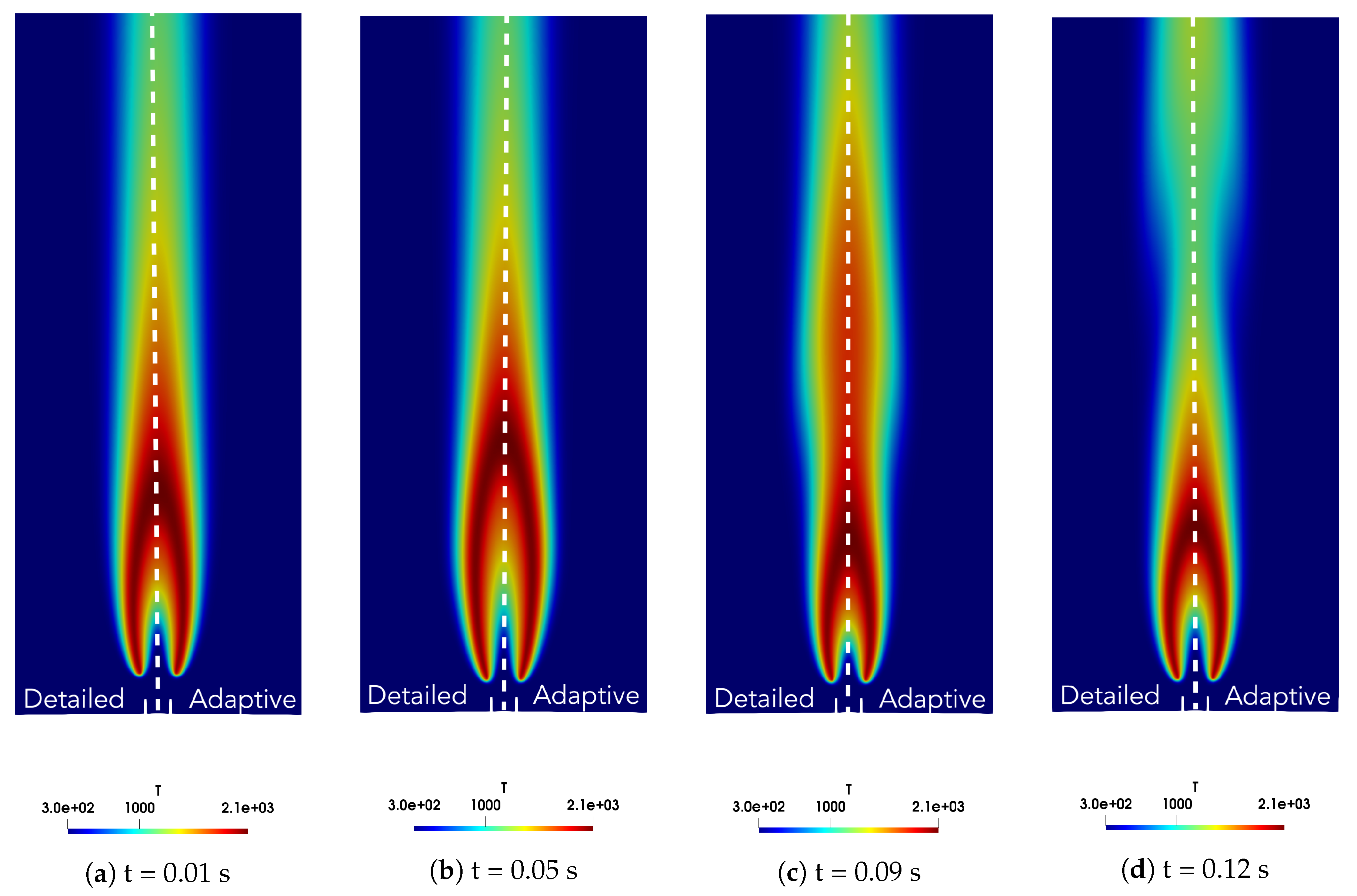
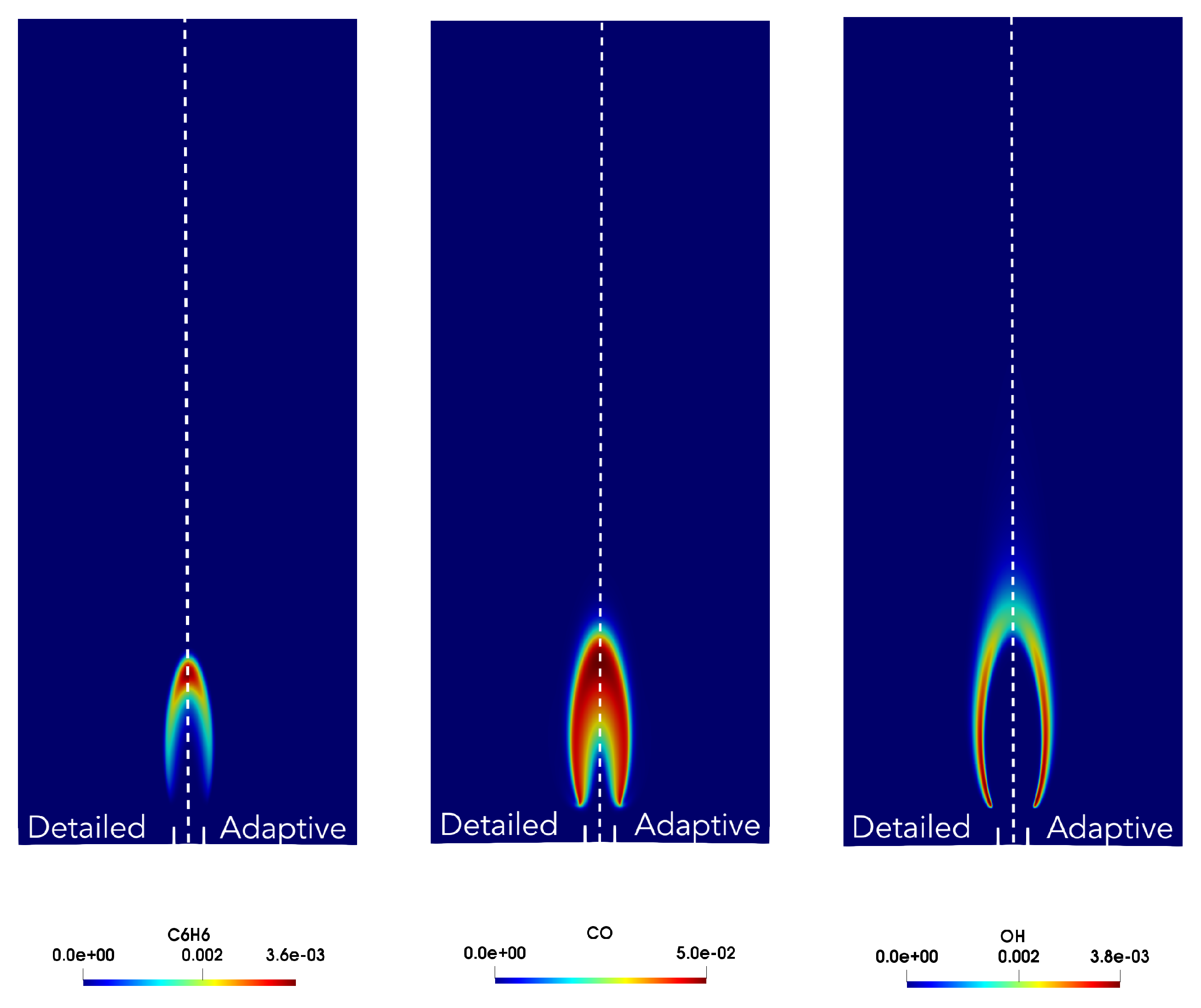
4. Results
4.1. Adaptive Simulation with SKM Partitioning
4.2. Adaptive Simulation with LPCA Partitioning
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lu, T.; Law, C.K. Toward accommodating realistic fuel chemistry in large-scale computations. Prog. Energy Combust. Sci. 2009, 35, 192–215. [Google Scholar] [CrossRef]
- Fang, X.; Huang, Z.; Qiao, X.; Ju, D.; Bai, X. Skeletal mechanism development for a 3-component jet fuel surrogate using semi-global sub-mechanism construction and mechanism reduction. Fuel 2018, 229, 53–59. [Google Scholar] [CrossRef]
- Chen, Y.; Mehl, M.; Xie, Y.; Chen, J.Y. Improved skeletal reduction on multiple gasoline-ethanol surrogates using a Jacobian-aided DRGEP approach under gasoline compression ignition (GCI) engine conditions. Fuel 2017, 210, 617–624. [Google Scholar] [CrossRef]
- Ranade, R.; Alqahtani, S.; Farooq, A.; Echekki, T. An extended hybrid chemistry framework for complex hydrocarbon fuels. Fuel 2019, 251, 276–284. [Google Scholar] [CrossRef]
- Liang, L.; Stevens, J.G.; Farrell, J.T. A dynamic adaptive chemistry scheme for reactive flow computations. Proc. Combust. Inst. 2009, 32, 527–534. [Google Scholar] [CrossRef]
- Contino, F.; Jeanmart, H.; Lucchini, T.; D’Errico, G. Coupling of in situ adaptive tabulation and dynamic adaptive chemistry: An effective method for solving combustion in engine simulations. Proc. Combust. Inst. 2011, 33, 3057–3064. [Google Scholar] [CrossRef]
- Zhou, L.; Wei, H. Chemistry acceleration with tabulated dynamic adaptive chemistry in a realistic engine with a primary reference fuel. Fuel 2016, 171, 186–194. [Google Scholar] [CrossRef]
- Komninos, N. An algorithm for the adaptive run-time reduction of chemical mechanisms during HCCI simulation. Fuel 2015, 140, 328–343. [Google Scholar] [CrossRef]
- Newale, A.S.; Liang, Y.; Pope, S.B.; Pepiot, P. A combined PPAC-RCCE-ISAT methodology for efficient implementation of combustion chemistry. Combust. Theory Model. 2019, 23, 1–33. [Google Scholar] [CrossRef]
