A Multiomics Assessment of Preoperative Exercise in Pancreatic Cancer Survivors Receiving Neoadjuvant Therapy: A Case Series
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants, Assessments, and Intervention
2.2. Plasma Samples
2.3. Metabolomics and Lipidomics Sample Preparation
2.4. Metabolomics and Lipidomics UHPLC-MS Data Acquisition and Processing
2.5. Metabolomics and Lipidomics Data Analysis
2.6. Inductively-Coupled Plasma (ICP) Mass Spectrometry
2.7. Proteomics Sample Preparation
2.8. Proteomics Data Acquisition and Processing
2.9. Feature Enrichment Analyses
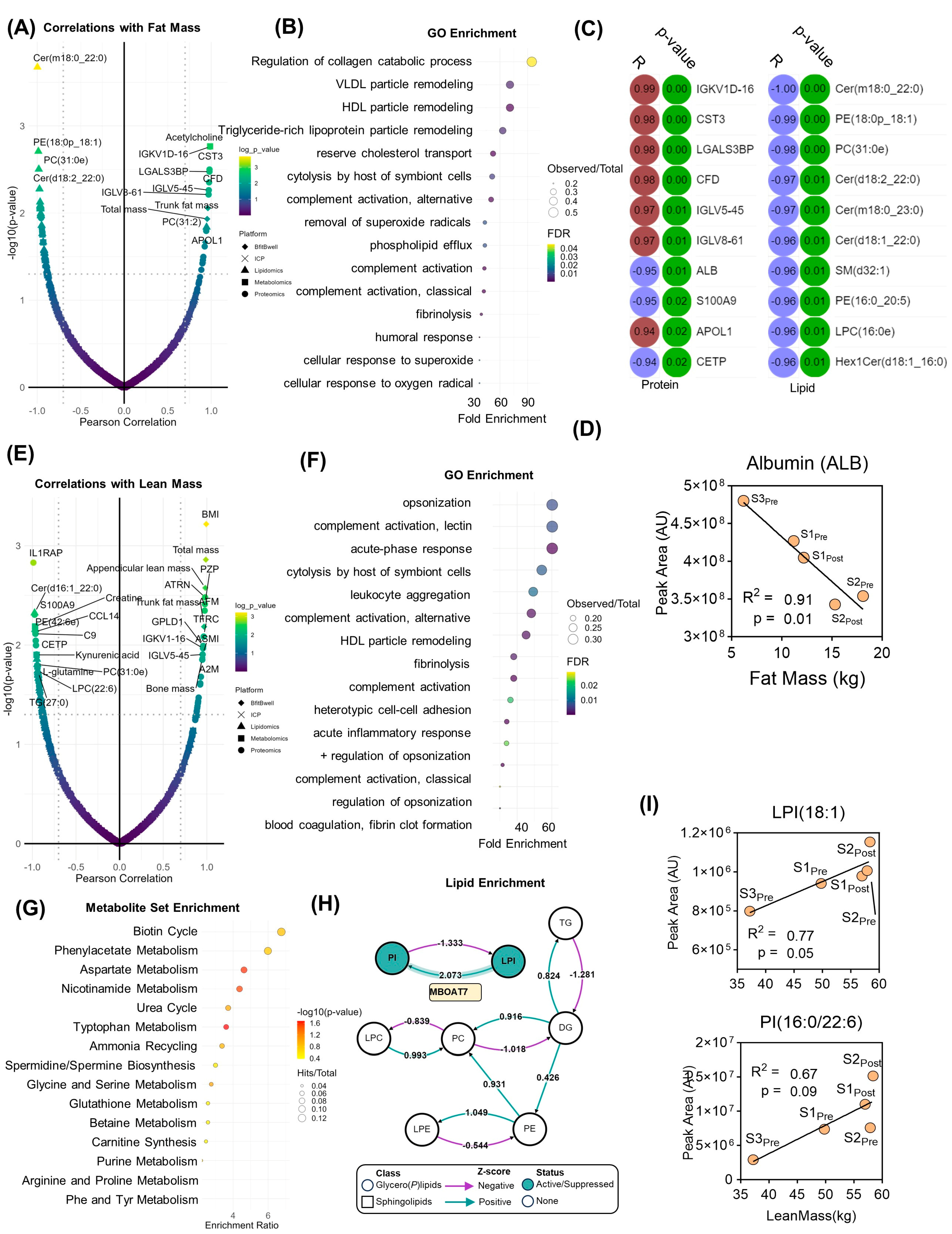
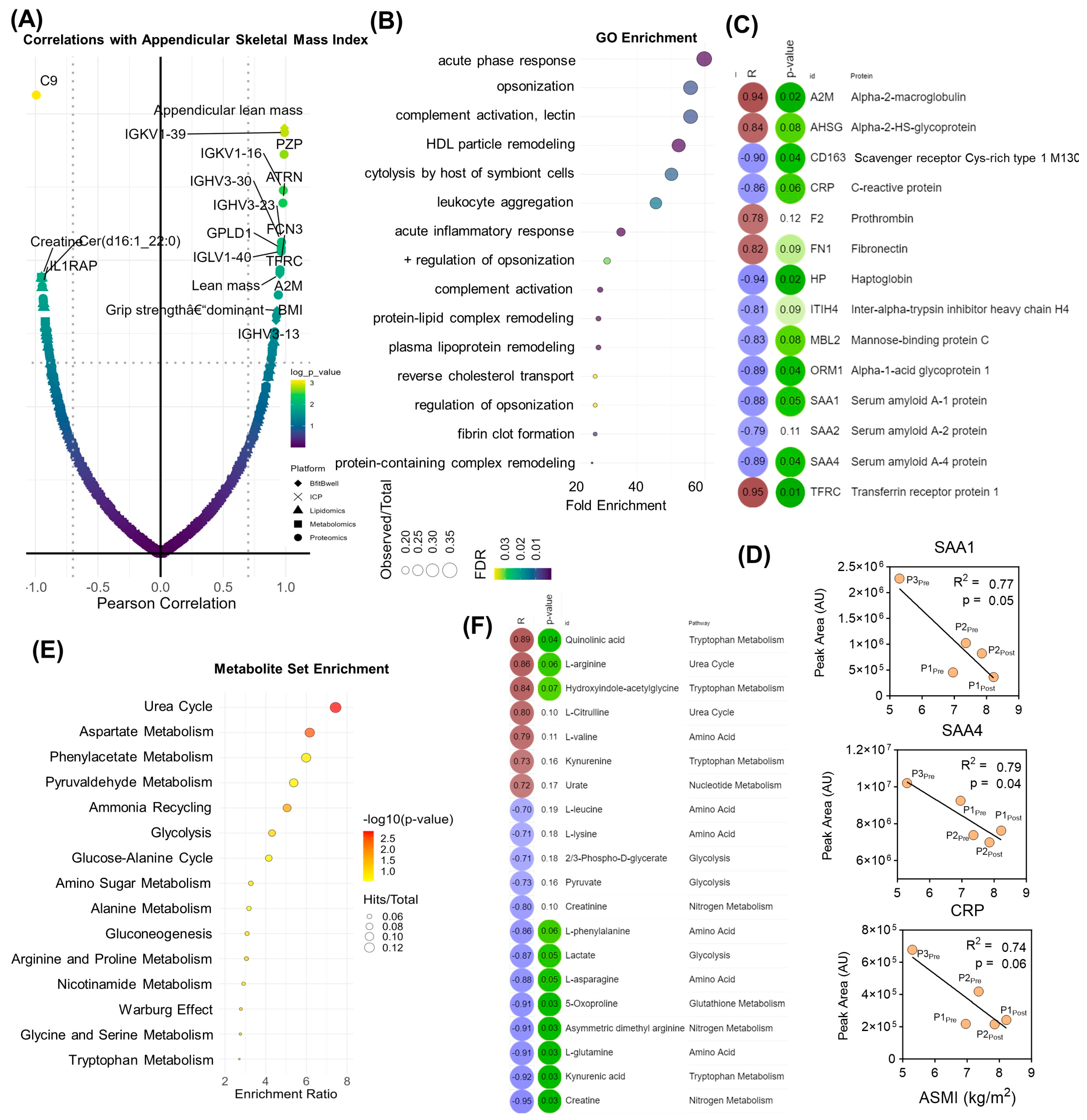
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A2M | Alpha-2-Macroglobulin |
| AHSG | Alpha-2-HS-Glycoprotein |
| AC | Acylcarnitine |
| BMI | Body Mass Index |
| CD14 | Cluster of Differentiation 14 |
| CRP | C-Reactive Protein |
| DBH | Dopamine Beta Hydroxylase |
| DXA | Dual-Energy X-Ray Absorptiometry |
| GO | Gene Ontology |
| HCA | Hierarchical Clustering Analysis |
| HPX | Hemopexin |
| HRNR | Hornerin |
| ICP-MS | Inductively Coupled Plasma Mass Spectrometry |
| IGHV3-20 | Immunoglobulin Heavy Variable 3-20 |
| IGLV5-39 | Immunoglobulin Lambda Variable 5-39 |
| KRT2 | Keratin 2 |
| KRT33B | Keratin 33B |
| KRT6A | Keratin 6A |
| LC-MS | Liquid Chromatography-Mass Spectrometry |
| MBOAT7 | Membrane Bound O-Acyltransferase Domain Containing 7 |
| MSEA | Metabolite Set Enrichment Analysis |
| MWT | Meter Walk Test |
| NAT | Neoadjuvant Treatment |
| NOS | Nitric Oxide Synthase |
| PASC | Post-Acute Sequalae of COVID19 |
| PC | Phosphocholine |
| PCA | Principal Component Analysis |
| PE | Phosphoethanolamine |
| PI | Phosphatidylinositol |
| PLA2G | Phospholipase A2 Group |
| PLS-DA | Partial Least Squares-Discriminant Analysis |
| PLXDC2 | Plexin Domain Containing 2 |
| QCs | Quality Controls |
| RPE | Rating of Perceived Exertion |
| SAA1 | Serum Amyloid A1 |
| SAA2 | Serum Amyloid A2 |
| SDS | Sodium Dodecyl Sulfate |
| SM | Sphingomyelin |
| SMPDB | Small Molecule Pathway Database |
| TEAB | Triethylammonium Bicarbonate |
| TLR4 | Toll Like Receptor 4 |
