Abstract
Molecular dynamics simulations have been performed for a model aqueous solution of mucin. As mucin is a central part of lubricin, a key component of synovial fluid, we investigate its ability to form cross-linked networks. Such network formation could be of major importance for the viscoelastic properties of the soft-matter system and crucial for understanding the lubrication mechanism in articular cartilage. Thus, the inter- and intra-molecular interaction energies between the residues of mucin are analyzed. The results indicate that the mucin concentration significantly impacts its cross-linking behavior. Between 160 g/L and 214 g/L, there seems to be a critical concentration above which crowding begins to alter intermolecular interactions and their energies. This transition is further supported by the mean squared displacement of the molecules. At a high concentration, the system starts to behave subdiffusively due to network development. We also calculate a sample mean squared displacement and p-variation tests to demonstrate how the statistical nature of the dynamics is likewise altered for different concentrations.
1. Introduction
The last 50 years has seen ongoing research into the mechanism of lubrication seen within joints [1,2,3]. Hyaluronic acid (HA), phospholipids (PL), and lubricin (proteoglycan 4 (PRG4)), the constituent components of synovial fluid [4], interact with a complicated multiscale, synergistic nature [2,5,6,7,8] to create a lubrication phenomenon that is still not well described. Thus, it is important to analyze the dynamics of each component, as well as the synergy between them. In [9,10,11], we focused on the tribological role of HA–PL interactions, an effect also investigated in [12] for its role in tribological surgical adjuvant. To continue building an understanding of synovial lubrication, we now study the inter- and intra-molecular properties of lubricin within synovial fluid. To such an end, we analyze the energy of inter- and intra-molecular interactions in the system.
It was shown by Chang et al. that solutions of HA-PRG4 play an important role in joint lubrication and wear resistance [13]. It was proven experimentally that lubricin alters the frictional behavior of model hydrophobic and hydrophilic surfaces. They conjectured that lubricin reduces wear by shielding the surfaces from direct contact, while HA does not adsorb and thus does not impact friction. Further, in their experiment, the frictional behavior of a physiologically-consistent mixture of lubricin and HA was similar to that of lubricin alone [13]. The significance extends to articular cartilage (AC) systems, whose components possess a dual hydrophobic-hydrophilic nature. The otherwise ordinary affinity of these nonpolar (NP) and polar (P) molecules for similar groups is ultimately what leads to the self-ordering seen in AC systems.
Mucin in joint fluid was first isolated by acetic acid precipitation as early as 1846 by Frerichs [14]. Mucins are large glycoproteins, which are very widely distributed throughout the different organs of the human body, e.g., in stomach, lungs, respiratory tract, gastrointestinal tract, liver, kidney, colon, eye, and ear [15]. They lubricate and protect a large range of epithelial surfaces by forming gel-like mucosae when secreted in a large enough concentration. The assumption is that mucin is similarly responsible for the lubricating properties of lubricin. Lubricin exhibits a bottle-like structure. Such a formation has been shown to prevent inter-chain penetration between sliding surfaces coated with this kind of molecule. This consequence is believed to be beneficial for the low friction coefficient in a natural joint systems [5]. Lubricin, however, is a heavily O-glycosylated protein macromolecule built from more than 1400 amino acids (AA). It is too complex to be modeled due to the lack of structural information on the glycosylation sites and the glycan chains, but in its small central region (about 100 AA), mucin-like domains are present [13,16,17,18].
Genetic sequencing distinguished 22 human mucin genes, denoted as MUC1–MUC22. Although synovial mucin differs from the mucins of epithelial origin [19], it shares many structural similarities with members of the mucin family, which contribute to lubricating properties [20]. In this paper, MUC1 was chosen as an example of the mucin structure as it is most widely distributed in humans. There were several recent studies, theoretical [21,22,23], as well as experimental [24], which analyzed the docking/binding and adhesive properties of mucin, which suggest that hydrogen bonds, in particular inter-molecular interactions, play an important role in many biological processes. How mucins cross-link to form networks and which atomic interactions govern this process would seem to be of particular importance also for its frictional attributes. The present paper focuses on this problem.
Most studies of protein structures and interactions are carried out in dilute environments. In vivo, however, proteins operate in environments in which other molecules can easily restrict their motion, with concentrations from 50 up to 400 g/L [25]. It is intuitive that this will affect their behavior. In [26], it was shown that the excluded volume effect, produced by a high density of macromolecules, alters the stability and the folding rate of globular proteins. The difference between crowded and diluted environments can be observed in the inter- and intra-molecular interactions, especially hydrogen bonds’ creation and annihilation, and the hydrophobic interactions, which are of major importance for self-arrangement and dynamic properties. Molecular crowding also affects the transport properties of proteins; molecules in a crowded system exhibit subdiffusion [27]. Additional studies on the crowding effect in protein systems have been performed in more recent years [26,27,28,29], but a description of how crowding affects interactions in protein networks and its consequences for friction are still worth further exploration. Such studies could be very important for biomedical application, e.g., to improve methods of detecting and healing of osteoarthritis [30].
In this paper, we consider a model water-based solution of mucin as a canonical ensemble, with quasi-infinite (periodic) boundary conditions. We present molecular dynamics simulations of mucin interactions, with attention to network formation. The simulations were performed for five different concentrations to locate the onset of crowding effects and the resulting effects on frictional properties of the environment.
The paper is structured as follows. In the next section (Section 2), we detail the molecular dynamics simulation and describe the calculation of all parameters used to explore the characteristics of protein network formation. Next (see Section 2.2), we describe a method to distinguish which stochastic model is best for describing the time evolution of the system. Section 3 presents results and their discussion in light of how they can contribute to the modification of the viscoelastic properties of the system. The paper closes with conclusions (Section 4).
2. Methods
In order to investigate the cross-linking abilities of the mucin, all-atom molecular dynamics simulations of five different concentrations of mucin solution in water were performed. An analysis of these results is used to describe protein network formation in (non-)crowded systems and its relation to friction in the system.
