Dissecting Complex Traits Using Omics Data: A Review on the Linear Mixed Models and Their Application in GWAS
Abstract
:1. Introduction
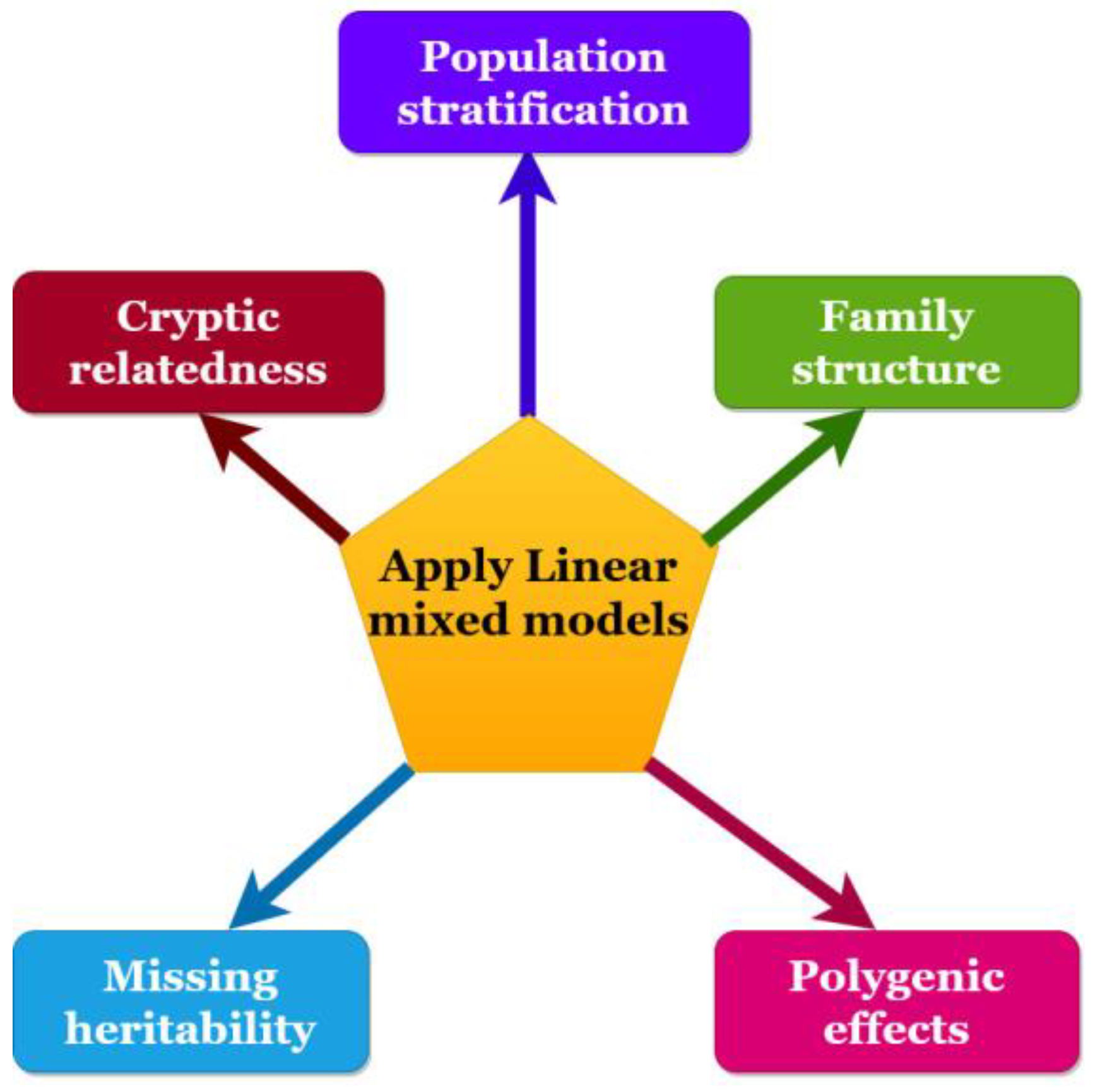
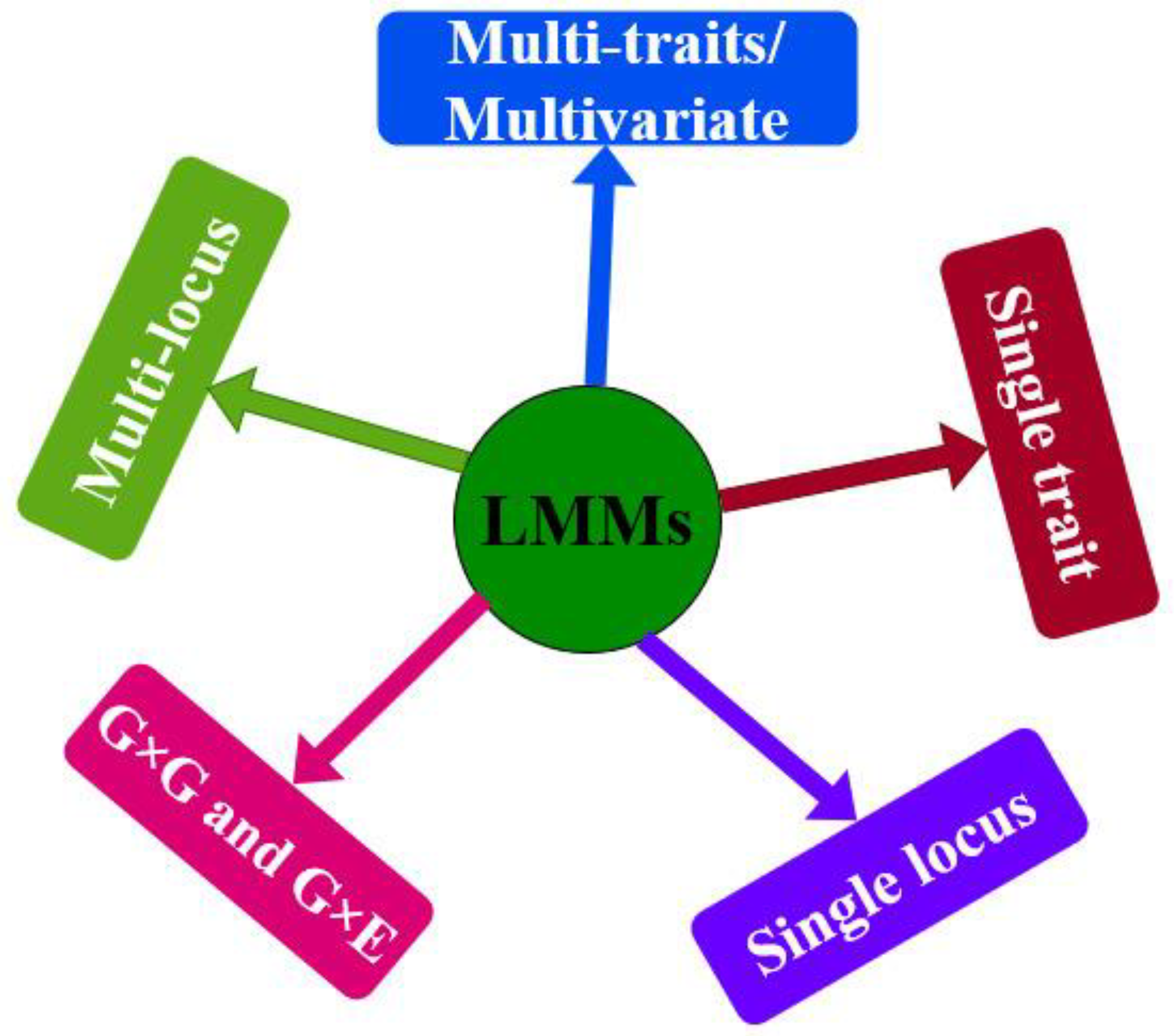
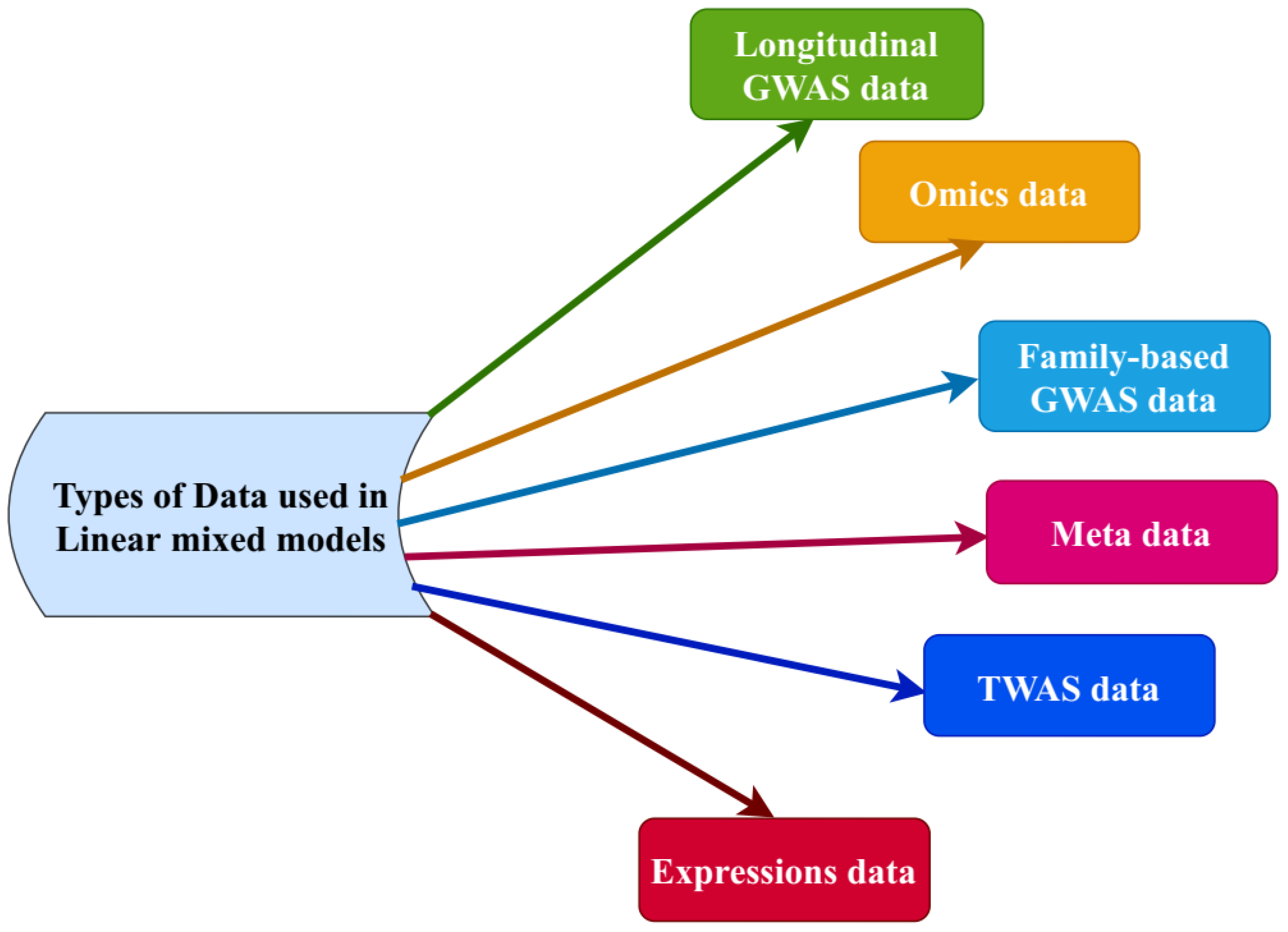
2. Linear Mixed Models
2.1. LMMs for Single Locus Analysis
2.2. LMMs for Multilocus Analysis
| Tool | Description | Link | Effect | Polygenic Background | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| a | d | α | a | d | α | ||||
| GRAMMAR | GRAMMAR is an alternate method to pedigree-founded QTL association mapping, which is quick and easy. It can handle millions of markers and is significantly faster than the evaluated genotype approach for association analysis. | ✓ | ✓ | [32] | |||||
| EMMA | EMMA is a fixed model edition of LMM used to control GWAS’s population structure and genetic relatedness. | http://mouse.cs.ucla.edu/emma/ (accessed on 20 October 2022) | ✓ | ✓ | [19] | ||||
| CMLM/P3D | CMLM (compressed MLM) diminished the sample size into groups using the clustering method, P3D (population parameters previously determined), which removes the re-calculation of variance components. The combined application of these two methods prominently abridged computing time and retained/enhanced statistical power. | https://www.maizegenetics.net/tassel (accessed on 20 October 2022) | ✓ | ✓ | [20] | ||||
| EMMAX | EMMAX is a variance component approach founded on the LMM method, which decreases the computational time for analysis of big GWAS data sets and is used for fixing sample structure in GWASs. | http://genetics.cs.ucla.edu/emmax/ (accessed on 20 October 2022) | ✓ | [21] | |||||
| FaST-LMM | FaST-LMM, an LMM-based method, used the subset of markers to manage the polygenic effect, resulting in accelerated speed and less required memory for GWAS. | https://github.com/fastlmm/FaST-LMM/ (accessed on 20 October 2022) | ✓ | ✓ | [27] | ||||
| FaST-LMM-Select | FaST-LMM-Select is a simple method that shows that wisely choosing a reduced number of SNPs consistently enhances power, expands standardization, and decreases computational time. | http://mscompbio.codeplex.com/ (accessed on 20 October 2022) | ✓ | ✓ | [37] | ||||
| GRAMMAR-Gamma | GRAMMAR-Gamma is an exceptionally fast variance component-based method that can be used for the massive human cohort in GWAS. It is established based on the analytical approximation within the context of the score test method. | http://www.genabel.org/ (accessed on 20 October 2022) | ✓ | ✓ | [38] | ||||
| WarpedLMM | WarpedLMM is a simplification of the ordinary LMM that estimates an ideal transformation from the monitored data for genetic study. Subsequently, this method’s power and accuracy will increase in GWAS. | http://github.com/pmbio/warpedLMM (accessed on 20 October 2022) | ✓ | ✓ | [25] | ||||
| ECMLM | ECMLM, enriched CMLM, uses various related algorithms and then selects the most effective mixture between the relationship algorithm and grouping algorithm resulting in increased power and can be applied for complex traits. | http://www.maizegenetics.net/gapit (accessed on 20 October 2022) | ✓ | ✓ | [36] | ||||
| SUPER | SUPER method intensely decreases the number of genetic markers utilized to define individual relationships, resulting in fast computation and increased statistical power despite utilizing the whole set of SNPs. | http://www.zzlab.net/GAPIT/ (accessed on 20 October 2022) | ✓ | ✓ | [39] | ||||
| RMLM | RMLM, random-SNP-effect MLM, treats the SNP-effect as random and uses Bonferroni correction to determine the p-value for significance. | ✓ | ✓ | [24] | |||||
| GMMAT | GMMAT is an R package for carrying out association tests using GLMMs in GWAS and sequencing association studies. | https://cran.r-project.org/web/packages/GMMAT/index.html (accessed on 20 October 2022) | ✓ | [40] | |||||
| LMM-Score | LMM-Score is a new method proposed to identify the genetic loci of complex traits. The simulation study showed that this method’s power increased and needed less computing time than the traditional LMM methods. | ✓ | ✓ | [1] | |||||
| Tool | Description | Link | Effect | Polygenic Background | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| a | d | α | a | d | α | ||||
| MLMM | MLMM, a multi-locus mixed-model, is an LMM-based method for complex traits, which is computationally effective and shows excellent performance regarding power and FDR compared with existing methods. | https://github.com/Gregor-Mendel-Institute/mlmm (accessed on 20 October 2022) | ✓ | ✓ | ✓ | ✓ | [42] | ||
| LMM-Lasso | LMM-Lasso links the benefits of LMM with Lasso regression, which is free of tuning parameters and efficiently corrects population structure. LMM-Lasso instantaneously detects potential causal variants and provides multi-marker-founded phenotype prediction from genotype. | https://github.com/BorgwardtLab/LMM-Lasso (accessed on 20 October 2022) | ✓ | ✓ | [45] | ||||
| Puma | PUMA, a standard model for utilizing a family of GWAS data, has been proposed to detect a weak association that the traditional methods cannot identify. It used a penalized maximum likelihood method utilizing a general linear model to take thousands of markers in a particular statistical method instantaneously. | http://mezeylab.cb.bscb.cornell.edu/Software.aspx (accessed on 20 October 2022) | ✓ | ✓ | [46] | ||||
| BOLT-LMM | BOLT-LMM is an efficient LMM that is computationally fast and gains power by demonstrating more accurate, non-infinitesimal genetic designs through a Bayesian admixture preceding marker impact. | http://www.hsph.harvard.edu/alkes-price/software/ (accessed on 20 October 2022) | ✓ | ✓ | [51] | ||||
| mrMLM | mrMLM (multi-locus RMLM) used markers selected from the RMLM method with a flexible selection criterion, and simulation results showed that the mrMLM is stronger in QTN discovery and more precise in QTN effect estimation than the RMLM and EMM. | https://cran.r-project.org/web/packages/mrMLM/index.html (accessed on 20 October 2022) | ✓ | ✓ | [24] | ||||
| FarmCPU | FarmCPU was formulated to control the confounding factors, significantly enhance statistical power, and decrease computing power. | https://www.zzlab.net/FarmCPU/ (accessed on 20 October 2022) | ✓ | ✓ | [44] | ||||
| FASTmrEMMA | FASTmrEMMA, a dominant multi-locus model widely used in QTN identification and model fit, has a lower bias in QTN effect calculation and needs a lower running time than existing single- and multi-locus methods. | https://cran.r-project.org/web/packages/mrMLM/index.html (accessed on 20 October 2022) | ✓ | ✓ | [26] | ||||
| StepLMM | StepLMM is a consistent, versatile, and computationally proficient method that can be applied to GS and GWAS. StepLMM has excellent efficiency in both GWAS and GS and is workable for agronomic breeding and human genomic studies. | ✓ | ✓ | [48] | |||||
| FASTmrMLM | FASTmrMLM is a multi-locus method, which is a fast and authentic algorithm in GWAS and assures superior statistical power, high accuracy of estimates, and low false-positive rate. | https://cran.r-project.org/web/packages/mrMLM/index.html (accessed on 20 October 2022) | ✓ | ✓ | [43] | ||||
| SGL-LMM | SGL-LMM, a multi-marker method, combined SGL and LMM for controlling confounding factors in GWAS. It includes the effect of multiple markers and integrates biological group info as preceding evidence in the model. | ✓ | ✓ | [50] | |||||
2.3. Multivariate/Multi-Traits LMMs
2.4. Linear Mixed Models in Epistasis (G × G) and Gene-Environment (G × E) Interaction
| Tool | Description | Link | Effect | Polygenic Background | Reference | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | d | α | ae | de | αe | a | d | α | ae | de | αe | ||||
