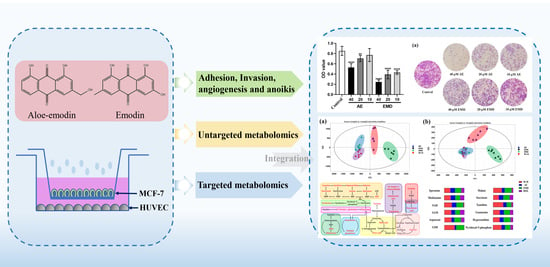
Cell Metabolomics Reveals the Potential Mechanism of Aloe Emodin and Emodin Inhibiting Breast Cancer Metastasis
Abstract
:1. Introduction
2. Results
2.1. Metabolomic Comparison of Single-Culture and Co-Culture MCF-7 Cells
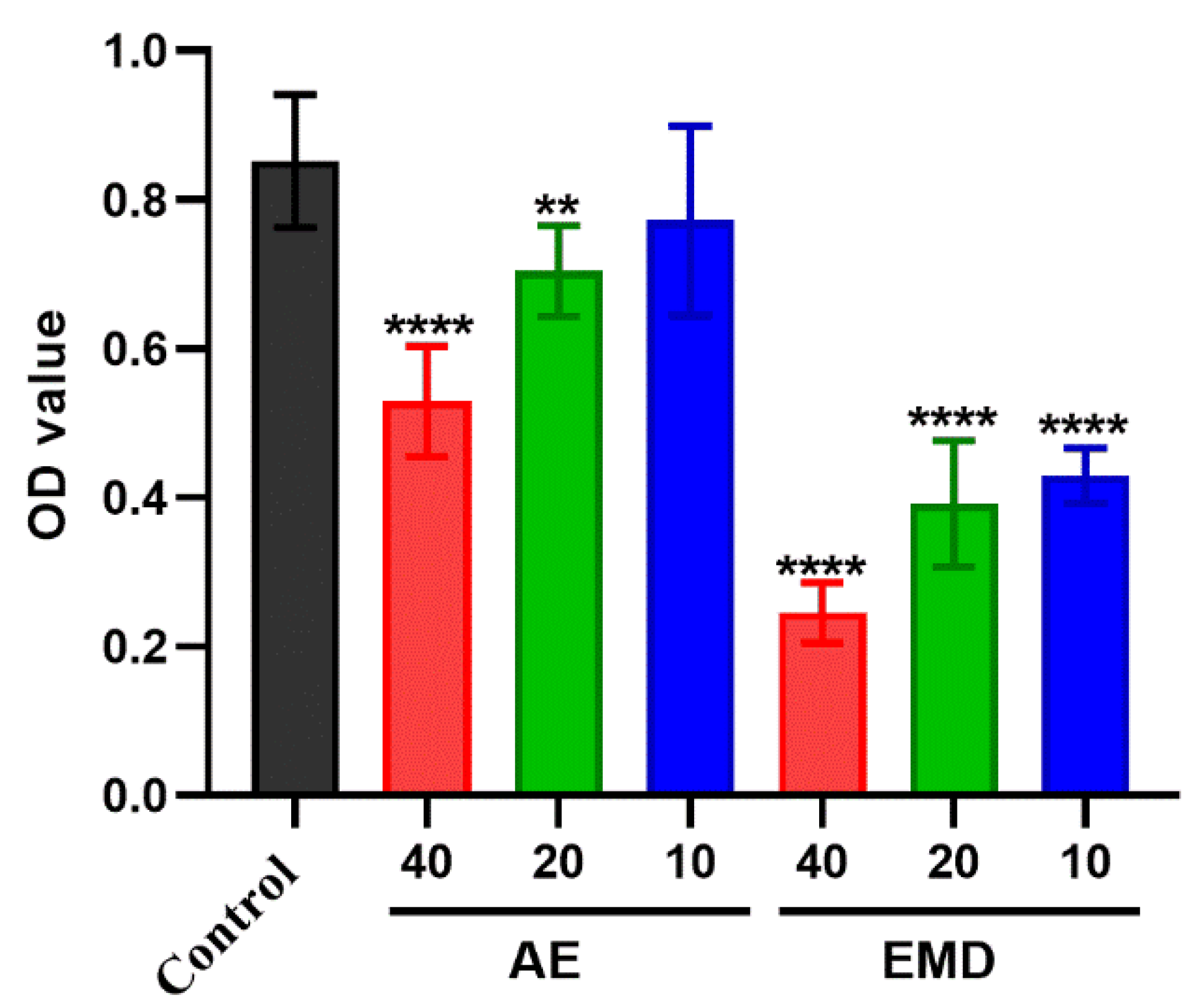
2.2. Cytotoxicity of AE and EMD
2.3. AE and EMD Reduce the Adhesion of MCF-7 Cells to HUVEC Cells
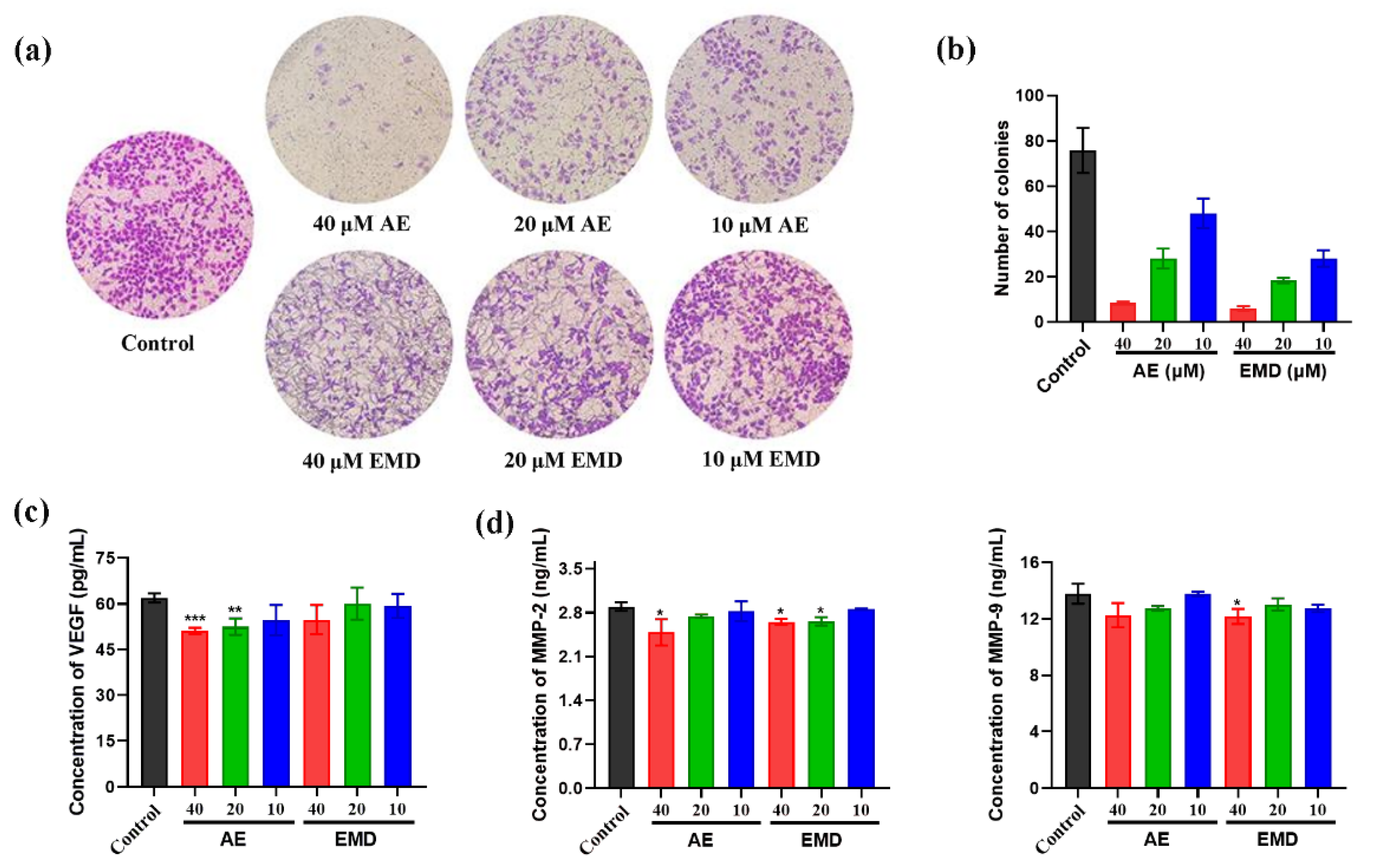
2.4. AE and EMD Inhibit the Invasion and Metastasis of MCF-7 Cells
2.5. Effects of AE and EMD on Metabolism of MCF-7 Cells
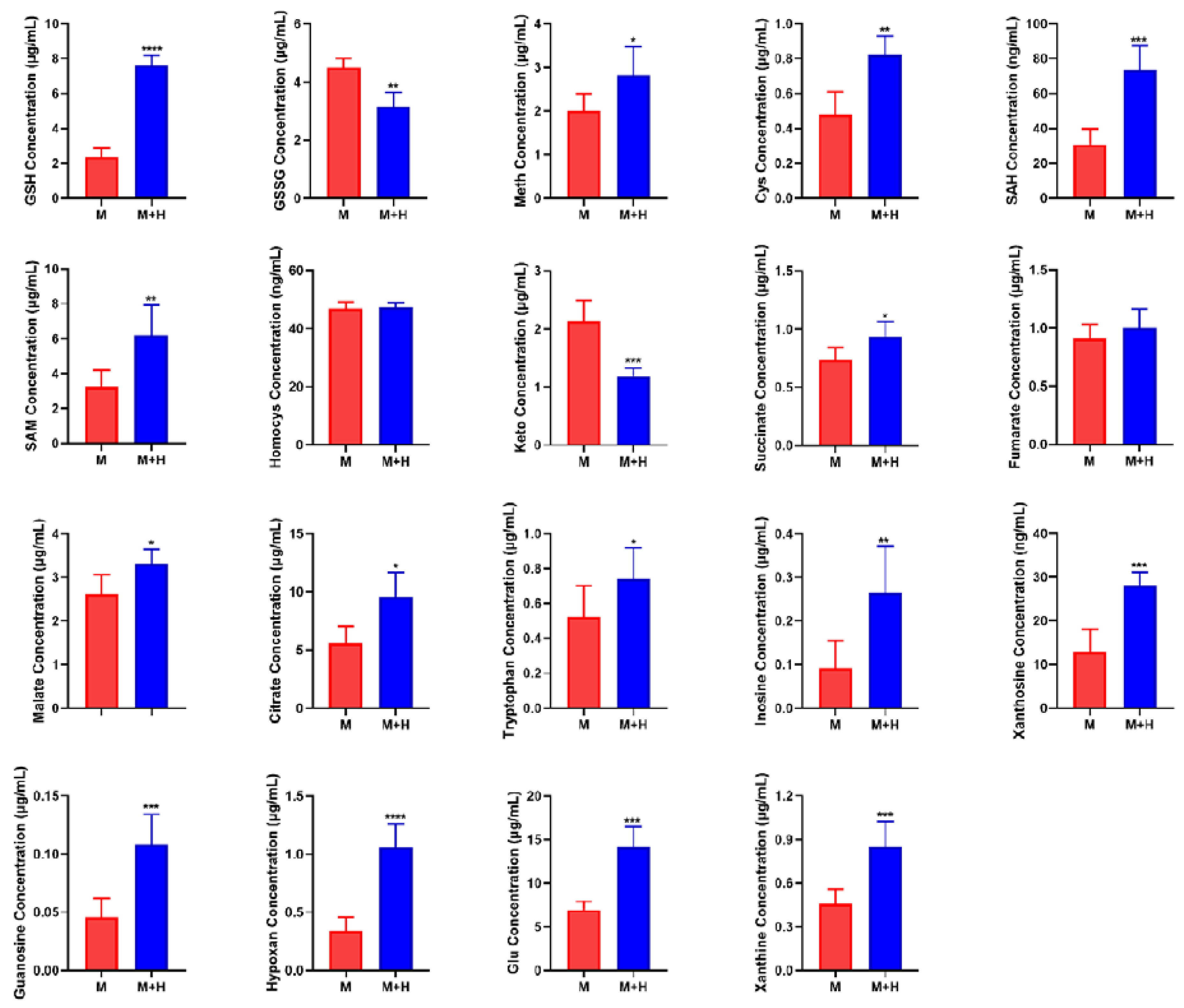
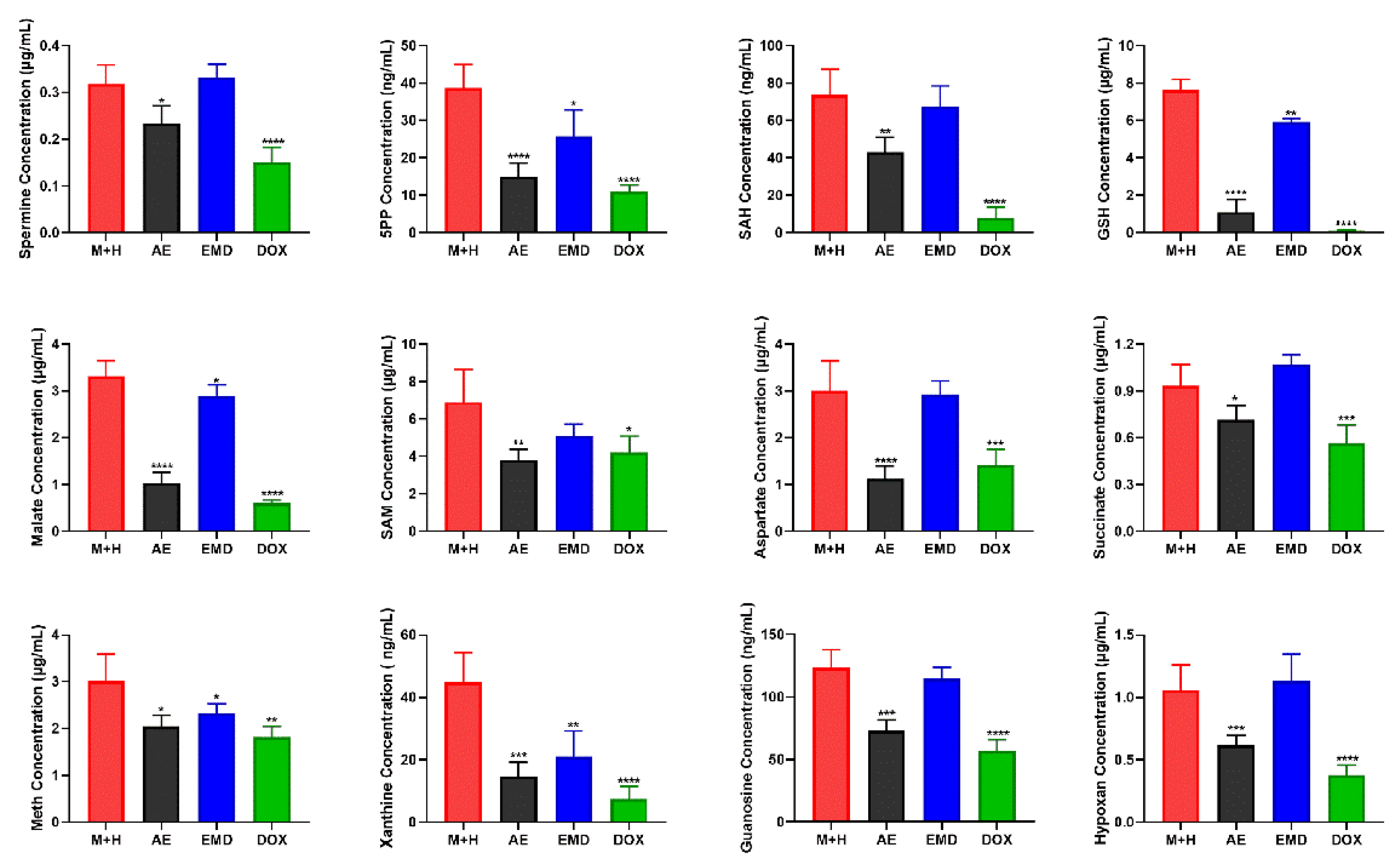
2.6. Quantitative Analysis of Biomarkers
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Cytotoxicity Assay
4.4. Adhesion Assay
4.5. Invasion Assay
4.6. Activity Determination of MMP-2, MMP-9, and VEGF
4.7. Soft Agar Colony Formation
4.8. Cell Sample Preparation for Metabolomic Analyses
4.9. UPLC-MS Conditions
4.10. Data Processing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fidler, I.J. The organ microenvironment and cancer metastasis. Differentiation 2002, 70, 498–505. [Google Scholar] [CrossRef]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef]
- Stroka, K.M.; Konstantopoulos, K. Physical Biology in Cancer. 4. Physical cues guide tumor cell adhesion and migration. Am. J. Physiol. Physiol. 2014, 306, C98–C109. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.; Ding, B.S.; Guo, P.; Lee, S.B.; Butler, J.M.; Casey, S.C.; Simons, M.; Tam, W.; Felsher, D.W.; Shido, K.; et al. Angiocrine factors deployed by tumor vascular niche induce B cell lymphoma invasiveness and chemoresistance. Cancer Cell 2014, 25, 350–365. [Google Scholar] [CrossRef] [Green Version]
- Maishi, N.; Hida, K. Tumor endothelial cells accelerate tumor metastasis. Cancer Sci. 2017, 108, 1921–1926. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.Y.; Chiang, S.Y.; Lin, J.G.; Ma, Y.S.; Liao, C.L.; Weng, S.W.; Lai, T.Y.; Chung, J.G. Emodin, aloe-emodin and rhein inhibit migration and invasion in human tongue cancer SCC-4 cells through the inhibition of gene expression of matrix metalloproteinase-9. Int. J. Oncol. 2010, 36, 1113–1120. [Google Scholar]
- Gu, J.; Cui, C.F.; Yang, L.; Wang, L.; Jiang, X.H. Emodin inhibits colon cancer cell invasion and migration by suppressing epithelial-mesenchymal transition via the Wnt/beta-catenin pathway. Oncol. Res. 2019, 27, 193–202. [Google Scholar] [CrossRef]
- Suboj, P.; Babykutty, S.; Gopi, D.R.V.; Nair, R.S.; Srinivas, P.; Gopala, S. Aloe emodin inhibits colon cancer cell migration/angiogenesis by downregulating MMP-2/9, RhoB and VEGF via reduced DNA binding activity of NF-kappa B. Eur. J. Pharm. Sci. 2012, 45, 581–591. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, X.F.; Zhou, Q.M.; Lu, Y.Y.; Zhang, H.; Chen, Q.L.; Zhao, M.; Su, S.B. Inhibitory effect of emodin on migration, invasion and metastasis of human breast cancer MDA-MB-231 cells in vitro and in vivo. Oncol. Rep. 2015, 33, 338–346. [Google Scholar] [CrossRef] [Green Version]