2.1. Simulation Details
Human mucin MUC1 (PDB ID: 2ACM—SEA domain of MUC1; see Figure 1) was taken from the Protein Data Bank (PDB)[31] and modified using the YASARA Structure Software (Vienna, Austria) [32] by adding missing hydrogen atoms. Its molecular mass was equal to 12 kDa.
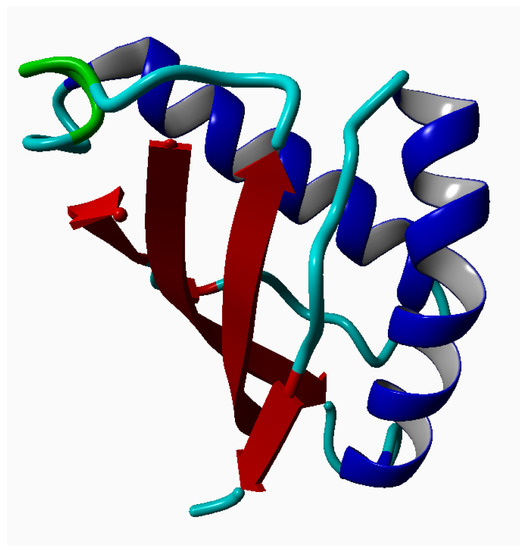
Figure 1.
Ribbon structure of the macromolecule MUC1 drawn using YASARA. It is a complex of two protein chains. The first is built of 66 AA and the second of 55 AA. The structure was taken from PDB.
The AMBER03 force field [33] was chosen to evaluate interactions between mucin molecules due to its proven performance in biophysical systems. It is given by:
where , , and are force constants. The first two sums accounted for the deviations from equilibrium bond length and bond angle , respectively. The third accounted for torsion from dihedral angles , with the phase taking a value of or . The final sum corresponded to short-range repulsion (), London dispersion (), and electrostatic forces (). Usually, the dielectric constant is taken as where the solvent is represented explicitly [33]. Parameters were as follows: temperature T = 310 K, pH = 7.0, and 0.9% NaCl aqueous solution (using the four-site model (TIP3P) of water [34]), with a time step of 2 fs. Berendsen’s method with a relaxation time of 1 fs maintained constant temperature and pressure. The simulation box was given sides of , , , and we applied periodic boundary conditions. The mucin molecules were placed in the simulation box in the folded form, and after the addition of water molecules, the system was minimized for steps with a time step of 2 fs.
To investigate crowding in the system, increasing concentrations of mucin were chosen: g/L (5 mucin molecules), g/L (10 mucin molecules), g/L (15 mucin molecules), g/L (20 mucin molecules), and g/L (25 mucin molecules). Each case was repeated 20 times to obtain more statistically-reliable information. The simulation runs had the same initial conditions, but differed only slightly in simulation temperature (at the third decimal digit), which in YASARA also served as the random number seed.
Thus, the simulated system probed concentrations in a range encompassing what is present in living organisms. The chosen range allowed the model system to accurately simulate mucin both without crowding (too few particles to form a network) and with obvious crowding (stable network formation) in the time range of 0–20 ns.
2.1.1. Simulation Parameters
The radius of gyration of a (mucin) molecule can be written as:
where is the number of atoms in the molecule, is the position vector of i atom, and is the position vector of the center of mass of the molecule.
The mean squared displacement (MSD) of a molecule at time step t is defined as:
where is the number of molecules in the simulation box and is the position vector of geometrical center of a molecule i.
2.1.2. Diffusion Coefficient
The time evolution of the MSD of the protein particles (cf., Equation (3)) can characterize the viscoelastic properties of the system [35]. It is related to the mechanical response functions of the environment through the generalized Stokes–Einstein relation [36]. In 3D Euclidean space, the MSD should follow the relation [37,38]:
where is a generalized self-diffusion coefficient (a constant that does not depend on time and is of the dimension of ) [39,40]. An exponent represents anomalous diffusion ( represents normal diffusion). In normal diffusion, the diffusion coefficient obeys the Stokes–Einstein law,
where is the characteristic thermal energy and is the friction coefficient, which depends on particle radius viscosity as . In anomalous diffusion, the quantity can still be identified with diffusion as the “apparent” diffusion coefficient , but it will scale in time (or length) as [41]:
2.2. Statistical Tests
Macromolecules present complex dynamics that can be modeled by stochastic processes. Several stochastic models bear consideration for the anomalous dynamics frequently seen in macromolecular systems and characterized by Equation (4). We will distinguish between continuous-time random walk (CTRW) and fractional Brownian motion (FBM) or the more general fractional Levy -stable motion (FLSM), to determine which optimally describes the evolution of macromolecules with non-Markovian evolution [42].
We performed statistical tests that can help discriminate between FBM, FLSM, and CTRW processes. First is the sample mean squared displacement. If we consider stochastic discrete process , then the sample mean squared displacement (sample MSD) for a characteristic time lag is defined as [43]:
Parameter is a random variable that yields valuable information when we consider only small , i.e., when is much smaller than the length of the series N. If the process , which underlies the recorded signal, follows from a continuous-time random walk, then , where denotes a similar distribution. By taking different time dependencies, the sample MSD will reveal whether our process is better modeled as a CTRW or FLSM [43].
A p-variational test was also used in this work. For given time and , the p-variation is defined as [44]:
where (unrelated to the force constant from Equation (1)) is given by [43]:
where X is a parameter calculated from the MD simulations, taking the form of a single time series , , …, . In practice, sample p-variation can be obtained according to [45]:
Observations of as a function of n can yield further information on the origins of time series.
3. Results and Discussion
3.1. Geometrical Properties of the Model Protein System
The molecular initial (a,c) and final (b,d) structures of mucin solutions, for two representative concentrations ( and ), are presented in Figure 2. For the lowest concentration, the proteins were able to move freely in the simulation box, following a near random walk until meeting. Three mucin molecules eventually formed an easily-breakable aggregate (having few intermolecular interactions, presented in the next Subsection). For the highest concentration, , initial and final arrangements (Subfigures c and d) looked very similar due to the creation of a more stable network, whose formation was enhanced due to crowding in the system. Differences between the initial and the final structures can be better investigated through the parameters discussed in the previous section.