- Pepiot-Desjardins, P.; Pitsch, H. An efficient error-propagation-based reduction method for large chemical kinetic mechanisms. Combust. Flame 2008, 154, 67–81. [Google Scholar] [CrossRef]
- D’Alessio, G.; Parente, A.; Stagni, A.; Cuoci, A. Adaptive chemistry via pre-partitioning of composition space and mechanism reduction. Combust. Flame 2020, 211, 68–82. [Google Scholar] [CrossRef]
- Ren, Z.; Pope, S.B. Second-order splitting schemes for a class of reactive systems. J. Comput. Phys. 2008, 227, 8165–8176. [Google Scholar] [CrossRef]
- Cuoci, A.; Frassoldati, A.; Faravelli, T.; Ranzi, E. Numerical modeling of laminar flames with detailed kinetics based on the operator-splitting method. Energy Fuels 2013, 27, 7730–7753. [Google Scholar] [CrossRef]
- Kohonen, T. The self-organizing map. Proc. IEEE 1990, 78, 1464–1480. [Google Scholar] [CrossRef]
- Kohonen, T.; Oja, E.; Simula, O.; Visa, A.; Kangas, J. Engineering applications of the self-organizing map. Proc. IEEE 1996, 84, 1358–1384. [Google Scholar] [CrossRef]
- Vesanto, J.; Alhoniemi, E. Clustering of the self-organizing map. IEEE Trans. Neural Netw. 2000, 11, 586–600. [Google Scholar] [CrossRef]
- Garcia, H.L.; Gonzalez, I.M. Self-organizing map and clustering for wastewater treatment monitoring. Eng. Appl. Artif. Intell. 2004, 17, 215–225. [Google Scholar] [CrossRef]
- Parente, A.; Sutherland, J.C. Principal component analysis of turbulent combustion data: Data pre-processing and manifold sensitivity. Combust. Flame 2013, 160, 340–350. [Google Scholar] [CrossRef]
- Lloyd, S. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability; University of California: Oakland, CA, USA, 1967; Volume 1, pp. 281–297. [Google Scholar]
- Davies, D.L.; Bouldin, D.W. A cluster separation measure. IEEE Trans. Pattern Anal. Mach. Intell. 1979, PAMI-1, 224–227. [Google Scholar] [CrossRef]
- Caliński, T.; Harabasz, J. A dendrite method for cluster analysis. Commun. Stat. Theory Methods 1974, 3, 1–27. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Tibshirani, R.; Walther, G.; Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. Ser. B Stat. Methodol. 2001, 63, 411–423. [Google Scholar] [CrossRef]