- He, Z.H.; Huang, Y.Q.; Weng, S.F.; Tan, Y.R.; He, T.P.; Qin, Y.M.; Liang, N.C. Effect of Aloe emodin on invasion and metastasis of high metastatic breast cancer MDA-MB-231 cells. J. Chin. Med. Mater. 2013, 36, 1481–1485. [Google Scholar]
- Lin, M.L.; Lu, Y.C.; Chung, J.G.; Wang, S.G.; Lin, H.T.; Kang, S.E.; Tang, C.H.; Ko, J.L.; Chen, S.S. Down-Regulation of MMP-2 Through the p38 MAPK-NF-kappa B-Dependent Pathway by Aloe-Emodin Leads to Inhibition of Nasopharyngeal Carcinoma Cell Invasion. Mol. Carcinog. 2010, 49, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Manu, K.A.; Shanmugam, M.K.; Ong, T.H.; Subramaniam, A.; Siveen, K.S.; Perumal, E.; Samy, R.P.; Bist, P.; Lim, L.H.K.; Kumar, A.P.; et al. Emodin Suppresses Migration and Invasion through the Modulation of CXCR4 Expression in an Orthotopic Model of Human Hepatocellular Carcinoma. PLoS ONE 2013, 8, e57015. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanderson, S.M.; Gao, X.; Dai, Z.; Locasale, J.W. Methionine metabolism in health and cancer: A nexus of diet and precision medicine. Nat. Rev. Cancer 2019, 19, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.P.; Yashinskie, J.J.; Koche, R.; Chandwani, R.; Tian, S.; Chen, C.C.; Baslan, T.; Marinkovic, Z.S.; Sanchez-Rivera, F.J.; Leach, S.D.; et al. alpha-Ketoglutarate links p53 to cell fate during tumour suppression. Nature 2019, 573, 595–599. [Google Scholar] [CrossRef]