| UHPLC-MS | Ultra-High Performance Liquid Chromatography-Mass Spectrometry |
| VASN | Vasorin |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Alamo, J.M.; Marin, L.M.; Suarez, G.; Bernal, C.; Serrano, J.; Barrera, L.; Gomez, M.A.; Muntane, J.; Padillo, F.J. Improving outcomes in pancreatic cancer: Key points in perioperative management. World J. Gastroenterol. 2014, 20, 14237–14245. [Google Scholar] [CrossRef] [PubMed]
- Tempero, M.A.; Malafa, M.P.; Behrman, S.W.; Benson, A.B., III; Casper, E.S.; Chiorean, E.G.; Chung, V.; Cohen, S.J.; Czito, B.; Engebretson, A.; et al. Pancreatic adenocarcinoma, version 2.2014: Featured updates to the NCCN guidelines. J. Natl. Compr. Cancer Netw. 2014, 12, 1083–1093. [Google Scholar] [CrossRef]
- Jack, S.; West, M.A.; Raw, D.; Marwood, S.; Ambler, G.; Cope, T.M.; Shrotri, M.; Sturgess, R.P.; Calverley, P.M.; Ottensmeier, C.H.; et al. The effect of neoadjuvant chemotherapy on physical fitness and survival in patients undergoing oesophagogastric cancer surgery. Eur. J. Surg. Oncol. 2014, 40, 1313–1320. [Google Scholar] [CrossRef]
- Cooper, A.B.; Slack, R.; Fogelman, D.; Holmes, H.M.; Petzel, M.; Parker, N.; Balachandran, A.; Garg, N.; Ngo-Huang, A.; Varadhachary, G.; et al. Characterization of Anthropometric Changes that Occur During Neoadjuvant Therapy for Potentially Resectable Pancreatic Cancer. Ann. Surg. Oncol. 2015, 22, 2416–2423. [Google Scholar] [CrossRef]
- Chandrabalan, V.V.; McMillan, D.C.; Carter, R.; Kinsella, J.; McKay, C.J.; Carter, C.R.; Dickson, E.J. Pre-operative cardiopulmonary exercise testing predicts adverse post-operative events and non-progression to adjuvant therapy after major pancreatic surgery. HPB 2013, 15, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Loughney, L.; West, M.A.; Kemp, G.J.; Grocott, M.P.; Jack, S. Exercise intervention in people with cancer undergoing neoadjuvant cancer treatment and surgery: A systematic review. Eur. J. Surg. Oncol. 2016, 42, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Padilha, C.S.; Marinello, P.C.; Galvao, D.A.; Newton, R.U.; Borges, F.H.; Frajacomo, F.; Deminice, R. Evaluation of resistance training to improve muscular strength and body composition in cancer patients undergoing neoadjuvant and adjuvant therapy: A meta-analysis. J. Cancer Surviv. 2017, 11, 339–349. [Google Scholar] [CrossRef]