- Brentan, B.; Meirelles, G.; Luvizotto, E., Jr.; Izquierdo, J. Hybrid SOM+ k-Means clustering to improve planning, operation and management in water distribution systems. Environ. Model. Softw. 2018, 106, 77–88. [Google Scholar] [CrossRef]
- Kambhatla, N.; Leen, T.K. Dimension reduction by local principal component analysis. Neural Comput. 1997, 9, 1493–1516. [Google Scholar] [CrossRef]
- Jolliffe, I. Principal Component Analysis; Springer: Berlin, Germany, 2011. [Google Scholar]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: Berlin, Germany, 2006. [Google Scholar]
- Parente, A.; Sutherland, J.; Dally, B.; Tognotti, L.; Smith, P. Investigation of the MILD combustion regime via principal component analysis. Proc. Combust. Inst. 2011, 33, 3333–3341. [Google Scholar] [CrossRef]
- Parente, A.; Sutherland, J.C.; Tognotti, L.; Smith, P.J. Identification of low-dimensional manifolds in turbulent flames. Proc. Combust. Inst. 2009, 32, 1579–1586. [Google Scholar] [CrossRef]
- Aversano, G.; Bellemans, A.; Li, Z.; Coussement, A.; Gicquel, O.; Parente, A. Application of reduced-order models based on PCA & Kriging for the development of digital twins of reacting flow applications. Comput. Chem. Eng. 2019, 121, 422–441. [Google Scholar]
- Turányi, T.; Tomlin, A.S. Analysis of Kinetic Reaction Mechanisms; Springer: Berlin, Germany, 2014. [Google Scholar]
- Rabitz, H.; Kramer, M.; Dacol, D. Sensitivity analysis in chemical kinetics. Annu. Rev. Phys. Chem. 1983, 34, 419–461. [Google Scholar] [CrossRef]
- Tomlin, A.S.; Pilling, M.J.; Merkin, J.H.; Brindley, J.; Burgess, N.; Gough, A. Reduced mechanisms for propane pyrolysis. Ind. Eng. Chem. Res. 1995, 34, 3749–3760. [Google Scholar] [CrossRef]
- Brown, N.J.; Li, G.; Koszykowski, M.L. Mechanism reduction via principal component analysis. Int. J. Chem. Kinet. 1997, 29, 393–414. [Google Scholar] [CrossRef]
- Vajda, S.; Turányi, T. Principal component analysis for reducing the Edelson-Field-Noyes model of the Belousov-Zhabotinskii reaction. J. Phys. Chem. 1986, 90, 1664–1670. [Google Scholar] [CrossRef]