- Platten, M.; Wick, W.; Van den Eynde, B.J. Tryptophan catabolism in cancer: Beyond IDO and tryptophan depletion. Cancer Res. 2012, 72, 5435–5440. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Yao, Y.; Scott, A.J.; Wilder-Romans, K.; Dresser, J.J.; Werner, C.K.; Sun, H.; Pratt, D.; Sajjakulnukit, P.; Zhao, S.G.; et al. Purine metabolism regulates DNA repair and therapy resistance in glioblastoma. Nat. Commun. 2020, 11, 3811. [Google Scholar] [CrossRef]
- Chavarrıa, T.; Sanchez-Jimenez, F.; Quesada, A.R.; Medina, M.A. Homocysteine inhibits the proliferation and invasive potential of HT-1080 human fibrosarcoma cells. Biochem. Biophys. Res. Commun. 2003, 301, 540–544. [Google Scholar] [CrossRef]
- Ciccarone, F.; Vegliante, R.; Di Leo, L.; Ciriolo, M.R. The TCA cycle as a bridge between oncometabolism and DNA transactions in cancer. Semin. Cancer Biol. 2017, 47, 50–56. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vacchelli, E.; Michels, J.; Garcia, P.; Kepp, O.; Senovilla, L.; Vitale, I.; Kroemer, G. Effects of vitamin B6 metabolism on oncogenesis, tumor progression and therapeutic responses. Oncogene 2013, 32, 4995–5004. [Google Scholar] [CrossRef] [Green Version]
- Pedley, A.M.; Benkovic, S.J. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci. 2017, 42, 141–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Du, W.; Cao, S.; Wu, Y.; Dong, N.; Wang, Y.; Xu, Y. Systematic analyses of glutamine and glutamate metabolisms across different cancer types. Chin. J. Cancer 2017, 36, 88. [Google Scholar] [CrossRef] [PubMed]
