Multiple Antimicrobial Resistance and Heavy Metal Tolerance of Biofilm-Producing Bacteria Isolated from Dairy and Non-Dairy Food Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Specimen Collection and Processing
2.3. Isolation and Characterization of Bacteria
2.4. Antimicrobial Resistance Testing
2.5. Biofilm Formation Assay
2.6. Heavy Metal Tolerance Testing
2.7. Statistical Analysis
3. Results
3.1. Microbial Count in Different Food Types
3.2. Distribution of Bacterial Isolates in Food Products
3.3. Antimicrobial Resistance of Bacterial Isolates
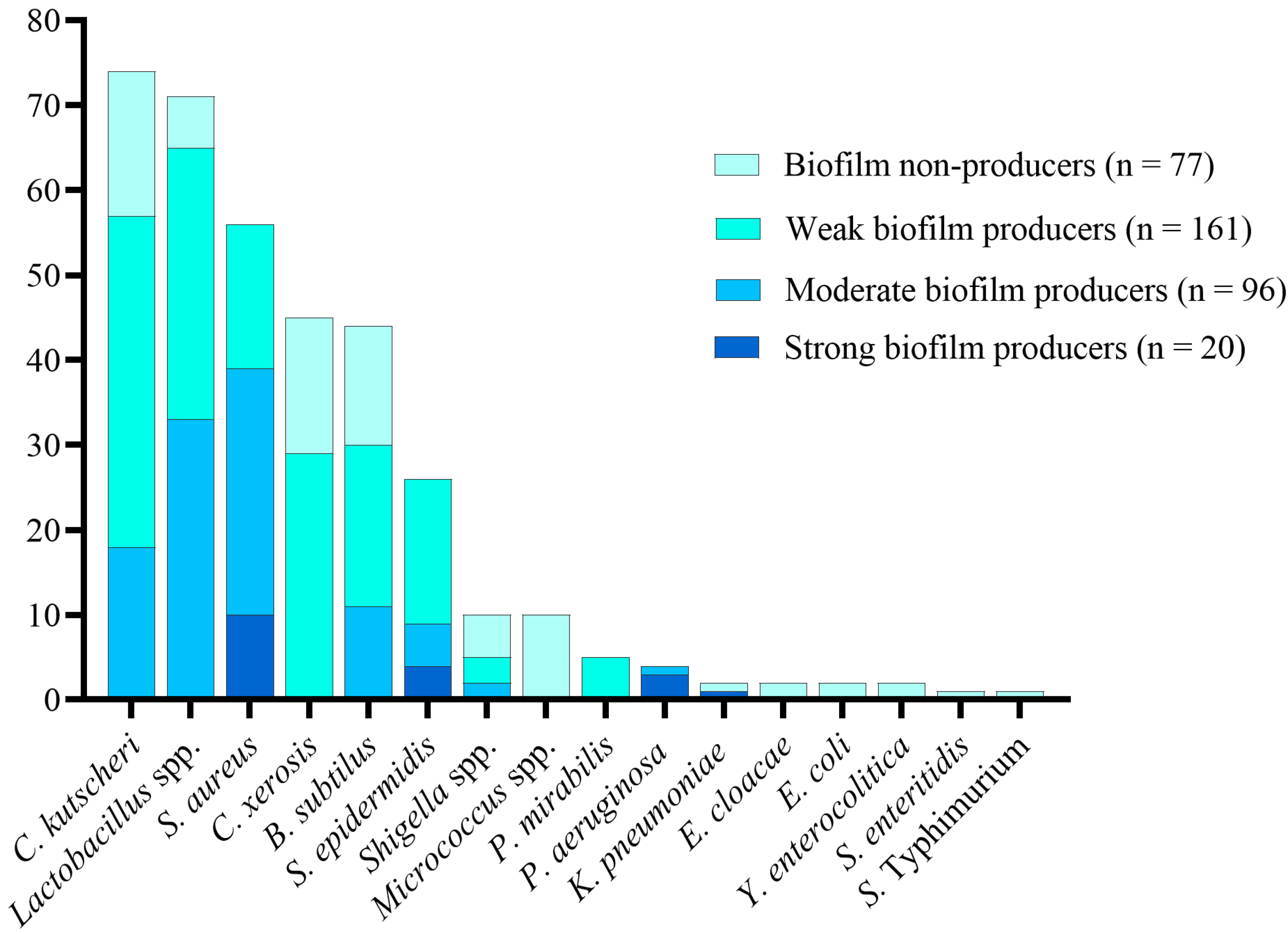
3.4. Biofilm Production in Bacterial Isolates
3.5. Relationship between AMR and Biofilm-Producing and Biofilm-Non-Producing Isolates
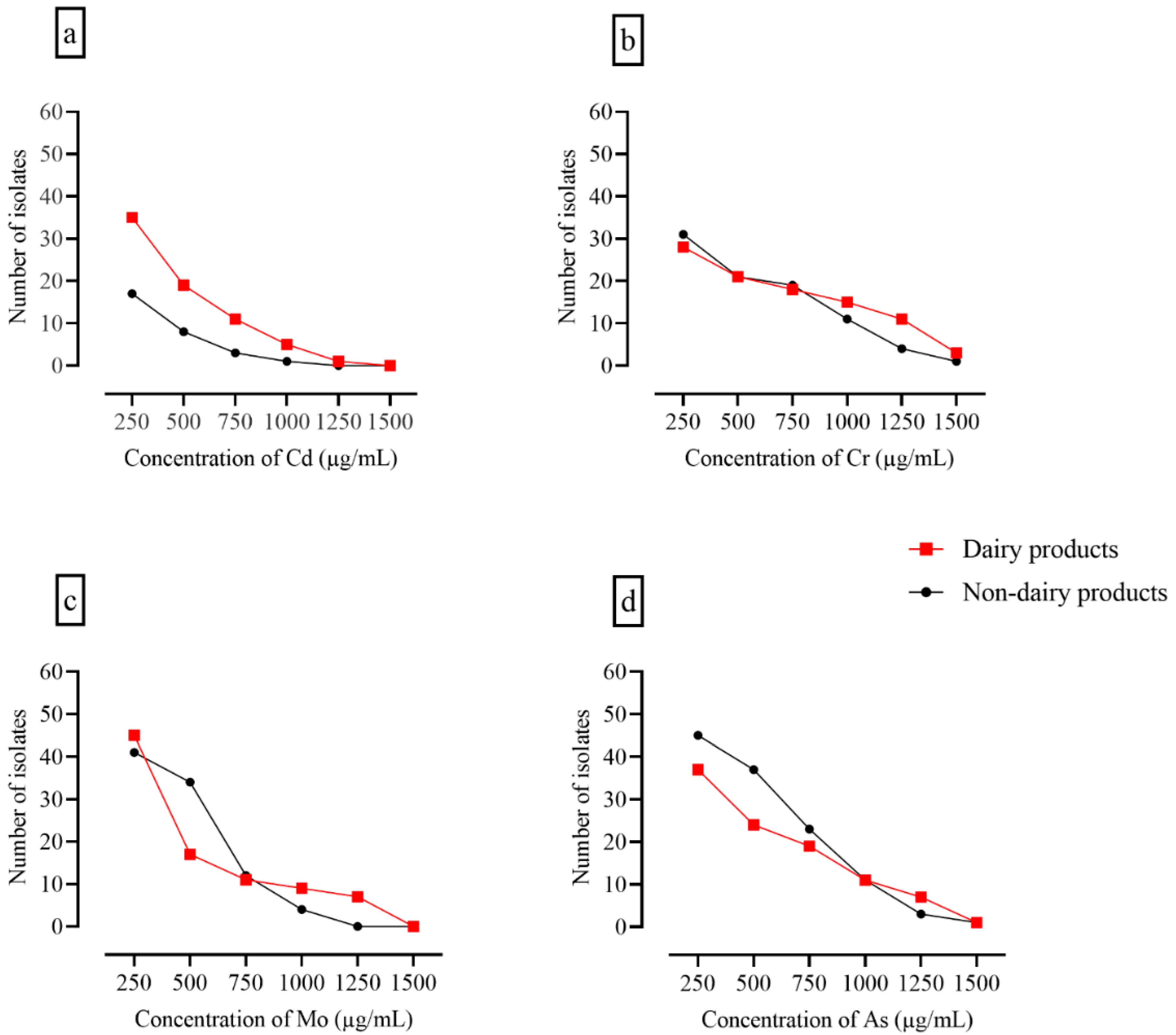
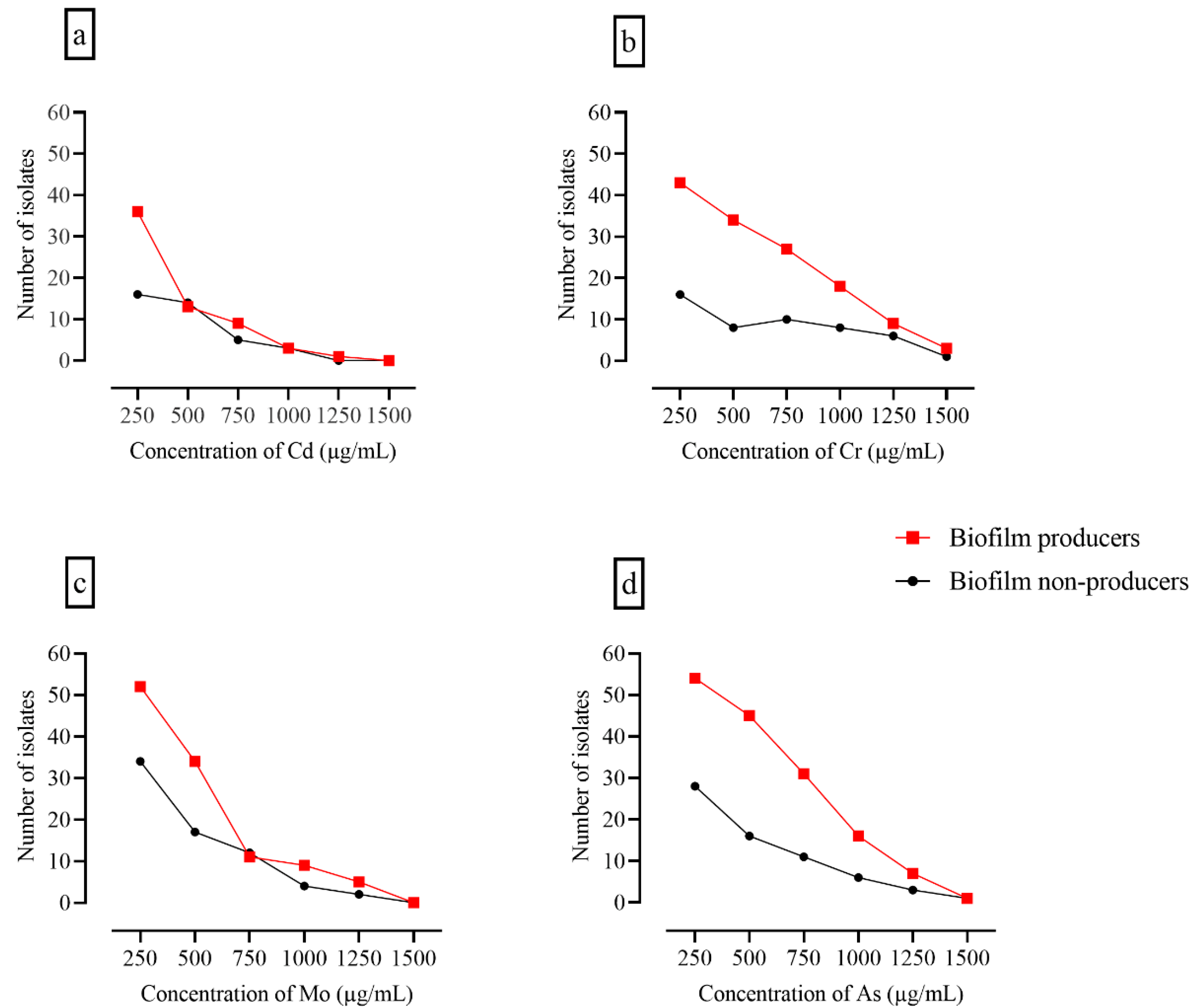
3.6. Assessment of Heavy Metal Tolerance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bedasa, S.; Shiferaw, D.; Abraha, A.; Moges, T. RETRACTED ARTICLE: Occurrence and antimicrobial susceptibility profile of Escherichia coli O157:H7 from food of animal origin in Bishoftu town, Central Ethiopia. Int. J. Food Contam. 2018, 5, 2. [Google Scholar] [CrossRef]
- WHO. Food Safety. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 23 August 2022).