- Lu, T.; Law, C.K. A directed relation graph method for mechanism reduction. Proc. Combust. Inst. 2005, 30, 1333–1341. [Google Scholar] [CrossRef]
- Zheng, X.; Lu, T.; Law, C.K. Experimental counterflow ignition temperatures and reaction mechanisms of 1, 3-butadiene. Proc. Combust. Inst. 2007, 31, 367–375. [Google Scholar] [CrossRef]
- Niemeyer, K.E.; Sung, C.J.; Raju, M.P. Skeletal mechanism generation for surrogate fuels using directed relation graph with error propagation and sensitivity analysis. Combust. Flame 2010, 157, 1760–1770. [Google Scholar] [CrossRef]
- Dijkstra, E.W. A note on two problems in connexion with graphs. Numer. Math. 1959, 1, 269–271. [Google Scholar] [CrossRef]
- Mohammed, R.; Tanoff, M.; Smooke, M.; Schaffer, A.; Long, M. Computational and experimental study of a forced, time-varying, axisymmetric, laminar diffusion flame. Symp. Int. Combust. 1998, 27, 693–702. [Google Scholar] [CrossRef]
- Ranzi, E.; Frassoldati, A.; Stagni, A.; Pelucchi, M.; Cuoci, A.; Faravelli, T. Reduced kinetic schemes of complex reaction systems: Fossil and biomass?derived transportation fuels. Int. J. Chem. Kinet. 2014, 46, 512–542. [Google Scholar] [CrossRef]
- Caruana, R.; Lawrence, S.; Giles, C.L. Overfitting in neural nets: Backpropagation, conjugate gradient, and early stopping. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2001; pp. 402–408. [Google Scholar]
- Wick, A.; Attili, A.; Bisetti, F.; Pitsch, H. DNS-driven analysis of the Flamelet/Progress Variable model assumptions on soot inception, growth, and oxidation in turbulent flames. Combust. Flame 2020, 214, 437–449. [Google Scholar] [CrossRef]
- Swaminathan, N. Physical Insights on MILD combustion from DNS. Front. Mech. Eng. 2019, 5, 59. [Google Scholar] [CrossRef]
- Luo, Z.; Yoo, C.S.; Richardson, E.S.; Chen, J.H.; Law, C.K.; Lu, T. Chemical explosive mode analysis for a turbulent lifted ethylene jet flame in highly-heated coflow. Combust. Flame 2012, 159, 265–274. [Google Scholar] [CrossRef]