- Casero, R.A.; Murray Stewart, T.; Pegg, A.E. Polyamine metabolism and cancer: Treatments, challenges and opportunities. Nat. Rev. Cancer 2018, 18, 681–695. [Google Scholar] [CrossRef]
- Wang, Z.; Yip, L.Y.; Lee, J.H.J.; Wu, Z.; Chew, H.Y.; Chong, P.K.W.; Teo, C.C.; Ang, H.Y.; Peh, K.L.E.; Yuan, J.; et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med. 2019, 25, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.B.; Luengo, A.; Danai, L.V.; Bush, L.N.; Diehl, F.F.; Hosios, A.M.; Lau, A.N.; Elmiligy, S.; Malstrom, S.; Lewis, C.A.; et al. Aspartate is an endogenous metabolic limitation for tumour growth. Nat. Cell Biol. 2018, 20, 782–788. [Google Scholar] [CrossRef] [PubMed]
- Mierke, C.T. Cancer Cells Regulate Biomechanical Properties of Human Microvascular Endothelial Cells. J. Biol. Chem. 2011, 286, 40025–40037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, A.; Ma, J.Y. Piceatannol-3-O-beta-d-glucopyranoside (PG) exhibits in vitro anti-metastatic and anti-angiogenic activities in HT1080 malignant fibrosarcoma cells. Phytomedicine 2019, 57, 95–104. [Google Scholar] [CrossRef]
- Deryugina, E.I.; Quigley, J.P. Tumor angiogenesis: MMP-mediated induction of intravasation- and metastasis-sustaining neovasculature. Matrix Biol. 2015, 44–46, 94–112. [Google Scholar] [CrossRef]