| WOMBAT | WOMBAT is a software package that analyzes multiple quantitative traits using REML. It is well-fitted to investigate big GWAS data sets and assure both computational effectiveness and accurate boosting of the likelihood function. | http://didgeridoo.une.edu.au/km/wombat.php (accessed on 21 October 2022) | ✓ | ✓ | [70] | ||||||||||
| GEMMA | GEMMA (genome-wide efficient mixed-model association) is used to calculate precise values of test statistics and is constructed on EMMA software. It can handle three types of models such as univariate and multivariate LMM and Bayesian sparse LMM. | http://www.xzlab.org/software.html (accessed on 21 October 2022) | ✓ | ✓ | [23] | ||||||||||
| MTMM | MTMM is an LMM method for associated phenotypes considering both between and within-trait variance components concurrently for multiple traits for adjusting population stratification in GWAS. | https://github.com/arthurkorte/MTMM (accessed on 21 October 2022) | ✓ | ✓ | ✓ | [57] | |||||||||
| FaST-LMM-Set | FaST-LMM-Set, a novel approach for set tests, can handle the confounding problem. It is based on the LMM and uses two random effects: the first random effect is used to capture the set association signal, and the second is used to control confounding factors. | http://mscompbio.codeplex.com (accessed on 21 October 2022) | ✓ | ✓ | [74] | ||||||||||
| mtSet | Set tests are an effective approach for genome-wide association essaying among groups of genetic variants and a single quantitative trait. mtSet is an application of effective set test algorithms for combined analysis across multiple traits, which can explain confounding factors, including relatedness and single and multiple traits that can be used for GWAS. | https://github.com/PMBio/mtSet (accessed on 21 October 2022) | ✓ | ✓ | [75] | ||||||||||
| LIMIX | LIMIX, a simple and effective LMM-based software, can execute a wide range of genetic analyses for multi-trait using GWAS data. It can handle diverse functions, including single-locus and interaction association studies and variance decomposition studies with LMMs. | https://limix.readthedocs.io/en/s/ (accessed on 21 October 2022) | ✓ | ✓ | ✓ | [73] | |||||||||
| BOLT-REML | BOLT-REML uses the RELM approach to estimate the variance parameters for models, taking multiple variance components and traits that solve computational problems that make it impossible to analyze large data sets. | https://www.hsph.harvard.edu/alkes-price/software/ (accessed on 21 October 2022) | ✓ | ✓ | [82] | ||||||||||
| mvLMM | mvLMM (matrix-variate linear mixed model) is a multiple-trait association mapping approach, which needs less computational time to execute inference in a multi-trait model by using data transformation and a ten-fold computational speed increase for large cohort analysis. | http://genetics.cs.ucla.edu/mvLMM (accessed on 21 October 2022) | ✓ | ✓ | [67] | ||||||||||
| GAMMA | GAMMA, a multivariate method, can coincidentally analyze numerous phenotypes and adjust for population structure. GAMMA is a more advanced method than others, which either cannot find true effects or have a higher false positive rate. | http://genetics.cs.ucla.edu/GAMMA/ (accessed on 21 October 2022) | ✓ | ✓ | [71] | ||||||||||
| LiMMBo | LiMMBo is a very easy and flexible method based on LMMs for multi-dimensional GWAS data with hundreds of phenotypes. It combines LMMs and bootstrapping for estimates of large trait covariance matrices. | https://github.com/HannahVMeyer/limmbo (accessed on 21 October 2022) | ✓ | ✓ | [85] | ||||||||||
| SGL-LMM | SGL-LMM combined SGL (sparse group lasso) and LMM for multivariate GWAS analysis. Results showed that the SGL-LMM improved the power to detect marker association in various settings. | ✓ | ✓ | [50] | |||||||||||
| SMMAT | SMMAT is a computationally effective variant test for continuous and binary traits. SMMAT can be used in structured and related samples with various possible origins of correlations from large-scale whole-genome sequencing studies. | https://github.com/hanchenphd/GMMAT (accessed on 21 October 2022) | ✓ | ✓ | [79] | ||||||||||
2.5. Linear Mixed Models in Transcriptome-Wide Association Studies (TWAS) and Longitudinal GWAS
| Tool | Description | Link | Effect | Polygenic Background | Reference | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | d | α | e | aa/aae/ae | ad/ ade | da/ dae /de | dd/dde | qqe | a | d | α | e | aa/ ae | ad | da/ de | dd | ||||
| FAM-MDR | FAM-MDR, a novel family-based and compromising epistasis finding exploration method, provides better results than the existing method PGMDR (Pedigree-based Generalized MDR) in terms of power, and it sufficiently contracts with numerous testing in epistasis tests. | http://www.statgen.be/ (accessed on 21 October 2022) | ✓ | ✓ | [97] | |||||||||||||||
| QTXNetwork | QTXNetwork is an LMM-based software that uses GPU to analyze diverse genetic effects concurrently. It can be used for calculating main genetic effects, G × G and G × E interaction effects on big omics data for complex traits and for calculating the heritability of specific genetic component effects. | http://ibi.zju.edu.cn/software/QTXNetwork (accessed on 21 October 2022) | ✓ | ✓ | ✓ | ✓ | ✓/✓ | ✓/✓ | ✓/✓ | ✓/✓ | ✓ | [96] | ||||||||
| iSet | The interaction set test, iSet, is an LMMs-based method that explains the polygenic effects and has more power to detect the interaction between environment and variants. | https://github.com/limix/limix (accessed on 21 October 2022) | ✓ | ✓ | [98] | |||||||||||||||
| REMMA | REMMA has been proposed to overcome the computational efficiency problem for handling epistatic effects in GWAS. It is more computationally efficient, has a lower type I error rate, and has higher QTL discovery power than other existing models. | https://github.com/chaoning/REMMA (accessed on 21 October 2022) | ✓ | ✓ | [100] | |||||||||||||||
| GxEMM | GxEMM is an integrative mixed model for polygenic interactions to disseminate the total effect of small G × E effects throughout the genome. | https://github.com/andywdahl/gxemm (accessed on 21 October 2022) | ✓ | ✕/✓ | ✕/✓ | [104] | ||||||||||||||
| StructLMM | StructLMM (structured linear mixed model) is a computationally effective method to detect and illustrate loci that relate to one or more environments. Hundreds of environmental variables can be used to study interactions using this model. | https://mybinder.org/v2/gh/limix/limix-tutorials/master?filepath=struct-lmm.ipynb (accessed on 21 October 2022) | ✓ | ✕/✓ | ✓ | [103] | ||||||||||||||
| Grid-LMM | Grid-LMM is a scalable algorithm for frequently suiting complex LMMs that can include heterogeneity, including additive and dominance genetic variance, uneven distribution of traits, and G × E interactions. | https://github.com/deruncie/GridLMM (accessed on 21 October 2022) | ✓ | ✕/✓ | [107] | |||||||||||||||
| FFselect | FFselect is an LMM based advanced method for the analysis of GWAS data incorporating shared environmental effects in the model. This method demonstrated enhanced power, controlled FDR (false discovery rate), and simultaneously adapted to environmental factors to enhance GWAS’s effectiveness. | https://github.com/NicholSchultz/FFselect (accessed on 21 October 2022) | ✓ | ✓ | [108] | |||||||||||||||
| REMMAX | REMMAX, REMMA eXpedited, is a proficient method for GWAS by adjusting numerous polygenic effects, and the time complexity is almost linear with the population size. | https://github.com/chaoning/GMAT (accessed on 21 October 2022) | ✓ | ✓ | ✓ | ✓ | ✓ | Polygenic background with normal distribution | [101] | |||||||||||
| 3VmrMLM | 3VmrMLM, a three-variance-component mixed model, was incorporated with the mrMLM method. It has more power and accuracy to discover all kinds of loci and give an unbiased estimation of their effects. | ✓ | ✓ | ✓ | ✓/✕ /✓ | ✓/✕ | ✓/✕/✓ | ✓/✕ | ✓ | ✓ | ✓ | ✓/✓ | ✓ | ✓/✓ | ✓ | [110] | ||||