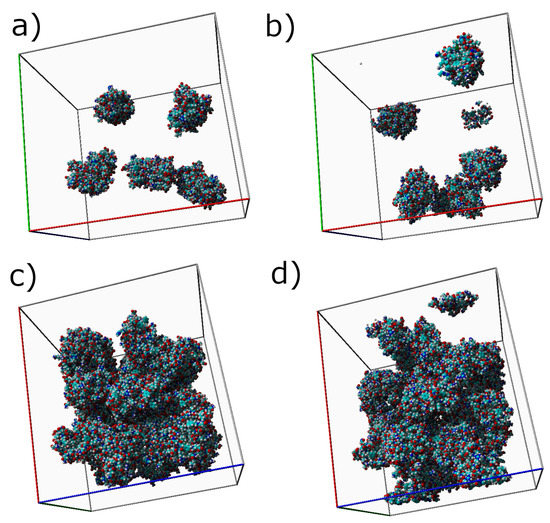
Figure 2.
Initial (a,c) and final (b,d) structures of the mucin network. (a,b) show simulation boxes for , and (c,d) show simulation boxes for . Light blue atoms represent carbon, dark blue nitrogen, red oxygen, yellow sulfur, and white hydrogen. Note that the proteins are fragmented due to the periodic boundary conditions on each wall.
According to Equation (2), the average radius of gyration, , of a mucin as a function of time is presented in Figure 3. This offers some information on the overall geometric changes occurring to a chain throughout the simulation.
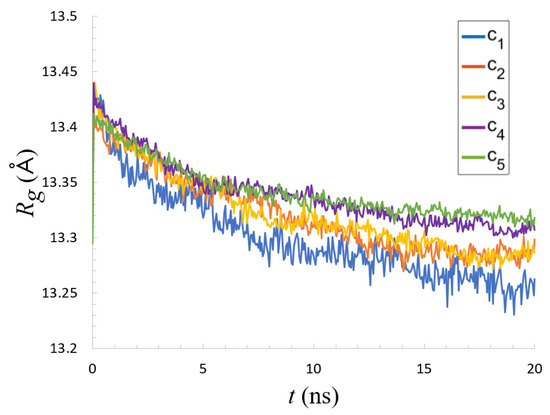
Figure 3.
Radius of gyration (averaged over all molecules originating across all simulation runs) as a function of time.
The initial mucin radius of gyration was about Å. In all cases, decreased with time. The different concentrations all followed a very similar trend. There was, however, a slight difference between the crowded and non-crowded systems. For high concentrations, declined marginally more slowly than for low concentrations where the decrease was more visible, though still only about 1.2%. For the uncrowded system, this suggests that, despite the initial folded state, some conformational changes still occurred, and mainly due to the force field presence, the molecule collapsed. In crowded conditions, the presence of other proteins in close proximity can prevent internal conformational changes due to the lack of physical space for the unfolding and refolding to occur.
3.2. Inter- and Intra-Molecular Interactions
The total energy of interactions, computed based on the last two parts of Equation (1) and averaged per mucin molecule, are presented in Figure 4.
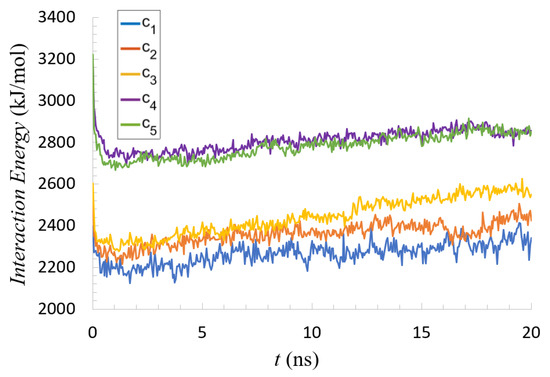
Figure 4.
Interaction energies (per one mucin molecule originating across all simulation runs) as a function of time.
All five cases appeared similar, but the trend of increasing energy with concentration revealed a crowding effect; it was not simply linear. Despite the mucin concentration increasing in even steps, there was a clear transition between and . Interesting features can also be seen in the binding tendencies between individual AAs. Maps of these interaction energies are depicted in Figure 5.
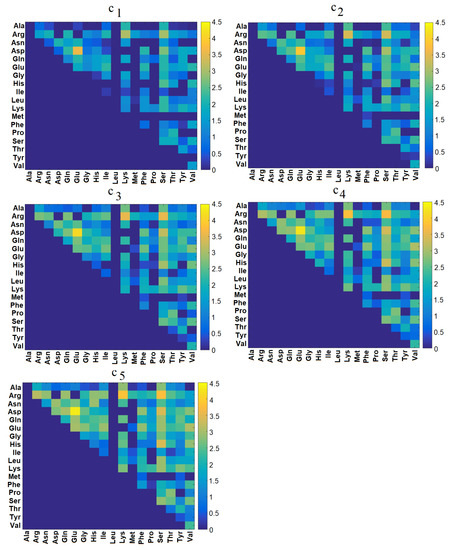
Figure 5.
Maps of interaction energies (kJ/mol) between each pair of AA in mucin.
The highest energies appeared between Asp-Glu, Arg-Lys, and Arg-Ser. The first two were pairs of AAs with opposing charges, allowing the formation of salt bridges. Ser is neutral and polar, but can still form hydrogen bonds with Arg. In all three, the energy was about 3–4 kJ/mol. The increased concentrations led to the association of additional strongly-interacting AA pairs. Arg, Ser, and Glu were most prominent on the interaction maps for all cases, though for and , two new pairs began to display considerable interactions: Asp-Ser and His-Ser (energies of about 3–4.5 kJ/mol).
Interaction energies can be further separated according to their location (inter- or intra-molecular) and origin (hydrogen bonds (HBO) or hydrophobic-polar (HP)) to see which prevails for network formation.