- Yao, Z.; Yuan, T.; Wang, H.; Yao, S.; Zhao, Y.; Liu, Y.; Jin, S.; Chu, J.; Xu, Y.; Zhou, W.; et al. MMP-2 together with MMP-9 overexpression correlated with lymph node metastasis and poor prognosis in early gastric carcinoma. Tumor Biol. 2017, 39, 700411. [Google Scholar] [CrossRef] [Green Version]
- Zong, L.; Xing, J.; Liu, S.; Liu, Z.; Song, F. Cell metabolomics reveals the neurotoxicity mechanism of cadmium in PC12 cells. Ecotoxicol. Environ. Saf. 2018, 147, 26–33. [Google Scholar] [CrossRef]
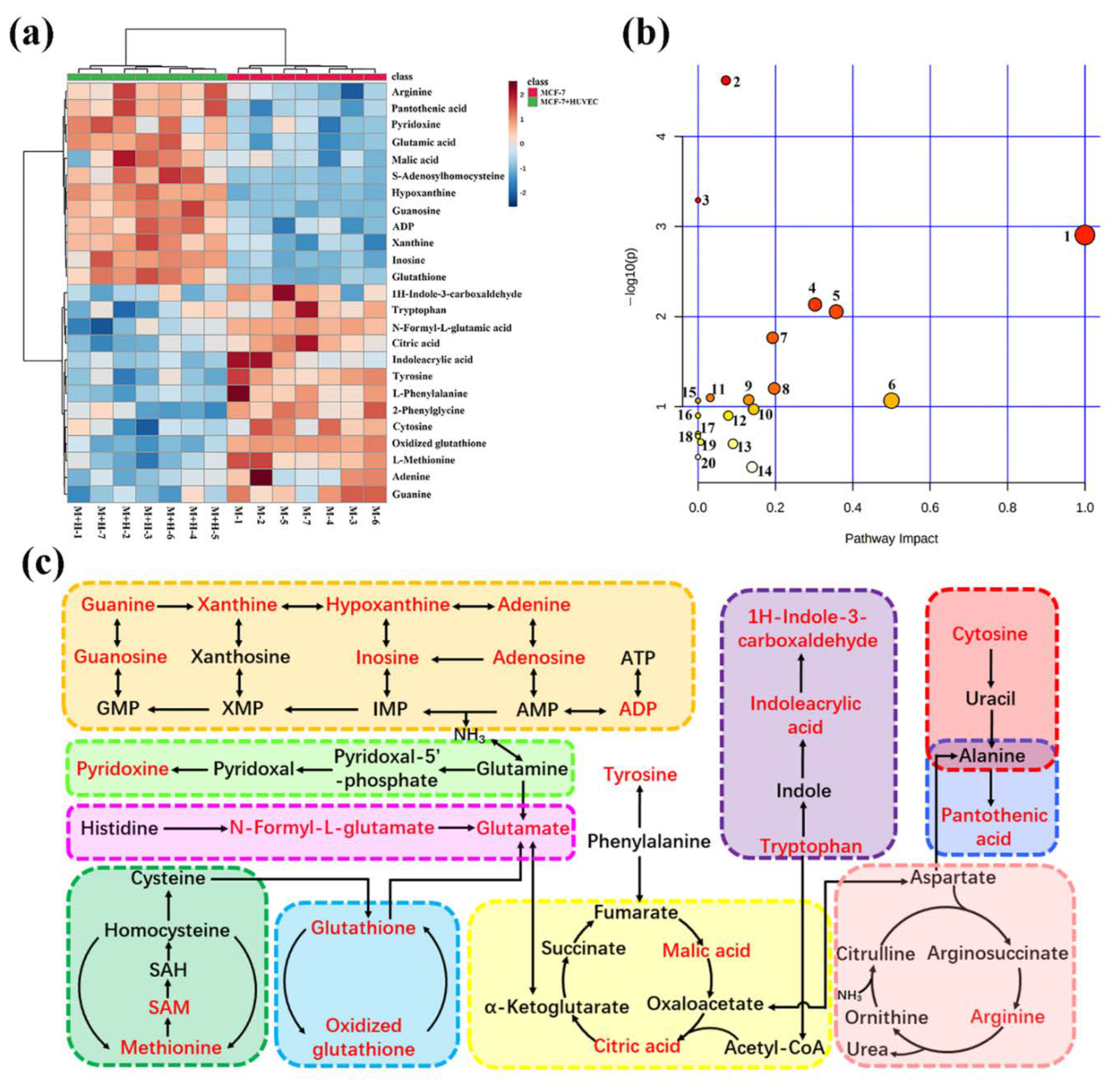


| Mode | Metabolites | RT (min) | Measured Mass (Da) | Mass Error (ppm) | AE | EMD | DOX | Pathway |
|---|---|---|---|---|---|---|---|---|
| ESI+ | Spermine | 0.50 | 203.2238 | 1.0 | ↓ * | - | ↓ *** | Polyamine metabolism |
| Arginine | 0.65 | 174.1128 | 6.3 | - | - | ↓ ** | Arginine biosynthesis | |
| Creatine | 0.45 | 132.1000 | −8.3 | ↓ * | - | ↓ ** | Glycine, serine, and threonine metabolism | |
| Methionine | 1.12 | 150.0589 | 0.0 | ↓ * | - | ↓ ** | Methionine cycle | |
| Glutathione | 1.39 | 308.0916 | 0.0 | ↓ ** | ↓ * | ↓ ** | Glutathione metabolism | |
| Isoleucine | 1.93 | 132.1031 | 4.5 | ↑ *** | - | ↑ *** | Valine, leucine, and isoleucine degradation | |
| 2-phenylglycine | 2.16 | 152.0700 | 4.8 | - | - | ↑ ** | - | |
| Pyridoxine | 2.16 | 170.0825 | 4.7 | ↑ * | - | ↑ *** | Vitamin B6 metabolism | |
| Adenine | 1.26 | 136.0630 | 5.1 | ↓ ** | - | ↓ *** | Purine metabolism | |
| ADP | 4.47 | 428.0375 | 0.7 | ↓ * | - | ↓ * | Purine metabolism | |