2.6. LMM-Based Packages in GWAS
| Tool | Description | Link | Effect | Polygenic Background | Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| a | d | α | e | a | d | α | e | ||||
| SMART | SMART is based on the extension of LMM that utilizes various corresponding annotations matched to diverse approaches and algorithms. SMART can be applied to construct useful SNP set experiments and decide novel trait-tissue related and useful annotations concerning trait-tissue associations. | http://www.xzlab.org/software.html (accessed on 21 October 2022) | ✓ | ✓ | [115] | ||||||
| LSMM | LSMM incorporates both genic and cell-type targeted functional annotations in GWAS. It uses the EM algorithm for parameter estimations and statistical implications. The power increased compared with current methods to detect the risk variants (SNPs) and cell-type targeted functional observations by the LSMM approach. | https://github.com/mingjingsi/LSMM (accessed on 21 October 2022) | ✓ | ✓ | [114] | ||||||
| CoMM | CoMM, a collaborative mixed model, is to inquire about the recurring role of linked variants in complex traits. CoMM is computationally fast and statistically effective in analyzing genetic contributions to complex traits by maximizing information in transcriptome data. | https://github.com/gordonliu810822/CoMM (accessed on 21 October 2022) | ✓ | ✓ | [111] | ||||||
| CoMM-S2 | CoMM-S2 uses summary statistics GWAS data to study the mechanism of genetic variants. This method uses similar approaches to CoMM, except for summary statistics data and simulation and real data analysis showed that the efficiency of CoMM-S2 is equivalent to CoMM and CoMM-S2 applied in the CoMM package. | https://github.com/gordonliu810822/CoMM (accessed on 21 October 2022) | ✓ | ✓ | [116] | ||||||
| KIN-LMEM | KIN-LMEM is a mixed-model-based approach for executing association mapping, which utilizes numerous phenotype measurements for each individual. | http://genetics.cs.ucla.edu/longGWAS/ (accessed on 21 October 2022) | ✓ | ✓ | [120] | ||||||
| Tool | Description | Link | Effect | Polygenic Background | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| a (aa/ad) | d (dd/da) | α | a | d | α | ||||
| DMU | DMU is a broadly employed package for analyzing MMM in quantitative genetics and genomics. It applies advanced tools to calculate variance components and fixed effects and predict random effects. | http://dmu.agrsci.dk (accessed on 21 October 2022) | ✓ | [124] | |||||
| ASREML | ASReml utilizes LMMs to analyze big and complex data, and many variance models for random effects are available in the LMM in the ASReml package. | https://www.vsni.co.uk/ (accessed on 21 October 2022) | ✓ | ✓ | [127] | ||||
| GenABEL | GenABEL is an R package GWAS, which applies an efficient GWA data storehouse and dealing, quick processes for verifying the quality of genetic data, statistical analysis, and representation of GWAS data. | https://mran.microsoft.com/snapshot/2018-05-12/web/packages/GenABEL/index.html (accessed on 21 October 2022) | [125] | ||||||
| lrgpr | lrgpr is very computationally powerful and efficient for analyzing big GWAS and NGS datasets. It provides a collaborative model conforming to assist exploratory data analysis from the perspective of the LMM. | http://lrgpr.r-forge.r-project.org/ (accessed on 21 October 2022) | ✓ | ✓ | [126] | ||||
| lme4qtl | lme4qtl, an extension of lme4, adds novel models for genetic studies and extends a flexible model for settings with numerous levels of connection and would be useful while covariance matrices are sparse. | https://github.com/variani/lme4qtl (accessed on 21 October 2022) | ✓ | ✓ | [128] | ||||
| Sci-LMM | SciLMM is a systematic model for analyzing the ancestries of millions of individuals. SciLMM uses LMM approaches in the presence of the dependencies encoded by matrices constructed by the model. This tool is adaptable, can be elongated in various ways, and is valuable for GWAS. | https://github.com/TalShor/SciLMM (accessed on 21 October 2022) | ✓ (✓/✓) | ✓ (✓/✓) | [129] | ||||
| Single-RunKing | Single-RunKing is a useful R package to speed up the computation in GWAS by using LMMs. It uses R/fastLmPure to numerically understand the genetic effects of screened SNPs and concentrate on significant SNPs found by the EMMAX algorithm. | https://rdrr.io/cran/RcppBlaze/man/fastLmPure.html (accessed on 21 October 2022) | ✓ | ✓ | [130] | ||||
| LiMMBo | LiMMBo is a very easy and flexible method based on LMMs for multi-dimensional GWAS data with hundreds of phenotypes. It combines LMMs and bootstrapping for estimates of large trait covariance matrices. | https://github.com/HannahVMeyer/limmbo (accessed on 21 October 2022) | ✓ | ✓ | [85] | ||||
| SGL-LMM | SGL-LMM combined SGL (sparse group lasso) and LMM for multivariate GWAS analysis, with improved power to detect marker association in various settings. | https://rdrr.io/cran/RcppBlaze/man/fastLmPure.html (accessed on 21 October 2022) | ✓ | ✓ | [50] | ||||
| SMMAT | SMMAT is a computationally effective variant test for continuous and binary traits. SMMAT can be used in structured and related samples with various possible origins of correlations from large-scale whole-genome sequencing studies. | https://rdrr.io/github/hanchenphd/GMMAT/man/SMMAT.html (accessed on 21 October 2022) | ✓ | ✓ | [79] | ||||
2.7. Web-Based Software/Server Tools Using Linear Mixed Models
| Tool | Description | Link | Effect | Polygenic Background | Reference | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | d | α | e | ae | aa (aae) | ad | da | dd | a | d | α | e | ||||
| QxPak | Qxpak is a mixed-model-based software that allows a very versatile tool for QTL mapping in various populations and can be used for multi-trait and multiQTL analysis in genomic studies. | ✓ | ✓ | [139] | ||||||||||||
| TASSEL | TASSEL is software that measures trait associations, evolutionary patterns, and LD calculation. Database browsing and importing are assisted by incorporated middleware. | https://www.maizegenetics.net/tassel (accessed on 21 October 2022) | ✓ | ✓ | [140] | |||||||||||
| QTLNetwork | QTLNetwork is software for mapping and displaying the genetic structure underlying complex traits for observational populations that came from a cross relating to dual inbred lines. QTLNetwork provides a GUI facility and can deal with data from diverse forms of observational populations. | http://ibi.zju.edu.cn/software/qtlnetwork (accessed on 21 October 2022) | ✓ | ✓ | ✓ | ✓ (✓) | [141] | |||||||||
| GCTA | GCTA, genome-wide complex trait analysis, is a widely used software incorporating many methods for analyzing complex traits using GWAS. | https://cnsgenomics.com/software/gcta/ (accessed on 21 October 2022) | ✓ | ✓ | [69] | |||||||||||
| GAPIT | GAPIT applies promoted statistical approaches, including the CMLM and CMLM-based CMLM-founded genomic prediction. | https://www.maizegenetics.net/GAPIT (accessed on 21 October 2022) | Several methods including EMMA, P3D/CMLM, ECMLM, MLMM, SUPER and FarmCPU implemented in GAPIT. See the effect and polygenic background in the respective methods tables. | [143] | ||||||||||||
| MASTOR | MASTOR is a mixed model-based approach for analyzing GWAS data using the score test for genetic association with a quantitative trait, where sample individuals are related. MASTOR attains high power by using full kinship information to integrate partly missing data in the investigation when adjusting for dependence. | http://www.stat.uchicago.edu/%7Emcpeek/software/MASTOR/index.html (accessed on 21 October 2022) | ✓ | ✓ | [144] | |||||||||||
| MMM | MMM, a software package, used LMM with one random effect whose covariance design can be easily assigned by the users for GWAS. It can handle more than 20,000 individuals and 500,000 genetic variants and use other data. | ✓ | ✓ | [65] | ||||||||||||