- Malveiro, C.; Correia, I.R.; Cargaleiro, C.; Magalhães, J.P.; de Matos, L.V.; Hilário, S.; Sardinha, L.B.; Cardoso, M.J. Effects of exercise training on cancer patients undergoing neoadjuvant treatment: A systematic review. J. Sci. Med. Sport 2023, 26, 586–592. [Google Scholar] [CrossRef]
- Parker, N.H.; Gorzelitz, J.; Ngo-Huang, A.; Caan, B.J.; Prakash, L.; Garg, N.; Petzel, M.Q.B.; Schadler, K.; Basen-Engquist, K.; Katz, M.H.G. The Role of Home-Based Exercise in Maintaining Skeletal Muscle During Preoperative Pancreatic Cancer Treatment. Integr. Cancer Ther. 2021, 20, 1534735420986615. [Google Scholar] [CrossRef]
- Parker, N.H.; Ngo-Huang, A.; Lee, R.E.; O’Connor, D.P.; Basen-Engquist, K.M.; Petzel, M.Q.B.; Wang, X.; Xiao, L.; Fogelman, D.R.; Schadler, K.L.; et al. Physical activity and exercise during preoperative pancreatic cancer treatment. Support. Care Cancer 2019, 27, 2275–2284. [Google Scholar] [CrossRef] [PubMed]
- Ngo-Huang, A.; Parker, N.H.; Bruera, E.; Lee, R.E.; Simpson, R.; O’Connor, D.P.; Petzel, M.Q.B.; Fontillas, R.C.; Schadler, K.; Xiao, L.; et al. Home-Based Exercise Prehabilitation During Preoperative Treatment for Pancreatic Cancer Is Associated With Improvement in Physical Function and Quality of Life. Integr. Cancer Ther. 2019, 18, 1534735419894061. [Google Scholar] [CrossRef] [PubMed]
- Bundred, J.R.; Kamarajah, S.K.; Hammond, J.S.; Wilson, C.H.; Prentis, J.; Pandanaboyana, S. Prehabilitation prior to surgery for pancreatic cancer: A systematic review. Pancreatology 2020, 20, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Marker, R.J.; Peters, J.C.; Purcell, W.T.; Jankowski, C.M. Effects of Preoperative Exercise on Physical Fitness and Body Composition in Pancreatic Cancer Survivors Receiving Neoadjuvant Therapy: A Case Series. Rehab Oncol. 2018, 36, E1–E9. [Google Scholar] [CrossRef]