| Oxidized glutathione | 4.47 | 613.1589 | −1.5 | ↑ * | ↑ * | ↑ * | Glutathione metabolism | |
| Phenylalanine | 4.70 | 166.0876 | 4.8 | ↑ *** | ↑ * | ↑ *** | Phenylalanine metabolism | |
| Guanine | 4.70 | 152.0580 | 5.3 | - | ↑ ** | - | Purine metabolism | |
| 1H-indole-3-carboxaldehyde | 5.29 | 146.0614 | 5.5 | ↑ * | - | ↑ *** | Tryptophan metabolism | |
| Indoleacrylic acid | 5.29 | 188.0723 | 5.8 | ↑ ** | - | ↑ *** | Tryptophan metabolism | |
| Phytosphingosine | 9.54 | 318.3009 | 0.3 | ↑ *** | ↑ * | ↑ *** | Sphingolipid metabolism | |
| ESI− | Glutamic acid | 0.73 | 146.0455 | 1.4 | - | - | ↓ *** | D-glutamine and D-glutamate metabolism |
| Tyrosine | 2.49 | 180.0662 | 0.6 | ↑ *** | - | ↑ *** | Tyrosine metabolism | |
| Xanthine | 2.49 | 151.0257 | 0.7 | ↓ *** | - | ↓ *** | Purine metabolism | |
| Glutathione | 1.39 | 306.0759 | −0.3 | ↓ * | ↓ * | ↓ *** | Glutathione metabolism | |
| Oxidized glutathione | 4.47 | 611.1439 | −0.3 | ↑ * | - | ↓ * | Glutathione metabolism | |
| Malic acid | 0.88 | 133.0130 | −5.3 | ↓ *** | ↓ * | ↓ *** | Citrate cycle (TCA cycle) | |
| Phenylalanine | 4.62 | 164.0720 | 4.8 | ↑ ** | ↑ * | ↑ *** | Phenylalanine metabolism | |
| Hypoxanthine | 1.93 | 135.0308 | 0.7 | ↓ *** | ↓ * | ↓ *** | Purine metabolism | |
| Tryptophan | 5.29 | 203.0820 | −0.5 | ↑ ** | - | ↑ *** | Tryptophan metabolism | |
| Pantothenic acid | 4.96 | 218.1029 | 0.5 | ↑ *** | ↓ * | ↑ *** | Pantothenate and CoA biosynthesis | |
| Azelaic acid | 7.12 | 187.0970 | 0.0 | ↑ * | - | ↑ * | - | |