According to YASARA’s definition of HBO, a bond is formed between two oxygen atoms if: (i) the hydrogen was within of each oxygen; and (ii) the oxygens were within of each other. Additionally, the bond energy must be greater than one fourth the optimal value or 6.25 kJ/mol (or 1.5 kcal/mol). Thus, only strong bonds (up to ) were considered. The HBO energy (as well as HP interaction strength between hydrophobic atoms) was calculated by the algorithm described previously [12]. Both results presented information about the strength of the protein network. Intramolecular HP interactions can be identified with conformational stability of an individual molecule, while backbone intermolecular HBO interactions are related to network stability [46].
A large difference between the number of inter- and intra-molecular interactions for both HP and HBO can be seen (see Figure 6 and Figure 7).
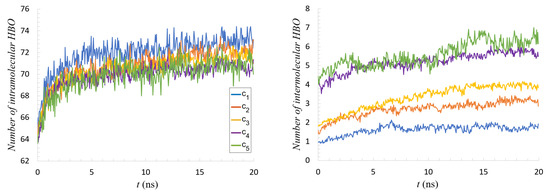
Figure 6.
Number of hydrogen bonds per single mucin molecule. Two cases are presented: (left) intramolecular; (right) intermolecular.
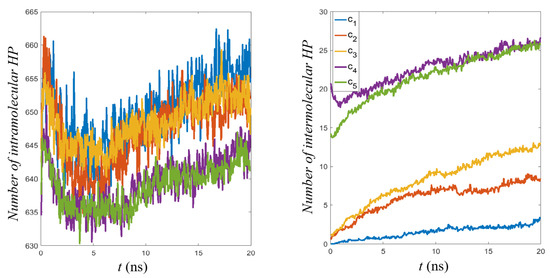
Figure 7.
Number of hydrophobic contacts per single mucin molecule. Two cases are presented: (left) intramolecular; (right) intermolecular.
For example, the relative number of intermolecular HBO interactions to total HBO interactions was about 2% for , 4% for , 5% for , 7% for , and 8% for . This is due to the fact that the conformation taken from PDB was the protein’s folded state, so the number of intramolecular interactions was maximized at the start of the simulation.
As the proteins aggregated in the course of the simulation, the number of intermolecular interactions increased, but more time would probably be needed for this to reach a maximum. The crowding in higher concentrations likely prevented molecules from finding their lowest energy conformations, while the extra freedom in low concentrations allowed for more fluctuations. That is why for (and also ) stabilization began after 10 ns for both parameters (see Figure 6). Weak networking seemed to appear for the first time at concentration as evidenced by the continued increase in intermolecular interactions, but would still be broken with relatively small amounts of energy. This continued increase mirrored those seen for high concentrations, just for a smaller absolute number of interactions (a downward shift). The cases and were very similar with almost the same number of HBO and HP contacts by the end of the simulation (cf. Figure 6 and Figure 7). Thus, increasing the concentration even further would likely not impact the network mechanical properties.
There was a marked decrease (about ) in the number of intramolecular HP interactions during the first 5 ns of simulation (see Figure 7, left). In the same time, increases in the number of intramolecular HP interactions (see Figure 7, right) and also HBO of both types were seen. This points to a conformational reorganization inside individual protein chains that was similar across the concentrations. One also notices that the relation between parameters inverted when moving from intra- to inter-molecular interactions. Increasing concentration favors interactions between molecules while slightly mitigating intramolecular interactions. This, again, was evidence of the competition for bonds resulting from increased crowding.
In all cases, one can notice that the first 10 ns of simulation were crucial for network formation: the dynamics of contact creation was initially very fast, but after this time, the process slowed.
In all the interactions charts (cf., Figure 6 and Figure 7), a conspicuous gap between concentrations was visible. Namely, between and , there was a critical concentration above which a transition seemed to occur. More refined concentration values would be necessary to find the optimum conditions for strong network formation in this range, sparse enough to allow conformations to occur, but dense enough to provide the necessary cross-linking.
3.3. MSD of Model Proteins
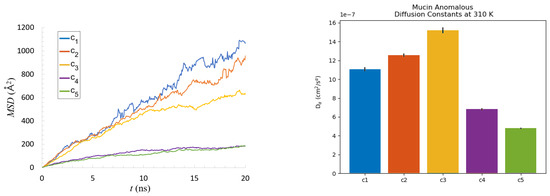
Figure 8.
Evolution of the protein MSD in time (left) and fitting values of diffusion coefficient (right) for five concentrations of mucin’s solutions, averaged over all molecules for all iterations of the simulation. Black lines show the STD of the fitting parameters.
Analyzing the MSD of the particles is important due to its relation to the mechanical response functions of the system through the generalized Stokes–Einstein relation (see Equation (2) in [36]). It is interesting to consider of what type of motion the MSD is representative. A detailed analysis is presented in the next paragraph (cf. Section 2.2). Because macromolecular systems often exhibit anomalous behavior, we tested if the throughout as in Equation (4). Values of the fitting parameter are shown in Table 2 and values of generalized (or fractional) diffusion coefficient are depicted in Figure 8 right. The fitting was done using the Python MDAnalysis Toolkit [47].

Table 2.
parameters for different concentrations of mucin based on the fitting function presented in Equation (4).
We can clearly see discrepancies between the results for the – and – cases. The parameter for was close to one. It could suggest a common diffusion-like movement, but knowing that the environment was viscoelastic, we can deduce that the function should take a different, more complex form (cf., Figures 2 and 3 in [36]) with three regimes: short times, intermediate times, when plateau of MSD is obtained, and large times [48].
The parameters for and were lower than one; this concentration range also behaved similarly to the viscoelastic regime, though favoring the elastic contribution. Small aggregates were created, and transient networks appeared. Proteins in high concentrations, , tended to attach to each other and replace intra HBO and HP interactions with their intermolecular counterparts. This resulted in abnormal diffusion ( was closest to , i.e., subdiffusion) due to the confinement of mucin created by the more stable network. In this case, the soft-matter system exhibited quite solid-like behavior (elasticity). It should be noted that for intracellular media, elastic forces could arise not just from the polymer network, but also from the forces due to the activity of molecular motors [35].