- Nakajima, H.; Yokoyama, Y.; Inoue, T.; Nagaya, M.; Mizuno, Y.; Kadono, I.; Nishiwaki, K.; Nishida, Y.; Nagino, M. Clinical Benefit of Preoperative Exercise and Nutritional Therapy for Patients Undergoing Hepato-Pancreato-Biliary Surgeries for Malignancy. Ann. Surg. Oncol. 2019, 26, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Fearon, K.; Strasser, F.; Anker, S.D.; Bosaeus, I.; Bruera, E.; Fainsinger, R.L.; Jatoi, A.; Loprinzi, C.; MacDonald, N.; Mantovani, G.; et al. Definition and classification of cancer cachexia: An international consensus. Lancet Oncol. 2011, 12, 489–495. [Google Scholar] [CrossRef]
- Contrepois, K.; Wu, S.; Moneghetti, K.J.; Hornburg, D.; Ahadi, S.; Tsai, M.-S.; Metwally, A.A.; Wei, E.; Lee-McMullen, B.; Quijada, J.V.; et al. Molecular Choreography of Acute Exercise. Cell 2020, 181, 1112–1130.e16. [Google Scholar] [CrossRef]
- Jaguri, A.; Al Thani, A.A.; Elrayess, M.A. Exercise Metabolome: Insights for Health and Performance. Metabolites 2023, 13, 694. [Google Scholar] [CrossRef]
- Schranner, D.; Kastenmüller, G.; Schönfelder, M.; Römisch-Margl, W.; Wackerhage, H. Metabolite Concentration Changes in Humans After a Bout of Exercise: A Systematic Review of Exercise Metabolomics Studies. Sports Med. Open 2020, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Davison, G.; Vinaixa, M.; McGovern, R.; Beltran, A.; Novials, A.; Correig, X.; McClean, C. Metabolomic Response to Acute Hypoxic Exercise and Recovery in Adult Males. Front. Physiol. 2018, 9, 1682. [Google Scholar] [CrossRef] [PubMed]
- Nemkov, T.; Cendali, F.; Stefanoni, D.; Martinez, J.L.; Hansen, K.C.; San-Millán, I.; D’Alessandro, A. Metabolic Signatures of Performance in Elite World Tour Professional Male Cyclists. Sports Med. 2023, 53, 1651–1665. [Google Scholar] [CrossRef] [PubMed]
- Cendali, F.; D’Alessandro, A.; Nemkov, T. Dried blood spot characterization of sex-based metabolic responses to acute running exercise. Anal. Sci. Adv. 2023, 4, 37–48. [Google Scholar] [CrossRef]
- San-Millan, I.; Stefanoni, D.; Martinez, J.L.; Hansen, K.C.; D’Alessandro, A.; Nemkov, T. Metabolomics of Endurance Capacity in World Tour Professional Cyclists. Front. Physiol. 2020, 11, 578. [Google Scholar] [CrossRef] [PubMed]