| S-adenosylhomo-cysteine | 4.47 | 383.1134 | −1.0 | ↓ * | ↓ * | ↓ *** | Methionine cycle | |
| Cytosine | 0.88 | 110.0356 | 1.8 | ↓ * | - | ↓ *** | Pyrimidine metabolism | |
| N-formyl-L-glutamic acid | 1.26 | 174.0402 | 0.0 | ↓ *** | ↑ *** | ↓ *** | Histidine metabolism | |
| Folic acid | 5.47 | 440.1315 | −0.9 | ↑ ** | - | ↑ *** | Folate biosynthesis | |
| Inosine | 4.62 | 267.0735 | 2.2 | ↑ * | - | ↑ ** | Purine metabolism | |
| Citric acid | 1.72 | 191.0192 | 0.0 | ↑ * | - | ↑ * | Citrate cycle (TCA cycle) | |
| Guanosine | 4.62 | 282.0839 | 0.4 | ↓ * | ↓ * | ↓ * | Purine metabolism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, G.; Liu, Z.; Zheng, Z.; Song, F.; Zhuang, X.; Liu, S. Cell Metabolomics Reveals the Potential Mechanism of Aloe Emodin and Emodin Inhibiting Breast Cancer Metastasis. Int. J. Mol. Sci. 2022, 23, 13738. https://doi.org/10.3390/ijms232213738
Cheng G, Liu Z, Zheng Z, Song F, Zhuang X, Liu S. Cell Metabolomics Reveals the Potential Mechanism of Aloe Emodin and Emodin Inhibiting Breast Cancer Metastasis. International Journal of Molecular Sciences. 2022; 23(22):13738. https://doi.org/10.3390/ijms232213738
Chicago/Turabian StyleCheng, Guorong, Zhiqiang Liu, Zhong Zheng, Fengrui Song, Xiaoyu Zhuang, and Shu Liu. 2022. "Cell Metabolomics Reveals the Potential Mechanism of Aloe Emodin and Emodin Inhibiting Breast Cancer Metastasis" International Journal of Molecular Sciences 23, no. 22: 13738. https://doi.org/10.3390/ijms232213738