How macromolecular crowding affects cellular processes such as protein folding and assembly was studied in [49]. The authors showed experimentally that crowding led to anomalous subdiffusive of cytoplasmic macromolecules. Moreover, they proposed that the anomaly in the diffusion (manifested by parameter) could be used as a quantifiable measure for the crowdedness. Similar approaches can be seen in many other publications (e.g., [26,27,41,50]). Banks et al. studied the diffusion of tracer proteins in highly-concentrated (up to 400 g/L) random-coil polymer and globular protein solutions. Furthermore, noticing the subdiffusion, they computed an apparent diffusion coefficient and observed its relation with concentration (see Figure 3 in [41]). They showed a lowering of in the course of the concentration. We can also see a similar effect when looking at Figure 9.
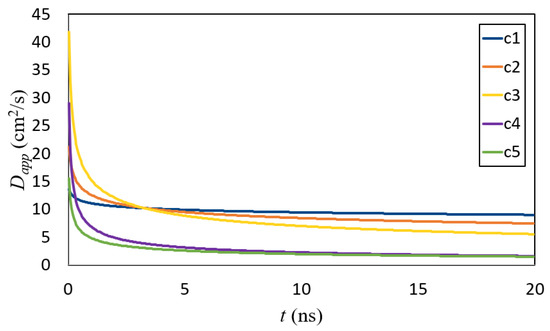
Figure 9.
Apparent diffusion coefficient as a function of time.
The was computed based on Equation (6) where the parameters and were found earlier by fitting (cf., Table 2 and Figure 8). Two different regimes emerged in Figure 9: initial (up to about 5 ns) and long times (above it). The concentration had a different influence on the structure formation in each of the regimes.
A similar approach to diffusion in (non-)crowded protein solutions was found in [48]. The authors presented molecular dynamics simulations of villin headpiece solutions at various concentrations. They analyzed how diffusion depends on protein–protein contacts. Although they considered the results (mainly MSD) on three separate time scales, their results were still in accordance with our own. Namely, they claimed that internal protein dynamics remained largely unaltered as the protein concentration increased, but translational diffusion slowed. The decrease in diffusion was interpreted resulting from the transient formation of protein clusters.
3.4. Determining a Stochastic Model
In order to frame the dynamics from a statistical point of view, calculations of sample MSD and p-variational tests were performed. The first test, sample MSD, indicated that the relationship between and was almost linear for small for all HBO time series calculated in our simulations. Directional parameters for the function differed from one. For all concentrations, the number of intramolecular HBO interactions (-) yielded a much steeper relation than for the corresponding number of intermolecular HBO interactions. In Figure 10, we show the results of the test for concentration as an example. All results of the sample MSD suggested that a CTRW did not underlie the process, as this would produce sublinear behavior.
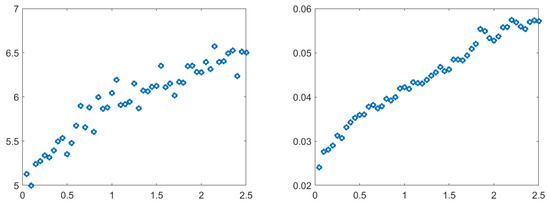
Figure 10.
Calculated sample MSD as a function of time lag () for concentration . Two cases are presented: (left) intermolecular; (right) intramolecular.
Next, we performed the p-variational test according to Equation (10). This test again excluded a CTRW for modeling changes in the number of inter- and intra-molecular HBO interactions for all concentrations. These changes of HBO should be modeled as FBM or FLSM, but discrimination between these two requires an additional statistical test. The most interesting results are presented in Figure 11, Figure 12 and Figure 13.
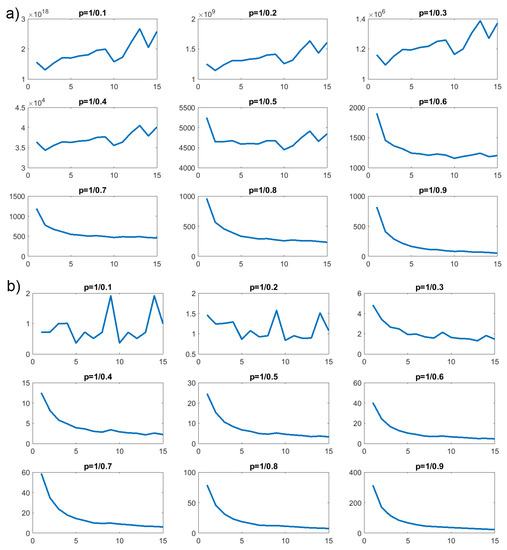
Figure 11.
p-variation as a function of n for HBO in concentration : (a) intermolecular; (b) intramolecular.
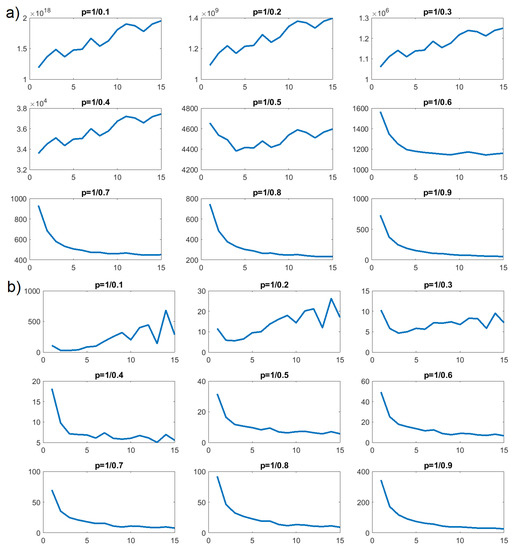
Figure 12.
p-variation as a function of n for HBO in concentration : (a) intermolecular; (b) intramolecular.