- Trisjono, P.; Pitsch, H. Systematic analysis strategies for the development of combustion models from DNS: A review. Flow Turbul. Combust. 2015, 95, 231–259. [Google Scholar] [CrossRef]
- D’Alessio, G.; Attili, A.; Cuoci, A.; Pitsch, H.; Parente, A. Analysis of turbulent reacting jets via Principal Component Analysis. In Data Analysis for Direct Numerical Simulations of Turbulent Combustion; Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar]
- Roh, Y.; Heo, G.; Whang, S.E. A survey on data collection for machine learning: A big data-ai integration perspective. IEEE Trans. Knowl. Data Eng. 2019. [Google Scholar] [CrossRef]
- Najafabadi, M.M.; Villanustre, F.; Khoshgoftaar, T.M.; Seliya, N.; Wald, R.; Muharemagic, E. Deep learning applications and challenges in big data analytics. J. Big Data 2015, 2, 1. [Google Scholar] [CrossRef]
- Bach, S.H.; He, B.; Ratner, A.; Ré, C. Learning the structure of generative models without labeled data. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 7–9 August 2017; Volume 70, pp. 273–282. [Google Scholar]


| 8 | 44 | 37 | 48 | 0.093 |
| k | ||||
|---|---|---|---|---|
| 8 | 42 | 34 | 49 | 0.076 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Alessio, G.; Cuoci, A.; Aversano, G.; Bracconi, M.; Stagni, A.; Parente, A. Impact of the Partitioning Method on Multidimensional Adaptive-Chemistry Simulations. Energies 2020, 13, 2567. https://doi.org/10.3390/en13102567
D’Alessio G, Cuoci A, Aversano G, Bracconi M, Stagni A, Parente A. Impact of the Partitioning Method on Multidimensional Adaptive-Chemistry Simulations. Energies. 2020; 13(10):2567. https://doi.org/10.3390/en13102567
Chicago/Turabian StyleD’Alessio, Giuseppe, Alberto Cuoci, Gianmarco Aversano, Mauro Bracconi, Alessandro Stagni, and Alessandro Parente. 2020. "Impact of the Partitioning Method on Multidimensional Adaptive-Chemistry Simulations" Energies 13, no. 10: 2567. https://doi.org/10.3390/en13102567
APA StyleD’Alessio, G., Cuoci, A., Aversano, G., Bracconi, M., Stagni, A., & Parente, A. (2020). Impact of the Partitioning Method on Multidimensional Adaptive-Chemistry Simulations. Energies, 13(10), 2567. https://doi.org/10.3390/en13102567