- Huemer, M.T.; Bauer, A.; Petrera, A.; Scholz, M.; Hauck, S.M.; Drey, M.; Peters, A.; Thorand, B. Proteomic profiling of low muscle and high fat mass: A machine learning approach in the KORA S4/FF4 study. J. Cachexia Sarcopenia Muscle 2021, 12, 1011–1023. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Hou, Y.; Yin, M.; Sun, F.; Zhang, T.; Zhou, X.; Li, H.; Zheng, J.; Chen, X.; Li, C.; Ning, X.; et al. A metabolomics approach for predicting the response to neoadjuvant chemotherapy in cervical cancer patients. Mol. Biosyst. 2014, 10, 2126–2133. [Google Scholar] [CrossRef] [PubMed]
- Reisz, J.A.; Zheng, C.; D’Alessandro, A.; Nemkov, T. Untargeted and Semi-targeted Lipid Analysis of Biological Samples Using Mass Spectrometry-Based Metabolomics. Methods Mol. Biol. 2019, 1978, 121–135. [Google Scholar] [CrossRef]
- He, L.; Diedrich, J.; Chu, Y.Y.; Yates, J.R., III. Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Anal. Chem. 2015, 87, 11361–11367. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, D.; Nemkov, T.; Qadri, S.M.; Sheffield, W.P.; D’Alessandro, A. Inductively-Coupled Plasma Mass Spectrometry-Novel Insights From an Old Technology Into Stressed Red Blood Cell Physiology. Front. Physiol. 2022, 13, 828087. [Google Scholar] [CrossRef] [PubMed]
- Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; Hill, D.P. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar]
- Gaud, C.; Sousa, B.C.; Nguyen, A.; Fedorova, M.; Ni, Z.; O’Donnell, V.B.; Wakelam, M.J.; Andrews, S.; Lopez-Clavijo, A.F. BioPAN: A web-based tool to explore mammalian lipidome metabolic pathways on LIPID MAPS. F1000Research 2021, 10, 4. [Google Scholar] [CrossRef]
- Luo, H.; Galvão, D.A.; Newton, R.U.; Lopez, P.; Tang, C.; Fairman, C.M.; Spry, N.; Taaffe, D.R. Exercise Medicine in the Management of Pancreatic Cancer: A Systematic Review. Pancreas 2021, 50, 280–292. [Google Scholar] [CrossRef]
- Kurz, E.; Hirsch, C.A.; Dalton, T.; Shadaloey, S.A.; Khodadadi-Jamayran, A.; Miller, G.; Pareek, S.; Rajaei, H.; Mohindroo, C.; Baydogan, S.; et al. Exercise-induced engagement of the IL-15/IL-15Rα axis promotes anti-tumor immunity in pancreatic cancer. Cancer Cell 2022, 40, 720–737.e5. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Hodgman, C.F.; Schadler, K.L.; LaVoy, E.C. Effect of exercise on pancreatic cancer patients during treatment: A scoping review of the literature. Support. Care Cancer 2022, 30, 5669–5690. [Google Scholar] [CrossRef] [PubMed]