| OmicABEL | OmicABEL is freely accessible software that carries out fast mixed-model-based GWAS. It can handle single and multi-trait and uses CLAK-C HOL to explore significant complex traits, and CLAK-E IG is used for investigating the genomic control of various omics in GWAS. | http://www.genabel.org/packages/OmicABEL (accessed on 21 October 2022) | ✓ | [151] | ||||||||||||
| GAPIT Version 2 | GAPIT version 2 included some powerful LMMs, including FaST-LMM-Select, ECMLM, and SUPER. | https://www.zzlab.net/GAPIT/ (accessed on 21 October 2022) | GAPIT version 2 is an updated version of GAPIT. Several methods including FaST-LMM and FaST-LMM-Select along with others methods mentioned in the GAPIT implemented in GAPIT version 2. See the effect and polygenic background in the respective methods tables. | [34] | ||||||||||||
| PEPIS | PEPIS is a web-based tool for studying polygenic epistatic effects founded on an LMM employed to predict the functioning of hybrid rice. PEPIS was devotedly formulated to calculate epistatic effects and will help tackle the obstacles in genetic epistasis study. | http://bioinfo.noble.org/PolyGenic_QTL/ (accessed on 21 October 2022) | ✓ | ✓ | ✓(✕) | ✓ | ✓ | [146] | ||||||||
| MTG2 | MTG2 is an LMM-based software for analyzing complex traits using GWAS data. It incorporated AI algorithms and eigendecomposition, which is considerably faster than other REML methods. | https://sites.google.com/site/honglee0707/mtg2 (accessed on 21 October 2022) | ✓ | ✓ | [131] | |||||||||||
| PopPAnTe | PopPAnTe, an easy Java program based on the accurate LMM, allows a flexible permutation method to end the propagation of arbitrarily permuted samples. It could be used for the exact relationship between significant quantitative response and independent variables in family-based GWAS data. | https://sites.google.com/site/populationgenomics/poppante (accessed on 21 October 2022) | ✓ | ✓ | [145] | |||||||||||
| GCTB | GCTB is a software tool that includes a class of Bayesian LMMs for complex trait studies applying genome-wide SNPs for dissecting complex traits. It offers users many functions to reveal necessary signatures of evolution. | https://cnsgenomics.com/software/gctb/ (accessed on 21 October 2022) | ✓ | [148] | ||||||||||||
| OSCA | OSCA, a multipurpose software tool, manages omic data produced from high-throughput trials in big cohorts and helps analyze complex traits utilizing omic data. | https://cnsgenomics.com/software/osca/ (accessed on 21 October 2022) | ✓ | [149] | ||||||||||||
| fastGWA | fastGWA, an LMM model, is proposed for controlling population structure by PCA and relatedness by sparse GRM (genetic relationship matrix) for analyzing big data such as biobank-scale data in GWAS. | http://cnsgenomics.com/software/gcta/#fastGWA (accessed on 21 October 2022) | ✓ | ✓ | [150] | |||||||||||
3. Advantages and Weaknesses of Linear Mixed Models Used in GWAS
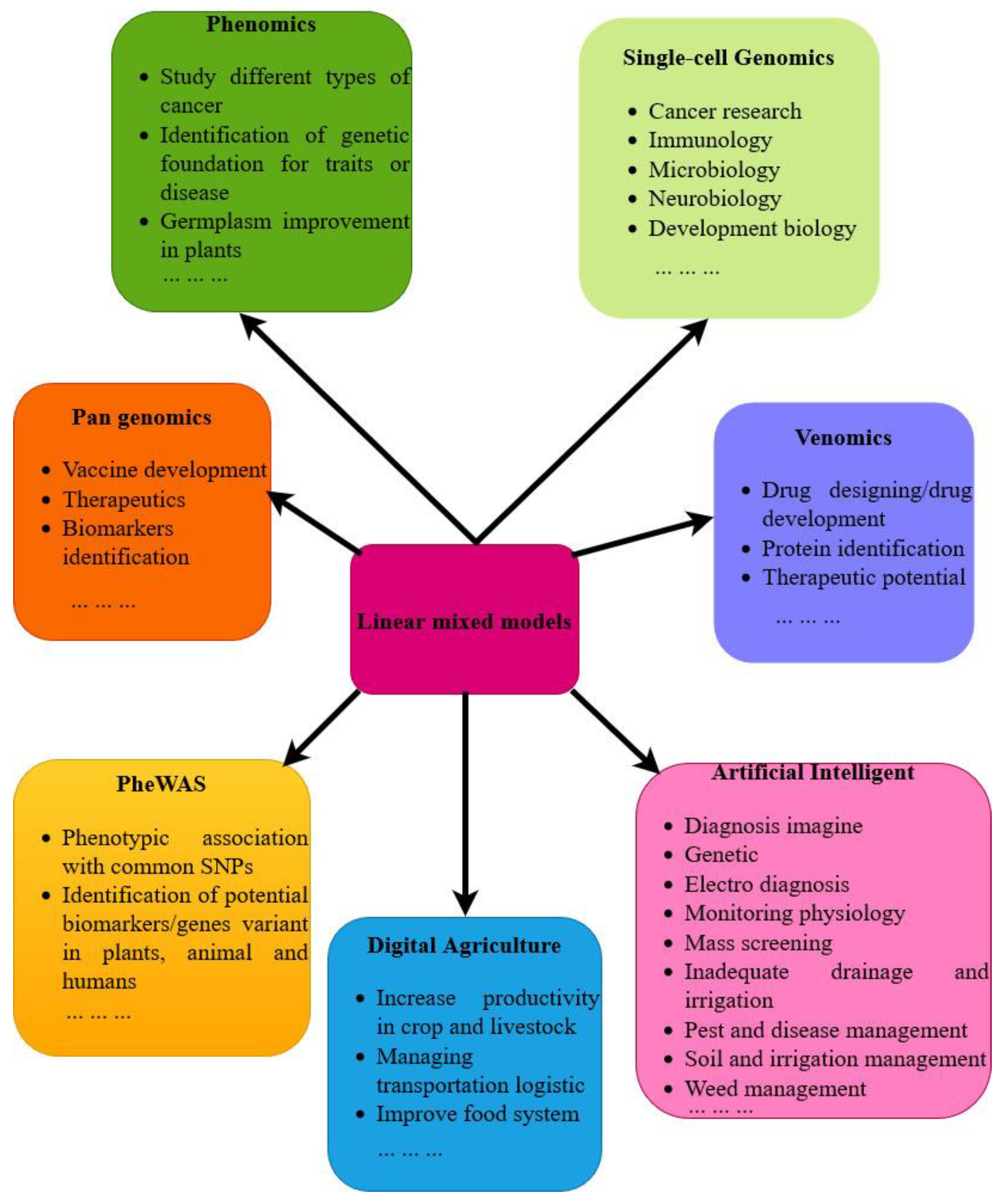
4. Future Perspective
5. Conclusions
Author Contributions
Funding
Institution Review Board Statements
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, T.; Wei, J.; Wang, X.; Miao, J.; Xu, L.; Zhang, L.; Gao, X.; Chen, Y.; Li, J.; Gao, H. A rapid and efficient linear mixed model approach using the score test and its application to GWAS. Livest. Sci. 2019, 220, 37–45. [Google Scholar] [CrossRef]
- Wang, Q.; Tang, J.; Han, B.; Huang, X. Advances in genome-wide association studies of complex traits in rice. TAG. Theor. Appl. Genet. Theor. Und Angew. Genet. 2020, 133, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Altshuler, D.; Daly, M.J.; Lander, E.S. Genetic mapping in human disease. Science 2008, 322, 881–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manolio, T.A. Cohort studies and the genetics of complex disease. Nat. Genet. 2009, 41, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Atwell, S.; Huang, Y.S.; Vilhjalmsson, B.J.; Willems, G.; Horton, M.; Li, Y.; Meng, D.; Platt, A.; Tarone, A.M.; Hu, T.T.; et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 2010, 465, 627–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, Y.; Ma, Y.; Zhou, Y.; Zhang, H.; Duan, L.; Chen, H.; Zeng, J.; Zhou, Q.; Wang, S.; Gu, W.; et al. Plant science. Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 2014, 346, 1084–1088. [Google Scholar] [CrossRef]
- Yang, W.; Guo, Z.; Huang, C.; Duan, L.; Chen, G.; Jiang, N.; Fang, W.; Feng, H.; Xie, W.; Lian, X.; et al. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 2014, 5, 5087. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Li, Y.X.; Shi, Y.S.; Song, Y.C.; Zhang, D.F.; Li, C.H.; Buckler, E.S.; Li, Y.; Zhang, Z.W.; Wang, T.Y. Joint-linkage mapping and GWAS reveal extensive genetic loci that regulate male inflorescence size in maize. Plant Biotechnol. J. 2016, 14, 1551–1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.; Zhou, G.F.; Shabala, S.; Chen, Z.H.; Cai, S.G.; Li, C.D.; Zhou, M.X. Genome-Wide Association Study Reveals a New QTL for Salinity Tolerance in Barley (Hordeum vulgare L.). Front. Plant Sci. 2016, 7, 946. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Chen, D.; Alqudah, A.M.; Roder, M.S.; Ganal, M.W.; Schnurbusch, T. Genome-wide association analyses of 54 traits identified multiple loci for the determination of floret fertility in wheat. New Phytol. 2017, 214, 257–270. [Google Scholar] [CrossRef]
- Matsuzaki, H.; Dong, S.; Loi, H.; Di, X.; Liu, G.; Hubbell, E.; Law, J.; Berntsen, T.; Chadha, M.; Hui, H.; et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat. Methods 2004, 1, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Gunderson, K.L.; Steemers, F.J.; Lee, G.; Mendoza, L.G.; Chee, M.S. A genome-wide scalable SNP genotyping assay using microarray technology. Nat. Genet. 2005, 37, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Altshuler, D.; Brooks, L.D.; Chakravarti, A.; Collins, F.S.; Daly, M.J.; Donnelly, P.; Gibbs, R.A.; Belmont, J.W.; Boudreau, A.; Leal, S.M.; et al. A haplotype map of the human genome. Nature 2005, 437, 1299–1320. [Google Scholar]
- de Bakker, P.I.; Yelensky, R.; Pe’er, I.; Gabriel, S.B.; Daly, M.J.; Altshuler, D. Efficiency and power in genetic association studies. Nat. Genet. 2005, 37, 1217–1223. [Google Scholar] [CrossRef]
- Hardy, J.; Singleton, A. Genomewide association studies and human disease. N. Engl. J. Med. 2009, 360, 1759–1768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, J.C.; Kiss, R.S.; Pertsemlidis, A.; Marcel, Y.L.; McPherson, R.; Hobbs, H.H. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 2004, 305, 869–872. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.M.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Kang, H.M.; Zaitlen, N.A.; Wade, C.M.; Kirby, A.; Heckerman, D.; Daly, M.J.; Eskin, E. Efficient control of population structure in model organism association mapping. Genetics 2008, 178, 1709–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.-Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Zaitlen, N.A.; Reich, D.; Patterson, N. New approaches to population stratification in genome-wide association studies. Nat. Rev. Genet. 2010, 11, 459–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.B.; Feng, J.Y.; Ren, W.L.; Huang, B.; Zhou, L.; Wen, Y.J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.M. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 2016, 6, 19444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fusi, N.; Lippert, C.; Lawrence, N.D.; Stegle, O. Warped linear mixed models for the genetic analysis of transformed phenotypes. Nat. Commun. 2014, 5, 4890. [Google Scholar] [CrossRef] [Green Version]
- Wen, Y.J.; Zhang, H.; Ni, Y.L.; Huang, B.; Zhang, J.; Feng, J.Y.; Wang, S.B.; Dunwell, J.M.; Zhang, Y.M.; Wu, R. Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Brief. Bioinform. 2017, 18, 906. [Google Scholar] [CrossRef] [Green Version]