- Zhao, X.; Zhong, J.; Wei, C.; Lin, C.W.; Ding, T. Current Perspectives on Viable but Non-culturable State in Foodborne Pathogens. Front. Microbiol. 2017, 8, 580. [Google Scholar] [CrossRef] [PubMed]
- Moosavy, M.; Mahmoudi, R.; Ghorbanpour, F.E.; Khatibi, S. Evaluation of microbial and physicochemical characteristics of raw cow milk delivered to pasteurized milk plants in Tabriz city, Iran. J. Food Res. 2018, 28, 183–196. [Google Scholar]
- Sugrue, I.; Tobin, C.; Ross, R.P.; Stanton, C.; Hill, C. Foodborne pathogens and zoonotic diseases. In Raw Milk; Elsevier: Amsterdam, The Netherlands, 2019; pp. 259–272. [Google Scholar]
- Moosavy, M.; Kordasht, H.K.; Khatibi, S.; Sohrabi, H. Assessment of the chemical adulteration and hygienic quality of raw cow milk in the northwest of Iran. Qual. Assur. Saf. Crops Foods 2019, 11, 491–498. [Google Scholar] [CrossRef]
- Van Hoorde, K.; Butler, F. Use of next-generation sequencing in microbial risk assessment. EFSA J. 2018, 16, e16086. [Google Scholar] [CrossRef]
- Lindqvist, R.; Sylvén, S.; Vågsholm, I. Quantitative microbial risk assessment exemplified by Staphylococcus aureus in unripened cheese made from raw milk. Int. J. Food Microbiol. 2002, 78, 155–170. [Google Scholar] [CrossRef]
- Gwida, M.M.; Al-Ashmawy, M.A. Culture versus PCR for Salmonella Species Identification in Some Dairy Products and Dairy Handlers with Special Concern to Its Zoonotic Importance. Vet. Med. Int. 2014, 2014, 502370. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Cerda-Leal, F.; Contreras, A.; Valenzuela-Riffo, N.; Rodríguez, A.; Aguirre, J. Cronobacter sakazakii and Microbiological Parameters in Dairy Formulas Associated with a Food Alert in Chile. Front. Microbiol. 2018, 9, 1708. [Google Scholar] [CrossRef]
- CDC. CDC and Food Safety. Available online: https://www.cdc.gov/foodsafety/cdc-and-food-safety.html#:~:text=Foodborne%20illness%20is%20common%2C%20costly,than%20%2415.6%20billion%20each%20year (accessed on 19 July 2022).
- Koutsoumanis, K.P.; Lianou, A.; Sofos, J.N. Food safety: Emerging pathogens. Encycl. Agric. Food Syst. 2014, 250–272. [Google Scholar]
- Ou, C.; Shang, D.; Yang, J.; Chen, B.; Chang, J.; Jin, F.; Shi, C. Prevalence of multidrug-resistant Staphylococcus aureus isolates with strong biofilm formation ability among animal-based food in Shanghai. Food Control 2020, 112, 107106. [Google Scholar] [CrossRef]
- Dutra, T.V.; Fernandes, M.d.S.; Perdoncini, M.R.F.G.; Anjos, M.M.d.; Abreu Filho, B.A.d. Capacity of Escherichia coli and Staphylococcus aureus to produce biofilm on stainless steel surfaces in the presence of food residues. J. Food Processing Preserv. 2018, 42, e13574. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Bhardwaj, D.K.; Taneja, N.K.; Shivaprasad, D.; Chakotiya, A.; Patel, P.; Taneja, P.; Sachdev, D.; Gupta, S.; Sanal, M.G. Phenotypic and genotypic characterization of biofilm forming, antimicrobial resistant, pathogenic Escherichia coli isolated from Indian dairy and meat products. Int. J. Food Microbiol. 2021, 336, 108899. [Google Scholar] [CrossRef] [PubMed]
- Srey, S.; Jahid, I.K.; Ha, S.-D. Biofilm formation in food industries: A food safety concern. Food Control 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Furukawa, S. Studies on formation, control and application of biofilm formed by food related microorganisms. Biosci. Biotechnol. Biochem. 2015, 79, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Yang, L.; Chen, G.; Li, B.; Chen, D.; Li, L.; Xu, Z. Pathogenic features and characteristics of food borne pathogens biofilm: Biomass, viability and matrix. Microb. Pathog. 2017, 111, 285–291. [Google Scholar] [CrossRef]