- Florez Bedoya, C.A.; Cardoso, A.C.F.; Parker, N.; Ngo-Huang, A.; Petzel, M.Q.; Kim, M.P.; Fogelman, D.; Romero, S.G.; Wang, H.; Park, M.; et al. Exercise during preoperative therapy increases tumor vascularity in pancreatic tumor patients. Sci. Rep. 2019, 9, 13966. [Google Scholar] [CrossRef]
- Basolo, A.; Ando, T.; Chang, D.C.; Hollstein, T.; Krakoff, J.; Piaggi, P.; Votruba, S. Reduced Albumin Concentration Predicts Weight Gain and Higher Ad Libitum Energy Intake in Humans. Front. Endocrinol. 2021, 12, 642568. [Google Scholar] [CrossRef] [PubMed]
- Ullman, N.A.; Burchard, P.R.; Dunne, R.F.; Linehan, D.C. Immunologic Strategies in Pancreatic Cancer: Making Cold Tumors Hot. J. Clin. Oncol. 2022, 40, 2789–2805. [Google Scholar] [CrossRef]
- Cormie, P.; Zopf, E.M.; Zhang, X.; Schmitz, K.H. The Impact of Exercise on Cancer Mortality, Recurrence, and Treatment-Related Adverse Effects. Epidemiol. Rev. 2017, 39, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Moik, F.; Ay, C. Hemostasis and cancer: Impact of haemostatic biomarkers for the prediction of clinical outcomes in patients with cancer. J. Thromb. Haemost. 2022, 20, 2733–2745. [Google Scholar] [CrossRef]
- Koivula, T.; Lempiäinen, S.; Rinne, P.; Rannikko, J.H.; Hollmén, M.; Sundberg, C.J.; Rundqvist, H.; Minn, H.; Heinonen, I. The effect of acute exercise on circulating immune cells in newly diagnosed breast cancer patients. Sci. Rep. 2023, 13, 6561. [Google Scholar] [CrossRef] [PubMed]
- Guntur, V.P.; Nemkov, T.; de Boer, E.; Mohning, M.P.; Baraghoshi, D.; Cendali, F.I.; San-Millán, I.; Petrache, I.; D’Alessandro, A. Signatures of Mitochondrial Dysfunction and Impaired Fatty Acid Metabolism in Plasma of Patients with Post-Acute Sequelae of COVID-19 (PASC). Metabolites 2022, 12, 1026. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Liang, L.; Gao, X.; Zhang, H.; Yao, P.; Hu, Y.; Ma, Y.; Wang, F.; Jin, Q.; Li, H.; et al. Early Prediction of Developing Type 2 Diabetes by Plasma Acylcarnitines: A Population-Based Study. Diabetes Care 2016, 39, 1563–1570. [Google Scholar] [CrossRef] [PubMed]