- Lippert, C.; Listgarten, J.; Liu, Y.; Kadie, C.M.; Davidson, R.I.; Heckerman, D. FaST linear mixed models for genome-wide association studies. Nat. Methods 2011, 8, 833–835. [Google Scholar] [CrossRef]
- Listgarten, J.; Lippert, C.; Kadie, C.M.; Davidson, R.I.; Eskin, E.; Heckerman, D. Improved linear mixed models for genome-wide association studies. Nat. Methods 2012, 9, 525–526. [Google Scholar] [CrossRef] [Green Version]
- Alamin, M.; Zhu, J.; Lou, X.; Xu, H. Dissecting Impacts of Nutrition on Epistasis and Ethnicity-Specific Effects of Calibrated Factor VIII Level in the Multiethnic Study of Atherosclerosis. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Harrison, X.A.; Donaldson, L.; Correa-Cano, M.E.; Evans, J.; Fisher, D.N.; Goodwin, C.E.D.; Robinson, B.S.; Hodgson, D.J.; Inger, R. A brief introduction to mixed effects modelling and multi-model inference in ecology. PeerJ 2018, 6, e4794. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.M.; Jia, Z.; Dunwell, J.M. Editorial: The Applications of New Multi-Locus GWAS Methodologies in the Genetic Dissection of Complex Traits. Front. Plant Sci. 2019, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Aulchenko, Y.S.; de Koning, D.J.; Haley, C. Genomewide rapid association using mixed model and regression: A fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics 2007, 177, 577–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.M.; Mao, Y.; Xie, C.; Smith, H.; Luo, L.; Xu, S. Mapping quantitative trait loci using naturally occurring genetic variance among commercial inbred lines of maize (Zea mays L.). Genetics 2005, 169, 2267–2275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Liu, X.; Wang, J.; Li, M.; Wang, Q.; Tian, F.; Su, Z.; Pan, Y.; Liu, D.; Lipka, A.E.; et al. GAPIT Version 2: An Enhanced Integrated Tool for Genomic Association and Prediction. Plant Genome 2016, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Xu, S. An expectation-maximization algorithm for the Lasso estimation of quantitative trait locus effects. Heredity 2010, 105, 483–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Liu, X.; Bradbury, P.; Yu, J.; Zhang, Y.M.; Todhunter, R.J.; Buckler, E.S.; Zhang, Z. Enrichment of statistical power for genome-wide association studies. BMC Biol. 2014, 12, 73. [Google Scholar] [CrossRef] [Green Version]
- Listgarten, J.; Lippert, C.; Heckerman, D. FaST-LMM-Select for addressing confounding from spatial structure and rare variants. Nat. Genet. 2013, 45, 470–471. [Google Scholar] [CrossRef]
- Svishcheva, G.R.; Axenovich, T.I.; Belonogova, N.M.; van Duijn, C.M.; Aulchenko, Y.S. Rapid variance components-based method for whole-genome association analysis. Nat. Genet. 2012, 44, 1166–1170. [Google Scholar] [CrossRef]
- Wang, Q.; Tian, F.; Pan, Y.; Buckler, E.S.; Zhang, Z. A SUPER powerful method for genome wide association study. PLoS ONE 2014, 9, e107684. [Google Scholar] [CrossRef]
- Chen, H.; Wang, C.; Conomos, M.P.; Stilp, A.M.; Li, Z.; Sofer, T.; Szpiro, A.A.; Chen, W.; Brehm, J.M.; Celedon, J.C.; et al. Control for Population Structure and Relatedness for Binary Traits in Genetic Association Studies via Logistic Mixed Models. Am. J. Hum. Genet. 2016, 98, 653–666. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Liu, H.; Chen, J.; Shi, T.; Zhang, C.; Sun, D.; He, Z.; Hao, Y.; Chen, W. Genome-Wide Association Studies of Free Amino Acid Levels by Six Multi-Locus Models in Bread Wheat. Front. Plant Sci. 2018, 9, 1196. [Google Scholar] [CrossRef]
- Segura, V.; Vilhjálmsson, B.J.; Platt, A.; Korte, A.; Seren, Ü.; Long, Q.; Nordborg, M. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 2012, 44, 825. [Google Scholar] [CrossRef] [Green Version]
- Tamba, C.L.; Zhang, Y.-M. A fast mrMLM algorithm for multi-locus genome-wide association studies. bioRxiv 2018, 341784. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef]
- Rakitsch, B.; Lippert, C.; Stegle, O.; Borgwardt, K. A Lasso multi-marker mixed model for association mapping with population structure correction. Bioinformatics 2013, 29, 206–214. [Google Scholar] [CrossRef]
- Hoffman, G.E.; Logsdon, B.A.; Mezey, J.G. PUMA: A unified framework for penalized multiple regression analysis of GWAS data. PLoS Comput. Biol. 2013, 9, e1003101. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Zhang, Y.W.; Xiang, Y.; Liu, M.H.; Zhang, Y.M. IIIVmrMLM: The R and C++ tools associated with 3VmrMLM, a comprehensive GWAS method for dissecting quantitative traits. Mol. Plant 2022, 15, 1251–1253. [Google Scholar] [CrossRef]
- Li, H.; Su, G.; Jiang, L.; Bao, Z. An efficient unified model for genome-wide association studies and genomic selection. Genet. Sel. Evol. 2017, 49, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.H.; Chen, Z.H. Extended Bayesian information criteria for model selection with large model spaces. Biometrika 2008, 95, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Wu, C.; Guo, M.; Zou, Q.; Liu, X.; Keinan, A. Combining Sparse Group Lasso and Linear Mixed Model Improves Power to Detect Genetic Variants Underlying Quantitative Traits. Front. Genet. 2019, 10, 271. [Google Scholar] [CrossRef]
- Loh, P.R.; Tucker, G.; Bulik-Sullivan, B.K.; Vilhjalmsson, B.J.; Finucane, H.K.; Salem, R.M.; Chasman, D.I.; Ridker, P.M.; Neale, B.M.; Berger, B.; et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015, 47, 284–290. [Google Scholar] [CrossRef]
- Jiang, C.; Zeng, Z.B. Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 1995, 140, 1111–1127. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.A.; Purcell, S.M. A multivariate test of association. Bioinformatics 2009, 25, 132–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Pei, Y.F.; Li, J.; Papasian, C.J.; Deng, H.W. Univariate/Multivariate Genome-Wide Association Scans Using Data from Families and Unrelated Samples. PLoS ONE 2009, 4, e6502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knott, S.A.; Haley, C.S. Multitrait least squares for quantitative trait loci detection. Genetics 2000, 156, 899–911. [Google Scholar] [CrossRef]
- Amos, C.I. Robust variance-components approach for assessing genetic linkage in pedigrees. Am. J. Hum. Genet. 1994, 54, 535–543. [Google Scholar]
- Korte, A.; Vilhjalmsson, B.J.; Segura, V.; Platt, A.; Long, Q.; Nordborg, M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat. Genet. 2012, 44, 1066–1071. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.H.; Yang, J.; Goddard, M.E.; Visscher, P.M.; Wray, N.R. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics 2012, 28, 2540–2542. [Google Scholar] [CrossRef] [Green Version]
- Vattikuti, S.; Guo, J.; Chow, C.C. Heritability and genetic correlations explained by common SNPs for metabolic syndrome traits. PLoS Genet. 2012, 8, e1002637. [Google Scholar] [CrossRef]
- Kruuk, L.E.B. Estimating genetic parameters in natural populations using the “animal model”. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2004, 359, 873–890. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Sohn, K.A.; Xing, E.P. A multivariate regression approach to association analysis of a quantitative trait network. Bioinformatics 2009, 25, i204–i212. [Google Scholar] [CrossRef] [Green Version]
- O’Reilly, P.F.; Hoggart, C.J.; Pomyen, Y.; Calboli, F.C.F.; Elliott, P.; Jarvelin, M.-R.; Coin, L.J.M. MultiPhen: Joint model of multiple phenotypes can increase discovery in GWAS. PLoS ONE 2012, 7, e34861. [Google Scholar] [CrossRef]
- Stephens, M. A unified framework for association analysis with multiple related phenotypes. PLoS ONE 2013, 8, e65245. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.M.; Abecasis, G.R. Family-based association tests for genomewide association scans. Am. J. Hum. Genet. 2007, 81, 913–926. [Google Scholar] [CrossRef] [Green Version]