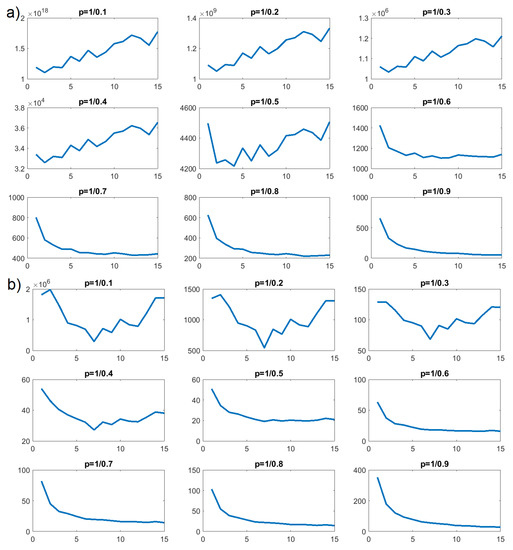
Figure 13.
p-variation as a function of n for HBO in concentration : (a) intermolecular; (b) intramolecular.
In Figure 12b, we can see that values of as a function of n increased for , , and , but decreased for and all further values of p. Such a property is characteristic of an FBM process. We performed the same tests on the number of intermolecular HBO interactions for concentration . The results of the p-variational are presented in Figure 12a and similarly confirmed that CTRW would be a poor stochastic model for describing the time evolution of intermolecular HBO interactions. Here, the clear increasing trend persisted until . Around , this trend changed and decreased for the remaining values; , , , and . This, again, is a property characteristic of FBM processes.
The p-variation tests for concentrations and indicated FBM processes for the number of intramolecular HBO interactions and the number of intermolecular HBO interactions in both cases. However, the shift in behavior for appeared at different values of the p parameter.
The same calculations were again performed for intramolecular HBO for concentrations and and are likewise presented on Figure 11b and Figure 13b. They indicated that these processes resembled an FBM model. Interestingly, Figure 11a and Figure 13a, representing intermolecular interactions, exhibit some differences from the rest. The first value () for and the first three for showed no clear trend, while smaller values of p generated definite decreases. For just these cases, we would conclude that an FLSM process would be most appropriate for modeling the behavior of intermolecular HBO formation and annihilation.
4. Conclusions
MD simulations of (non)crowded protein systems were performed at different concentrations. The results showed that between and (160 g/L and 214 g/L), a transition existed where crowding began affecting the dynamics of protein network formation. The crowding effect was clearly visible through interactions energy, the number of intermolecular HP and HBO interactions, and through the stabilization of the radius of gyration . Additionally, above some limiting concentration (within the same range as above), the system transitioned to subdiffusive behavior due to network formation, a feature that was shown through an analysis of the mean squared displacement. It would be interesting to further this narrow range of concentrations to determine the exact value where crowding occurs and the sharpness of the transition. The focus of our further research will be the problem of initial packing of the system and adding analysis of the influence of initial conditions on the network formation process. Additional work analyzing the complex modulus (G*) could tell us more about the viscoelastic properties of the system.
Similar behavior showing a threshold was noticed also in a recent experimental study, which analyzed more complex mucin solution (involving mucin (MUC2) oligomers, additional non-mucin proteins, and ions ); see Figure 2A in [51]. In their system, they noticed a threshold at a much lower concentration, between 5 and 10 g/L, when the system started to behave subdiffusively, showing in this way gel formation. The discrepancy was most probably caused by the differences in the system, mainly by the length of the mucin molecule: it is much longer, branched, and unfolded at low concentrations (see Figure 2C in [51]). Despite this, the vast similarity noticed indicated the usefulness of the molecular simulation technique in the studies within the polymer sciences area. Such a study can be used by the experimentalists to design molecular system with specific viscoelastic properties, which are important, e.g., in food and health sciences [52].
A novelty in our approach presented in the paper was the study of the nature of stochastic processes connected to the formation of a physically-cross-linked network in the mucin solution. Our calculations of sample MSD suggested that the CTRW model did not well describe the evolution of the number of HBO (inter- and intra-molecular) interactions in time, at least for the range of concentrations considered in this study. According to the above results, the nature of intermolecular HBO interaction dynamics also changed when we passed from to . For , an FBM process would seem to model the dynamics well, but for , an FLMS process appeared to be more appropriate. The same transition was evident from the intermolecular HBO dynamics when we passed from concentration to (a transition from an FLMS process to an FBM process). Overall, the statistical analysis of intramolecular HBO dynamics led us to conclude that FBM best described the process over the entire range of concentrations tested and demonstrated how alterations in bonding behavior (driven by increases in concentration) became evident in the statistical properties of the system.
Author Contributions
P.B. conceived of, designed, and performed the molecular dynamics simulations; N.K., M.K., and M.D. wrote scripts to prepare the data analysis; P.B., P.W., and N.K. analyzed the data; N.K., P.B., P.W., and S.Y. wrote the paper. All authors read, approved, and revised the final manuscript.
Funding
This research received no external funding.