- Leach, H.J.; Crisafio, M.E.; Howell, M.J.; Nicklawski, A.; Marker, R.J. A Group-Based, Videoconference-Delivered Physical Activity Program for Cancer Survivors. Transl. J. ACSM 2023, 8, e000221. [Google Scholar] [CrossRef] [PubMed]
- Wonders, K.Y.; Gnau, K.; Schmitz, K.H. Measuring the Feasibility and Effectiveness of an Individualized Exercise Program Delivered Virtually to Cancer Survivors. Curr. Sports Med. Rep. 2021, 20, 271–276. [Google Scholar] [CrossRef]
- Volani, C.; Malfertheiner, C.; Caprioli, G.; Fjelstrup, S.; Pramstaller, P.P.; Rainer, J.; Paglia, G. VAMS-Based Blood Capillary Sampling for Mass Spectrometry-Based Human Metabolomics Studies. Metabolites 2023, 13, 146. [Google Scholar] [CrossRef]
- Molloy, M.P.; Hill, C.; O’Rourke, M.B.; Chandra, J.; Steffen, P.; McKay, M.J.; Pascovici, D.; Herbert, B.R. Proteomic Analysis of Whole Blood Using Volumetric Absorptive Microsampling for Precision Medicine Biomarker Studies. J. Proteome Res. 2022, 21, 1196–1203. [Google Scholar] [CrossRef]
- Marasca, C.; Arana, M.E.B.; Protti, M.; Cavalli, A.; Mercolini, L.; Armirotti, A. Volumetric Absorptive Microsampling of Blood for Untargeted Lipidomics. Molecules 2021, 26, 262. [Google Scholar] [CrossRef]
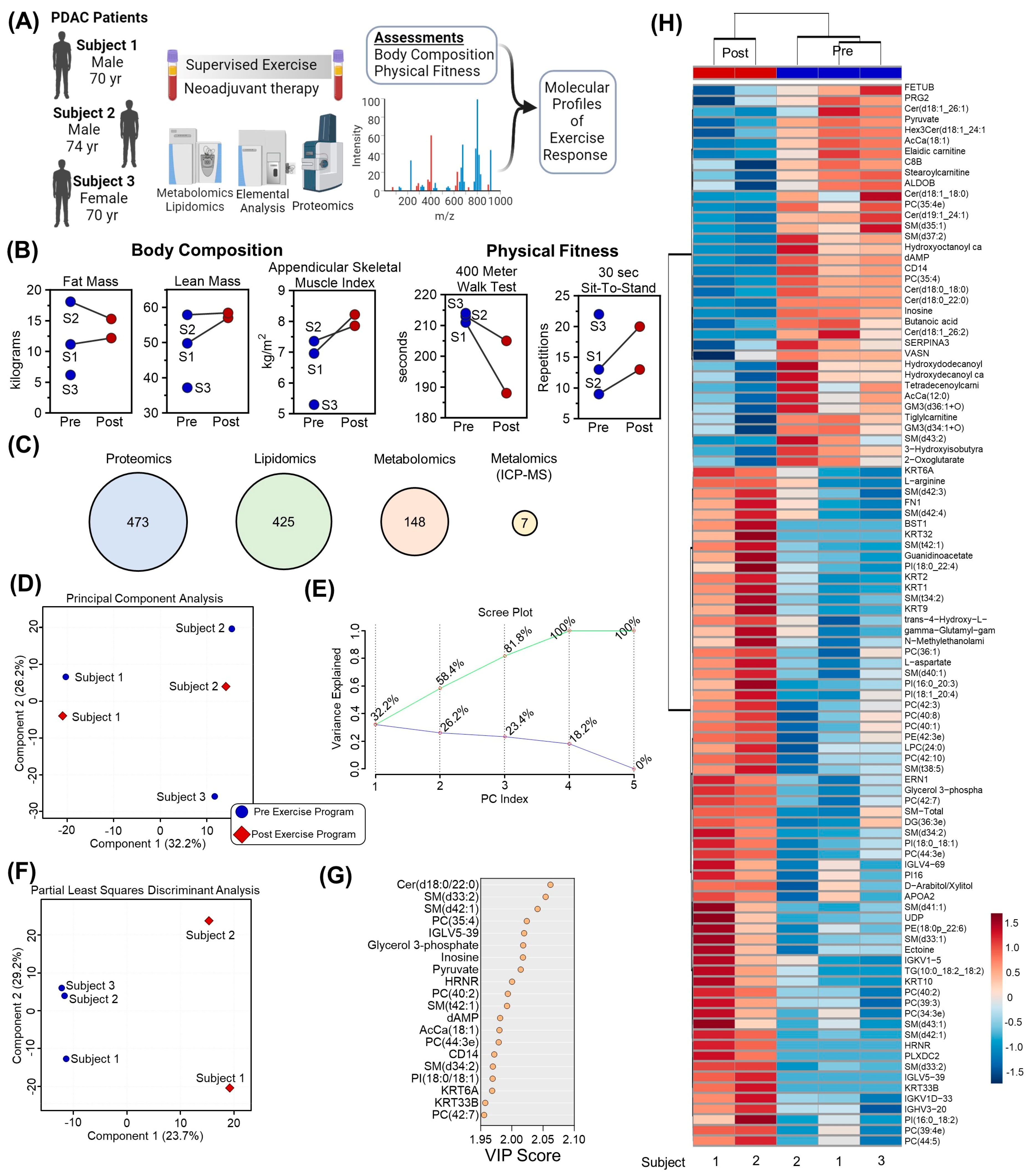


| Participant 1 | Participant 2 | Participant 3 | |
|---|---|---|---|
| Age | 70 years | 74 years | 70 years |
| Sex | Male | Male | Female |
| BMI | 20.8 | 24.8 | 16.3 |
| Length of exercise intervention (weeks) | 17 | 21 | 19 |
| Exercise sessions (N) | 28 | 55 | 46 |
| NAT Description | Four 2-week cycles of FOLFIRINOX followed by 5 treatments with SBRT | Three 4-week cycles of gemcitabine/ABRAXANE followed by 5 treatments with SBRT | Four 2-week cycles of FOLFIRINOX followed by 5 treatments with SBRT |
| Participant 1 | Participant 2 | Participant 3 | ||||||
|---|---|---|---|---|---|---|---|---|
| Baseline | Pre-Surgery | Post-Surgery | Baseline | Pre-Surgery | Post-Surgery | Baseline | Pre-Surgery | |