- Pirinen, M.; Donnelly, P.; Spencer, C.C. Efficient computation with a linear mixed model on large-scale data sets with applications to genetic studies. Ann. Appl. Stat. 2013, 7, 369–390. [Google Scholar] [CrossRef]
- Zhou, X.; Carbonetto, P.; Stephens, M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013, 9, e1003264. [Google Scholar] [CrossRef] [Green Version]
- Furlotte, N.A.; Eskin, E. Efficient Multiple-Trait Association and Estimation of Genetic Correlation Using the Matrix-Variate Linear Mixed Model. Genetics 2015, 200, 59-U112. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Stephens, M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 2014, 11, 407–409. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K. WOMBAT: A tool for mixed model analyses in quantitative genetics by restricted maximum likelihood (REML). J. Zhejiang Univ. Sci. B 2007, 8, 815–821. [Google Scholar] [CrossRef] [Green Version]
- Joo, J.W.; Kang, E.Y.; Org, E.; Furlotte, N.; Parks, B.; Hormozdiari, F.; Lusis, A.J.; Eskin, E. Efficient and Accurate Multiple-Phenotype Regression Method for High Dimensional Data Considering Population Structure. Genetics 2016, 204, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Zapala, M.A.; Schork, N.J. Statistical properties of multivariate distance matrix regression for high-dimensional data analysis. Front. Genet. 2012, 3, 190. [Google Scholar] [CrossRef] [PubMed]
- Lippert, C.; Casale, F.P.; Rakitsch, B.; Stegle, O. LIMIX: Genetic analysis of multiple traits. bioRxiv 2014, 003905. [Google Scholar] [CrossRef] [Green Version]
- Listgarten, J.; Lippert, C.; Kang, E.Y.; Xiang, J.; Kadie, C.M.; Heckerman, D. A powerful and efficient set test for genetic markers that handles confounders. Bioinformatics 2013, 29, 1526–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casale, F.P.; Rakitsch, B.; Lippert, C.; Stegle, O. Efficient set tests for the genetic analysis of correlated traits. Nat. Methods 2015, 12, 755–758. [Google Scholar] [CrossRef]
- Wu, M.C.; Kraft, P.; Epstein, M.P.; Taylor, D.M.; Chanock, S.J.; Hunter, D.J.; Lin, X. Powerful SNP-set analysis for case-control genome-wide association studies. Am. J. Hum. Genet. 2010, 86, 929–942. [Google Scholar] [CrossRef] [Green Version]
- Lippert, C.; Xiang, J.; Horta, D.; Widmer, C.; Kadie, C.; Heckerman, D.; Listgarten, J. Greater power and computational efficiency for kernel-based association testing of sets of genetic variants. Bioinformatics 2014, 30, 3206–3214. [Google Scholar] [CrossRef] [Green Version]
- Schifano, E.D.; Epstein, M.P.; Bielak, L.F.; Jhun, M.A.; Kardia, S.L.; Peyser, P.A.; Lin, X. SNP set association analysis for familial data. Genet. Epidemiol. 2012, 36, 797–810. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Huffman, J.E.; Brody, J.A.; Wang, C.; Lee, S.; Li, Z.; Gogarten, S.M.; Sofer, T.; Bielak, L.F.; Bis, J.C.; et al. Efficient Variant Set Mixed Model Association Tests for Continuous and Binary Traits in Large-Scale Whole-Genome Sequencing Studies. Am. J. Hum. Genet. 2019, 104, 260–274. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Benyamin, B.; McEvoy, B.P.; Gordon, S.; Henders, A.K.; Nyholt, D.R.; Madden, P.A.; Heath, A.C.; Martin, N.G.; Montgomery, G.W.; et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010, 42, 565–569. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Manolio, T.A.; Pasquale, L.R.; Boerwinkle, E.; Caporaso, N.; Cunningham, J.M.; de Andrade, M.; Feenstra, B.; Feingold, E.; Hayes, M.G.; et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 2011, 43, 519–525. [Google Scholar] [CrossRef]
- Loh, P.-R.; Bhatia, G.; Gusev, A.; Finucane, H.K.; Bulik-Sullivan, B.K.; Pollack, S.J.; de Candia, T.R.; Lee, S.H.; Wray, N.R.; Kendler, K.S. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet. 2015, 47, 1385. [Google Scholar] [CrossRef] [Green Version]
- Matilainen, K.; Mantysaari, E.A.; Lidauer, M.H.; Stranden, I.; Thompson, R. Employing a Monte Carlo algorithm in Newton-type methods for restricted maximum likelihood estimation of genetic parameters. PLoS ONE 2013, 8, e80821. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yang, C.; Shi, X.J.; Li, C.; Huang, J.; Zhao, H.Y.; Ma, S.G. Analyzing Association Mapping in Pedigree-Based GWAS Using a Penalized Multitrait Mixed Model. Genet. Epidemiol. 2016, 40, 382–393. [Google Scholar] [CrossRef] [Green Version]
- Hannah, M.V.; Casale, F.P.; Stegle, O.; Birney, E. LiMMBo: A simple, scalable approach for linear mixed models in high-dimensional genetic association studies. bioRxiv 2018, 255497. [Google Scholar] [CrossRef] [Green Version]
- Maki-Tanila, A.; Hill, W.G. Influence of gene interaction on complex trait variation with multilocus models. Genetics 2014, 198, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Eichler, E.E.; Flint, J.; Gibson, G.; Kong, A.; Leal, S.M.; Moore, J.H.; Nadeau, J.H. Missing heritability and strategies for finding the underlying causes of complex disease. Nat. Rev. Genet. 2010, 11, 446–450. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.H.; Hemani, G.; Haley, C.S. Detecting epistasis in human complex traits. Nat. Rev. Genet. 2014, 15, 722–733. [Google Scholar] [CrossRef]
- Hemani, G.; Shakhbazov, K.; Westra, H.J.; Esko, T.; Henders, A.K.; McRae, A.F.; Yang, J.; Gibson, G.; Martin, N.G.; Metspalu, A.; et al. Detection and replication of epistasis influencing transcription in humans. Nature 2014, 508, 249–253. [Google Scholar] [CrossRef] [Green Version]
- Herold, C.; Steffens, M.; Brockschmidt, F.F.; Baur, M.P.; Becker, T. INTERSNP: Genome-wide interaction analysis guided by a priori information. Bioinformatics 2009, 25, 3275–3281. [Google Scholar] [CrossRef] [Green Version]
- Hemani, G.; Theocharidis, A.; Wei, W.; Haley, C. EpiGPU: Exhaustive pairwise epistasis scans parallelized on consumer level graphics cards. Bioinformatics 2011, 27, 1462–1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schupbach, T.; Xenarios, I.; Bergmann, S.; Kapur, K. FastEpistasis: A high performance computing solution for quantitative trait epistasis. Bioinformatics 2010, 26, 1468–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kam-Thong, T.; Czamara, D.; Tsuda, K.; Borgwardt, K.; Lewis, C.M.; Erhardt-Lehmann, A.; Hemmer, B.; Rieckmann, P.; Daake, M.; Weber, F.; et al. EPIBLASTER-fast exhaustive two-locus epistasis detection strategy using graphical processing units. Eur. J. Hum. Genet. EJHG 2011, 19, 465–471. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, S.; Zou, F.; Wang, W. TEAM: Efficient two-locus epistasis tests in human genome-wide association study. Bioinformatics 2010, 26, i217–i227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, D.M.; Marchini, J.; Morris, A.P.; Cardon, L.R. Two-stage two-locus models in genome-wide association. PLoS Genet. 2006, 2, e157. [Google Scholar] [CrossRef]
- Zhang, F.T.; Zhu, Z.H.; Tong, X.R.; Zhu, Z.X.; Qi, T.; Zhu, J. Mixed Linear Model Approaches of Association Mapping for Complex Traits Based on Omics Variants. Sci. Rep. 2015, 5, 10298. [Google Scholar] [CrossRef] [Green Version]
- Cattaert, T.; Urrea, V.; Naj, A.C.; De Lobel, L.; De Wit, V.; Fu, M.; John, J.M.M.; Shen, H.; Calle, M.L.; Ritchie, M.D. FAM-MDR: A flexible family-based multifactor dimensionality reduction technique to detect epistasis using related individuals. PLoS ONE 2010, 5, e10304. [Google Scholar] [CrossRef] [Green Version]
- Casale, F.P.; Horta, D.; Rakitsch, B.; Stegle, O. Joint genetic analysis using variant sets reveals polygenic gene-context interactions. PLoS Genet. 2017, 13, e1006693. [Google Scholar] [CrossRef] [Green Version]
- Sul, J.H.; Bilow, M.; Yang, W.Y.; Kostem, E.; Furlotte, N.; He, D.; Eskin, E. Accounting for Population Structure in Gene-by-Environment Interactions in Genome-Wide Association Studies Using Mixed Models. PLoS Genet. 2016, 12, e1005849. [Google Scholar] [CrossRef]