Acknowledgments
This work is supported by the internal grants: BN-10/2019 of the Institute of Mathematics and Physics of the UTP University of Science and Technology in Bydgoszcz; No. BN-22/2019 of the Faculty of Telecommunications, Computer Science and Electrical Engineering of the UTP University of Science and Technology in Bydgoszcz.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AA | amino acid |
| AC | articular cartilage |
| CTRW | continuous-time random walk |
| FBM | fractional Brownian motion |
| FLMS | fractional Levy -stable motion |
| HA | hyaluronic acid |
| HP | hydrophobic-polar |
| HBO | hydrogen bond |
| MD | molecular dynamics |
| MSD | mean squared displacement |
| PL | phospholipids |
| PRG4 | lubricin |
References
- Wright, V.; Dowson, D. Lubrication and cartilage. J. Anat. 1976, 121, 107–118. [Google Scholar] [PubMed]
- Furey, M.J. Joint Lubrication. In The Biomedical Engineering Handbook; Bronzino, J.D., Ed.; CRC Press LLC: Boca Raton, FL, USA, 2000; Chapter 21; pp. 21-1–21-26. [Google Scholar]
- Katta, J.; Jin, Z.; Ingham, E.; Fisher, J. Biotribology of articular cartilage—A review of the recent advances. Med. Eng. Phys. 2000, 30, 1349–1363. [Google Scholar] [CrossRef] [PubMed]
- Tamer, T.M. Hyaluronan and synovial joint: Function, distribution and healing. Interdiscip. Toxicol. 2013, 6, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Dédinaité, A.; Claesson, P.M. Synergies in lubrication. Phys. Chem. Chem. Phys. 2017, 19, 23677–23689. [Google Scholar] [CrossRef] [PubMed]
- Dédinaité, A. Biomimetic lubrication. Soft Matter 2012, 8, 273–284. [Google Scholar] [CrossRef]
- Greene, G.W.; Banquy, X.; Lee, D.W.; Lowrey, D.D.; Yu, J.; Israelachvili, J. Adaptive mechanically controlled lubrication mechanism found in articular joints. Proc. Natl. Acad. Sci. USA 2011, 108, 5255–5259. [Google Scholar] [CrossRef] [PubMed]
- Gadomski, A.; Bełdowski, P.; Augé, W.K.; Hładyszowski, J.; Pawlak, Z.; Urbaniak, W. Toward a governing mechanism of nanoscale articular cartilage (physiologic) lubrication: Smoluchowski-type dynamics in amphiphile proton channels. Acta Phys. Pol. 2013, 44, 1801–1820. [Google Scholar] [CrossRef]
- Bełdowski, P.; Weber, P.; Dédinaité, A.; Claesson, P.; Gadomski, A. Physical crosslinking of hyaluronic acid in the presence of phospholipids in an aqueous nano-environment. Soft Matter 2018, 14, 8997–9004. [Google Scholar] [CrossRef]
- Bełdowski, P.; Kruszewska, N.; Yuvan, S.; Dendzik, Z.; Goudoulas, T.; Gadomski, A. Capstan-like mechanism in hyaluronan-phospholipid systems. Chem. Phys. Lipids 2018, 216, 17–24. [Google Scholar] [CrossRef]
- Bełdowski, P.; Weber, P.; Andrysiak, T.; Augé, W.K., II; De Leon, T.; Ledziński, D.; Gadomski, A. Anomalous behavior of hyaluronan crosslinking due to the presence of excess phospholipids in the articular cartilage system of osteoarthritis. Int. J. Mol. Sci. 2017, 18, 2779–2794. [Google Scholar]
- Siódmiak, J.; Bełdowski, P.; Augé, W.K.; Ledziński, D.; Śmigiel, S.; Gadomski, A. Molecular Dynamic Analysis of Hyaluronic Acid and Phospholipid Interaction in Tribological Surgical Adjuvant Design for Osteoarthritis. Molecules 2017, 22, 1436. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.P.; Abu-Lail, N.I.; Coles, J.M.; Guilak, F.; Jay, G.D.; Zauscher, S. Friction Force Microscopy of Lubricin and Hyaluronic Acid between Hydrophobic and Hydrophilic Surfaces. Soft Matter 2009, 5, 3438–3445. [Google Scholar] [CrossRef] [PubMed]
- Frerichs, F.T. Die Verdauung. In Handwörterbuch der Physiologie; Wagner, R., Ed.; Friedrich Vieweg und Sohn: Braunschweig, Germany, 1846; p. 463. [Google Scholar]
- Zappone, B.; Ruths, M.; Greene, G.W.; Jay, G.D.; Israelachvili, J.N. Adsorption, Lubrication, and Wear of Lubricin on Model Surfaces: Polymer Brush-Like Behavior of a Glycoprotein. Biophys. J. 2007, 92, 1693–1708. [Google Scholar] [CrossRef] [PubMed]
- Jay, G.D.; Waller, K.A. The biology of Lubricin: Near frictionless joint motion. Matrix Biol. 2014, 39, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Martin-Alarcon, L.; Schmidt, T.A. Rheological effects of macromolecular interactions in synovial fluid. Biorheology 2016, 53, 49–67. [Google Scholar] [CrossRef] [PubMed]
- Rimington, C. Synovial fluid mucin. Ann. Rheum. Dis. 1948, 8, 34–41. [Google Scholar] [CrossRef]
- Ropes, M.W.; Robertson, W.V.B.; Rossmeisl, E.C.; Perbody, R.B.; Bauer, W. Synovial fluid mucin. Acta Med. Scand. Suppl. 1947, 196, 700. [Google Scholar]
- Coles, J.M.; Chang, D.P.; Zauscher, S. Molecular mechanisms of aqueous boundary lubrication by mucinous glycoproteins. Curr. Opin. Colloid Interface Sci. 2010, 15, 406–416. [Google Scholar] [CrossRef]
- Pai, R.V.; Monpara, J.D.; Vavia, P.R. Exploring molecular dynamics simulation to predict binding with ocular mucin: An in silico approach for screening mucoadhesive materials for ocular retentive delivery systems. J. Control. Release 2019, 309, 190–202. [Google Scholar] [CrossRef]
- Lesitha Jeeva Kumari, J.; Jesu Jaya Sudan, R.; Sudandiradoss, C. Evaluation of peptide designing strategy against subunit reassociation in mucin 1: A steered molecular dynamics approach. PLoS ONE 2017, 12, 1–26. [Google Scholar] [CrossRef]
- Rhinehardt, K.L.; Srinivas, G.; Mohan, R.V. Molecular Dynamics Simulation Analysis of Anti-MUC1 Aptamer and Mucin 1 Peptide Binding. J. Phys. Chem. B 2015, 119, 6571–6583. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Jin, C.; Dédinaité, A.; Holgersson, J.; Karlsson, N.G.; Claesson, P.M. Influence of Glycosylation on Interfacial Properties of Recombinant Mucins: Adsorption, Surface Forces, and Friction. Langmuir 2017, 33, 4386–4395. [Google Scholar] [CrossRef] [PubMed]