| 400 MWT (s) | 211 | 188 (+11) | 195 (+8) | 213 | 205 (+4) | 213 (0) | 214 | 190 (+11) |
| 30 s Sit-to-Stand Test | 13 | 20 (+54) | 43.5 (+8) | 9 | 13 (+44) | 10 (+11) | 22 | 22 (0) |
| Total Mass (kg) | 63.5 | 71.8 (+13) | 64.7 (+2) | 78.8 | 76.5 (−3) | 69.2 (−12) | 45.2 | 47.3 (+4) |
| Lean Mass (kg) | 49.8 | 57.0 (+15) | 51.3 (+3) | 57.9 | 58.4 (+1) | 54.1 (−6) | 37.2 | 38.9 (+4) |
| Fat Mass (kg) | 11.2 | 12.2 (+9) | 10.8 (−4) | 18.1 | 15.3 (−15) | 12.5 (−31) | 6.2 | 6.6 (+6) |
| Appendicular Lean Mass (kg) | 21.7 | 25.6 (+18) | 22.7 (+5) | 23.9 | 25.6 (+7) | 22.7 (−5) | 15.1 | 15.5 (+3) |
| ASMI (kg/m2) | 6.96 * | 8.22 (+18) | 7.28 (+6) | 7.36 | 7.86 (+7) | 6.98 * (−5) | 5.3 * | 5.45 (+3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nemkov, T.; Cendali, F.; Dzieciatkowska, M.; Stephenson, D.; Hansen, K.C.; Jankowski, C.M.; D’Alessandro, A.; Marker, R.J. A Multiomics Assessment of Preoperative Exercise in Pancreatic Cancer Survivors Receiving Neoadjuvant Therapy: A Case Series. Pathophysiology 2024, 31, 166-182. https://doi.org/10.3390/pathophysiology31010013
Nemkov T, Cendali F, Dzieciatkowska M, Stephenson D, Hansen KC, Jankowski CM, D’Alessandro A, Marker RJ. A Multiomics Assessment of Preoperative Exercise in Pancreatic Cancer Survivors Receiving Neoadjuvant Therapy: A Case Series. Pathophysiology. 2024; 31(1):166-182. https://doi.org/10.3390/pathophysiology31010013
Chicago/Turabian StyleNemkov, Travis, Francesca Cendali, Monika Dzieciatkowska, Daniel Stephenson, Kirk C. Hansen, Catherine M. Jankowski, Angelo D’Alessandro, and Ryan J. Marker. 2024. "A Multiomics Assessment of Preoperative Exercise in Pancreatic Cancer Survivors Receiving Neoadjuvant Therapy: A Case Series" Pathophysiology 31, no. 1: 166-182. https://doi.org/10.3390/pathophysiology31010013
APA StyleNemkov, T., Cendali, F., Dzieciatkowska, M., Stephenson, D., Hansen, K. C., Jankowski, C. M., D’Alessandro, A., & Marker, R. J. (2024). A Multiomics Assessment of Preoperative Exercise in Pancreatic Cancer Survivors Receiving Neoadjuvant Therapy: A Case Series. Pathophysiology, 31(1), 166-182. https://doi.org/10.3390/pathophysiology31010013