- Ning, C.; Wang, D.; Kang, H.M.; Mrode, R.; Zhou, L.; Xu, S.Z.; Liu, J.F. A rapid epistatic mixed-model association analysis by linear retransformations of genomic estimated values. Bioinformatics 2018, 34, 1817–1825. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Tang, H.; Liu, J.F.; Xu, S.; Zhang, Q.; Ning, C. Rapid epistatic mixed-model association studies by controlling multiple polygenic effects. Bioinformatics 2020, 36, 4833–4837. [Google Scholar] [CrossRef]
- Robinson, M.R.; English, G.; Moser, G.; Lloyd-Jones, L.R.; Triplett, M.A.; Zhu, Z.; Nolte, I.M.; van Vliet-Ostaptchouk, J.V.; Snieder, H.; LifeLines Cohort, S.; et al. Genotype-covariate interaction effects and the heritability of adult body mass index. Nat. Genet. 2017, 49, 1174–1181. [Google Scholar] [CrossRef]
- Moore, R.; Casale, F.P.; Bonder, M.J.; Horta, D.; Franke, L.; Barroso, I.; Stegle, O.; Consortium, B. A linear mixed-model approach to study multivariate gene-environment interactions. Nat. Genet. 2019, 51, 180–186. [Google Scholar] [CrossRef]
- Dahl, A.; Nguyen, K.; Cai, N.; Gandal, M.J.; Flint, J.; Zaitlen, N. A Robust Method Uncovers Significant Context-Specific Heritability in Diverse Complex Traits. Am. J. Hum. Genet. 2020, 106, 71–91. [Google Scholar] [CrossRef]
- Dahl, A.; Cai, N.; Flint, J.; Zaitlen, N. GxEMM: Extending linear mixed models to general gene-environment interactions. bioRxiv 2018, 397638. [Google Scholar] [CrossRef]
- Wang, H.; Yue, T.; Yang, J.; Wu, W.; Xing, E.P. Deep mixed model for marginal epistasis detection and population stratification correction in genome-wide association studies. BMC Bioinform. 2019, 20, 656. [Google Scholar] [CrossRef]
- Runcie, D.E.; Crawford, L. Fast and flexible linear mixed models for genome-wide genetics. PLoS Genet. 2019, 15, e1007978. [Google Scholar] [CrossRef] [Green Version]
- Schultz, N.; Weigel, K. FFselect: An improved linear mixed model for genome-wide association study in populations featuring shared environments confounded by relatedness. bioRxiv 2020, 892455. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, E.; Matsunaga, H. Exploring efficient linear mixed models to detect quantitative trait locus-by-environment interactions. G3 2021, 11, jkab119. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Y.W.; Zhang, Z.C.; Xiang, Y.; Liu, M.H.; Zhou, Y.H.; Zuo, J.F.; Zhang, H.Q.; Chen, Y.; Zhang, Y.M. A compressed variance component mixed model for detecting QTNs and QTN-by-environment and QTN-by-QTN interactions in genome-wide association studies. Mol. Plant 2022, 15, 630–650. [Google Scholar] [CrossRef]
- Yang, C.; Wan, X.; Lin, X.; Chen, M.; Zhou, X.; Liu, J. CoMM: A collaborative mixed model to dissecting genetic contributions to complex traits by leveraging regulatory information. Bioinformatics 2019, 35, 1644–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albert, F.W.; Kruglyak, L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2015, 16, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Joehanes, R.; Chen, B.H.; Huan, T.; Ying, S.; Munson, P.J.; Johnson, A.D.; Levy, D.; O’Donnell, C.J. Identification of common genetic variants controlling transcript isoform variation in human whole blood. Nat. Genet. 2015, 47, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Ming, J.; Dai, M.; Cai, M.; Wan, X.; Liu, J.; Yang, C. LSMM: A statistical approach to integrating functional annotations with genome-wide association studies. Bioinformatics 2018, 34, 2788–2796. [Google Scholar] [CrossRef] [Green Version]
- Hao, X.; Zeng, P.; Zhang, S.; Zhou, X. Identifying and exploiting trait-relevant tissues with multiple functional annotations in genome-wide association studies. PLoS Genet. 2018, 14, e1007186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Shi, X.; Jiao, Y.; Huang, J.; Chen, M.; Zhou, X.; Sun, L.; Lin, X.; Yang, C.; Liu, J. CoMM-S2: A collaborative mixed model using summary statistics in transcriptome-wide association studies. Bioinformatics 2019, 36, 2009–2016. [Google Scholar] [CrossRef]
- Sabatti, C.; Service, S.K.; Hartikainen, A.L.; Pouta, A.; Ripatti, S.; Brodsky, J.; Jones, C.G.; Zaitlen, N.A.; Varilo, T.; Kaakinen, M.; et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat. Genet. 2009, 41, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Aulchenko, Y.S.; Ripatti, S.; Lindqvist, I.; Boomsma, D.; Heid, I.M.; Pramstaller, P.P.; Penninx, B.W.; Janssens, A.C.; Wilson, J.F.; Spector, T.; et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009, 41, 47–55. [Google Scholar] [CrossRef]
- Kamatani, Y.; Matsuda, K.; Okada, Y.; Kubo, M.; Hosono, N.; Daigo, Y.; Nakamura, Y.; Kamatani, N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 2010, 42, 210–215. [Google Scholar] [CrossRef]
- Furlotte, N.A.; Eskin, E.; Eyheramendy, S. Genome-wide association mapping with longitudinal data. Genet. Epidemiol. 2012, 36, 463–471. [Google Scholar] [CrossRef] [Green Version]
- Sikorska, K.; Rivadeneira, F.; Groenen, P.J.; Hofman, A.; Uitterlinden, A.G.; Eilers, P.H.; Lesaffre, E. Fast linear mixed model computations for genome-wide association studies with longitudinal data. Stat. Med. 2013, 32, 165–180. [Google Scholar] [CrossRef]
- Sikorska, K.; Montazeri, N.M.; Uitterlinden, A.; Rivadeneira, F.; Eilers, P.H.; Lesaffre, E. GWAS with longitudinal phenotypes: Performance of approximate procedures. Eur. J. Hum. Genet. EJHG 2015, 23, 1384–1391. [Google Scholar] [CrossRef]
- Sung, Y.; Feng, Z.; Subedi, S. A genome-wide association study of multiple longitudinal traits with related subjects. Stat 2016, 5, 22–44. [Google Scholar] [CrossRef] [Green Version]
- Madsen, P.; Sørensen, P.; Su, G.; Damgaard, L.H.; Thomsen, H.; Labouriau, R. DMU—A package for analyzing multivariate mixed models. In Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, Brazil, 13–18 August 2006. [Google Scholar]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, G.E.; Mezey, J.G.; Schadt, E.E. lrgpr: Interactive linear mixed model analysis of genome-wide association studies with composite hypothesis testing and regression diagnostics in R. Bioinformatics 2014, 30, 3134–3135. [Google Scholar] [CrossRef] [Green Version]
- Gilmour, A.; Gogel, B.; Cullis, B.; Thompson, R. ASReml User Guide Release 2.0; VSN International Ltd.: Hemel Hempstead, UK, 2006. [Google Scholar]
- Ziyatdinov, A.; Vazquez-Santiago, M.; Brunel, H.; Martinez-Perez, A.; Aschard, H.; Soria, J.M. lme4qtl: Linear mixed models with flexible covariance structure for genetic studies of related individuals. BMC Bioinform. 2018, 19, 68. [Google Scholar] [CrossRef]
- Shor, T.; Kalka, I.; Geiger, D.; Erlich, Y.; Weissbrod, O. Estimating variance components in population scale family trees. PLoS Genet. 2019, 15, e1008124. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Zhou, X.; Hao, Z.; Jiang, L.; Yang, R. Genome-wide barebones regression scan for mixed-model association analysis. Theor. Appl. Genet. 2020, 133, 51–58. [Google Scholar] [CrossRef]
- Lee, S.H.; van der Werf, J.H. MTG2: An efficient algorithm for multivariate linear mixed model analysis based on genomic information. Bioinformatics 2016, 32, 1420–1422. [Google Scholar] [CrossRef] [Green Version]
- Golan, D.; Lander, E.S.; Rosset, S. Measuring missing heritability: Inferring the contribution of common variants. Proc. Natl. Acad. Sci. USA 2014, 111, E5272–E5281. [Google Scholar] [CrossRef] [Green Version]
- Ge, T.; Chen, C.Y.; Neale, B.M.; Sabuncu, M.R.; Smoller, J.W. Phenome-wide heritability analysis of the UK Biobank. PLoS Genet. 2017, 13, e1006711. [Google Scholar] [CrossRef] [Green Version]