- Abriata, L.A.; Spiga, E.; Peraro, M.D. Molecular Effects of Concentrated Solutes on Protein Hydration, Dynamics, and Electrostatics. Biophys. J. 2016, 111, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Cino, E.A.; Karttunen, M.; Choy, W.-Y. Effects of Molecular Crowding on the Dynamics of Intrinsically Disordered Proteins. PLoS ONE 2012, 7, e49876. [Google Scholar] [CrossRef] [PubMed]
- Echeverria, C.; Kapral, R. Molecular crowding and protein enzymatic dynamics. Phys. Chem. Chem. Phys. 2012, 14, 6755–6763. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.M.; Kim, Y.C.; Mittal, J. Protein Composition Determines the Effect of Crowding on the Properties of Disordered Proteins. Biophys. J. 2016, 111, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Harada, R.; Sugita, Y.; Feig, M. Protein crowding affects hydration structure and dynamics. J. Am. Chem. Soc. 2012, 134, 4842–4849. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Petelska, A.; Bełdowski, P.; Augé, W.K.; Casey, T.; Walczak, D.; Lemke, K.; Gadomski, A. Hyaluronic acid and phospholipid interactions useful for repaired articular cartilage surfaces—A mini review toward tribological surgical adjuvants. Colloid Polym. Sci. 2017, 295, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Macao, B.; Johansson, D.G.A.; Hansson, G.C.; Hard, T. Solution structure of the SEA domain of human mucin 1 (MUC1). Nat. Struct. Mol. Biol. 2006, 13, 71–76. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015, 36, 996–1007. [Google Scholar] [CrossRef]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T.; et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/EWater Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Santamaría-Holek, I.; Rubí, J.M.; Gadomski, A. Thermokinetic approach of single particles and clusters involving anomalous diffusion under viscoelastic response. J. Phys. Chem. B 2007, 111, 2293–2298. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bellour, M.; Skouri, M.; Munch, J.-P.; Hébraud, P. Brownian motion of particles embedded in a solution of giant micelles. Eur. Phys. J. E 2002, 8, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Metzler, R.; Klafter, J. The random walk’s guide to anomalous diffusion: A fractional dynamics approach. Phys. Rep. 2000, 339, 1–77. [Google Scholar] [CrossRef]
- Metzler, R.; Jeon, J.-H.; Cherstvy, A.G.; Barkai, E. Anomalous diffusion models and their properties: Non-stationarity, non-ergodicity, and ageing at the centenary of single particle tracking. Phys. Chem. Chem. Phys. 2014, 16, 24128–24164. [Google Scholar] [CrossRef] [PubMed]
- Dechant, A.; Lutz, E.; Kessler, D.A.; Barkai, E. Scaling Green-Kubo relation and application to three aging systems. Phys. Rev. X 2014, 4, 011022. [Google Scholar] [CrossRef]
- Kneller, G.R.; Baczynski, K.; Pasenkiewicz-Gierula, M. Communication: Consistent picture of lateral subdiffusion in lipid bilayers: Molecular dynamics simulation and exact results. J. Chem. Phys. 2011, 135, 141105. [Google Scholar] [CrossRef]
- Banks, D.S.; Fradin, C. Anomalous Diffusion of Proteins Due to Molecular Crowding. Biophys. J. 2005, 89, 2960–2971. [Google Scholar] [CrossRef]
- Magdziarz, M.; Klafter, J. Detecting origins of subdiffusion: P-variation test for confined systems. Phys. Rev. E 2010, 82, 011129. [Google Scholar] [CrossRef]
- Ślȩzak, J.; Weron, K. Revisited approach to statistical analysis of ionic current fluctuations. Acta Phys. Pol. B 2012, 43, 1215–1226. [Google Scholar] [CrossRef]
- Magdziarz, M.; Weron, A.; Burnecki, K.; Klafter, J. Fractional Brownian Motion Versus the Continuous-Time Random Walk: A Simple Test for Subdiffusive Dynamics. Phys. Rev. Lett. 2009, 103, 180602. [Google Scholar] [CrossRef] [PubMed]
- Burnecki, K.; Weron, A. Fractional Lévy stable motion can model subdiffusive dynamics. Phys. Rev. E 2010, 82, 021130. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, A.W.; Knowles, T.P.; Waudby, C.A.; Vendruscolo, M. Inversion of the Balance between Hydrophobic and Hydrogen Bonding Interactions in Protein Folding and Aggregation. PLoS Comput. Biol. 2011, 7, e1002169. [Google Scholar] [CrossRef] [PubMed]
- Michaud-Agrawal, N.; Denning, E.J.; Woolf, T.B.; Beckstein, O. MDAnalysis: A Toolkit for the Analysis of Molecular Dynamics Simulations. J. Comput. Chem. 2011, 32, 2319–2327. [Google Scholar] [CrossRef]
- Nawrocki, G.; Wang, P.H.; Yu, I.; Sugita, Y.; Feig, M. Slow-Down in Diffusion in Crowded Protein Solutions Correlates with Transient Cluster Formation. J. Phys. Chem. B 2017, 121, 11072–11084. [Google Scholar] [CrossRef]
- Weiss, M.; Elsner, M.; Kartberg, F.; Nilsson, T. Anomalous Subdiffusion Is a Measure for Cytoplasmic Crowding in Living Cells. Biophys. J. 2004, 87, 3518–3524. [Google Scholar] [CrossRef]
- Wong, I.Y.; Gardel, M.L.; Reichman, D.R.; Weeks, E.R.; Valentine, M.T.; Bausch, A.R.; Weitz, D.A. Anomalous Diffusion Probes Microstructure Dynamics of Entangled F-Actin Networks. Phys. Rev. Lett. 2004, 92, 178101. [Google Scholar] [CrossRef]
- Meldrum, O.W.; Yakubov, G.E.; Bonilla, M.R.; Deshmukh, O.; McGuckin, M.A.; Gidley, M.J. Mucin gel assembly is controlled by a collective action of non-mucin proteins, disulfide bridges, Ca2+-mediated links, and hydrogen bonding. Sci. Rep. 2018, 8, 5802. [Google Scholar] [CrossRef]
- Bansil, R.; Turner, B.S. Mucin structure, aggregation, physiological functions and biomedical applications. Curr. Opin. Colloid Interface Sci. 2006, 11, 164–170. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).