- Weissbrod, O.; Flint, J.; Rosset, S. Estimating SNP-Based Heritability and Genetic Correlation in Case-Control Studies Directly and with Summary Statistics. Am. J. Hum. Genet. 2018, 103, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Speed, D.; Balding, D.J. MultiBLUP: Improved SNP-based prediction for complex traits. Genome Res. 2014, 24, 1550–1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golan, D.; Rosset, S. Effective Genetic-Risk Prediction Using Mixed Models. Am. J. Hum. Genet. 2014, 95, 383–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilhjalmsson, B.J.; Yang, J.; Finucane, H.K.; Gusev, A.; Lindstrom, S.; Ripke, S.; Genovese, G.; Loh, P.R.; Bhatia, G.; Do, R.; et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am. J. Hum. Genet. 2015, 97, 576–592. [Google Scholar] [CrossRef] [Green Version]
- Loh, P.R.; Kichaev, G.; Gazal, S.; Schoech, A.P.; Price, A.L. Mixed-model association for biobank-scale datasets. Nat. Genet. 2018, 50, 906–908. [Google Scholar] [CrossRef]
- Perez-Enciso, M.; Misztal, I. Qxpak: A versatile mixed model application for genetical genomics and QTL analyses. Bioinformatics 2004, 20, 2792–2798. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Yang, J.; Hu, C.; Hu, H.; Yu, R.; Xia, Z.; Ye, X.; Zhu, J. QTLNetwork: Mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 2008, 24, 721–723. [Google Scholar] [CrossRef] [Green Version]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [Green Version]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakobsdottir, J.; McPeek, M.S. MASTOR: Mixed-model association mapping of quantitative traits in samples with related individuals. Am. J. Hum. Genet. 2013, 92, 652–666. [Google Scholar] [CrossRef] [PubMed]
- Visconti, A.; Al-Shafai, M.; Al Muftah, W.A.; Zaghlool, S.B.; Mangino, M.; Suhre, K.; Falchi, M. PopPAnTe: Population and pedigree association testing for quantitative data. BMC Genom. 2017, 18, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Dai, X.; Wang, Q.; Xu, S.; Zhao, P.X. PEPIS: A Pipeline for Estimating Epistatic Effects in Quantitative Trait Locus Mapping and Genome-Wide Association Studies. PLoS Comput. Biol. 2016, 12, e1004925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abecasis, G.R.; Cardon, L.R.; Cookson, W.O. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000, 66, 279–292. [Google Scholar] [CrossRef] [Green Version]
- Zeng, J.; de Vlaming, R.; Wu, Y.; Robinson, M.R.; Lloyd-Jones, L.R.; Yengo, L.; Yap, C.X.; Xue, A.; Sidorenko, J.; McRae, A.F.; et al. Signatures of negative selection in the genetic architecture of human complex traits. Nat. Genet. 2018, 50, 746–753. [Google Scholar] [CrossRef]
- Zhang, F.T.; Chen, W.H.; Zhu, Z.H.; Zhang, Q.; Nabais, M.F.; Qi, T.; Deary, I.J.; Wray, N.R.; Visscher, P.M.; McRae, A.F.; et al. OSCA: A tool for omic-data-based complex trait analysis. Genome Biol. 2019, 20, 107. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.; Zheng, Z.; Qi, T.; Kemper, K.E.; Wray, N.R.; Visscher, P.M.; Yang, J. A resource-efficient tool for mixed model association analysis of large-scale data. Nat. Genet. 2019, 51, 1749–1755. [Google Scholar] [CrossRef]
- Fabregat-Traver, D.; Sharapov, S.; Hayward, C.; Rudan, I.; Campbell, H.; Aulchenko, Y.; Bientinesi, P. High-Performance Mixed Models Based Genome-Wide Association Analysis with omicABEL software. F1000Research 2014, 3, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Yang, T.; Zhou, Y.; Yin, S.; Li, P.; Liu, J.; Xu, S.; Yang, Z.; Xu, C. Genome-Wide Association Mapping of Starch Pasting Properties in Maize Using Single-Locus and Multi-Locus Models. Front. Plant Sci. 2018, 9, 1311. [Google Scholar] [CrossRef] [Green Version]
- Scheinfeldt, L.B.; Tishkoff, S.A. Recent human adaptation: Genomic approaches, interpretation and insights. Nat. Rev. Genet. 2013, 14, 692–702. [Google Scholar] [CrossRef] [Green Version]
- Hackinger, S.; Zeggini, E. Statistical methods to detect pleiotropy in human complex traits. Open Biol. 2017, 7, 170125. [Google Scholar] [CrossRef] [PubMed]
- Dudbridge, F.; Fletcher, O. Gene-environment dependence creates spurious gene-environment interaction. Am. J. Hum. Genet. 2014, 95, 301–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Weedon, M.N.; Purcell, S.; Lettre, G.; Estrada, K.; Willer, C.J.; Smith, A.V.; Ingelsson, E.; O’Connell, J.R.; Mangino, M.; et al. Genomic inflation factors under polygenic inheritance. Eur. J. Hum. Genet. EJHG 2011, 19, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Stahl, E.A.; Wegmann, D.; Trynka, G.; Gutierrez-Achury, J.; Do, R.; Voight, B.F.; Kraft, P.; Chen, R.; Kallberg, H.J.; Kurreeman, F.A.; et al. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet. 2012, 44, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, A.A.; Mathieson, I. Demographic history mediates the effect of stratification on polygenic scores. Elife 2020, 9, e61548. [Google Scholar] [CrossRef]
- Uffelmann, E.; Posthuma, D. Emerging Methods and Resources for Biological Interrogation of Neuropsychiatric Polygenic Signal. Biol. Psychiatry 2021, 89, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Primers 2021, 1, 59. [Google Scholar] [CrossRef]
- Guimaraes, L.C.; de Jesus, L.B.; Viana, M.V.C.; Silva, A.; Ramos, R.T.J.; Soares, S.D.; Azevedo, V. Inside the Pan-genome—Methods and Software Overview. Curr. Genom. 2015, 16, 245–252. [Google Scholar] [CrossRef] [Green Version]
- Snipen, L.; Almoy, T.; Ussery, D.W. Microbial comparative pan-genomics using binomial mixture models. BMC Genom. 2009, 10, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahaman, M.M.; Chen, D.; Gillani, Z.; Klukas, C.; Chen, M. Advanced phenotyping and phenotype data analysis for the study of plant growth and development. Front. Plant Sci. 2015, 6, 619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Poorter, H.; Dumschott, K.; Bolger, M.E.; Arend, D.; Osorio, S.; Gundlach, H.; Mayer, K.F.X.; Lange, M.; Scholz, U.; et al. Computational aspects underlying genome to phenome analysis in plants. Plant J. 2019, 97, 182–198. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.; Daly, N.L. Venomics: A Mini-Review. High Throughput 2018, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milward, E.A.; Daneshi, N.; Johnstone, D.M. Emerging real-time technologies in molecular medicine and the evolution of integrated ‘pharmacomics’ approaches to personalized medicine and drug discovery. Pharm. Ther. 2012, 136, 295–304. [Google Scholar] [CrossRef]
- Das, S.; Ghosh, I.; Banerjee, G.; Sarkar, U. Artificial Intelligence in Agriculture: A Literature Survey. Int. J. Sci. Res. Comput. Sci. Appl. Manag. Stud. 2018, 7, 1–6. [Google Scholar]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial intelligence in healthcare: Past, present and future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 2017, 18, 851–869. [Google Scholar] [CrossRef] [Green Version]
- Fountas, S.; Espejo-Garcia, B.; Kasimati, A.; Mylonas, N.; Darra, N. The Future of Digital Agriculture: Technologies and Opportunities. IT Prof. 2020, 22, 24–28. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alamin, M.; Sultana, M.H.; Lou, X.; Jin, W.; Xu, H. Dissecting Complex Traits Using Omics Data: A Review on the Linear Mixed Models and Their Application in GWAS. Plants 2022, 11, 3277. https://doi.org/10.3390/plants11233277
Alamin M, Sultana MH, Lou X, Jin W, Xu H. Dissecting Complex Traits Using Omics Data: A Review on the Linear Mixed Models and Their Application in GWAS. Plants. 2022; 11(23):3277. https://doi.org/10.3390/plants11233277
Chicago/Turabian StyleAlamin, Md., Most. Humaira Sultana, Xiangyang Lou, Wenfei Jin, and Haiming Xu. 2022. "Dissecting Complex Traits Using Omics Data: A Review on the Linear Mixed Models and Their Application in GWAS" Plants 11, no. 23: 3277. https://doi.org/10.3390/plants11233277
APA StyleAlamin, M., Sultana, M. H., Lou, X., Jin, W., & Xu, H. (2022). Dissecting Complex Traits Using Omics Data: A Review on the Linear Mixed Models and Their Application in GWAS. Plants, 11(23), 3277. https://doi.org/10.3390/plants11233277