- Qin, G.; Niu, Z.; Yu, J.; Li, Z.; Ma, J.; Xiang, P. Soil heavy metal pollution and food safety in China: Effects, sources and removing technology. Chemosphere 2021, 267, 129205. [Google Scholar] [CrossRef]
- Junaid, K.; Ejaz, H.; Asim, I.; Younas, S.; Yasmeen, H.; Abdalla, A.E.; Abosalif, K.O.A.; Alameen, A.A.M.; Ahmad, N.; Bukhari, S.N.A.; et al. Heavy Metal Tolerance Trend in Extended-Spectrum β-Lactamase Encoding Strains Recovered from Food Samples. Int J. Environ. Res. Public Health 2021, 18, 4718. [Google Scholar] [CrossRef]
- Dweba, C.C.; Zishiri, O.T.; El Zowalaty, M.E. Isolation and Molecular Identification of Virulence, Antimicrobial and Heavy Metal Resistance Genes in Livestock-Associated Methicillin-Resistant Staphylococcus aureus. Pathogens 2019, 8, 79. [Google Scholar] [CrossRef]
- Roberts, D.; Greenwood, M. Practical Food Microbiology; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Ema, F.A.; Shanta, R.N.; Rahman, M.Z.; Islam, M.A.; Khatun, M.M. Isolation, identification, and antibiogram studies of Escherichia coli from ready-to-eat foods in Mymensingh, Bangladesh. Vet. World 2022, 15, 1497. [Google Scholar] [CrossRef]
- Amare, A.; Worku, T.; Ashagirie, B.; Adugna, M.; Getaneh, A.; Dagnew, M. Bacteriological profile, antimicrobial susceptibility patterns of the isolates among street vended foods and hygienic practice of vendors in Gondar town, Northwest Ethiopia: A cross sectional study. BMC Microbiol. 2019, 19, 120. [Google Scholar] [CrossRef] [PubMed]
- Katz, S. Making a Streak Plate. Available online: https://asm.org/Protocols/The-Streak-Plate-Protocol (accessed on 30 August 2022).
- Holt, J.G. The Shorter Bergey’s Manual of Determinative Bacteriology; Williams & Wilkins Co.: Baltimore, MD, USA, 1977. [Google Scholar]
- Bari, A.; Zeeshan, F.; Zafar, A.; Ejaz, H.; Iftikhar, A.; Rathore, A.W. Childhood Acute Bacterial Meningitis: Clinical Spectrum, Bacteriological Profile and Outcome. J. Coll. Physicians Surg. Pak. 2016, 26, 822–826. [Google Scholar] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; Clinical and Laboratory Standard Institute (CLSI): Wayne, PA, USA, 2022; Volume CLSI supplement M100. [Google Scholar]
- Biswas, I.; Mettlach, J. A Simple Static Biofilm Assay for Acinetobacter baumannii. Methods Mol. Biol. 2019, 1946, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Ebert, C.; Tuchscherr, L.; Unger, N.; Pöllath, C.; Gladigau, F.; Popp, J.; Löffler, B.; Neugebauer, U. Correlation of crystal violet biofilm test results of Staphylococcus aureus clinical isolates with Raman spectroscopic read-out. J. Raman Spectrosc. 2021, 52, 2660–2670. [Google Scholar] [CrossRef]
- Stepanovic, S.; Vukovic, D.; Dakic, I.; Savic, B.; Svabic-Vlahovic, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Xu, Z.; Liang, Y.; Lin, S.; Chen, D.; Li, B.; Li, L.; Deng, Y. Crystal Violet and XTT Assays on Staphylococcus aureus Biofilm Quantification. Curr. Microbiol. 2016, 73, 474–482. [Google Scholar] [CrossRef]
- Adekanmbi, A.O.; Falodun, O.I. Heavy metals and antibiotics susceptibility profiles of Staphylococcus aureus isolated from several points receiving daily input from the Bodija abattoir in Ibadan, Oyo State, Nigeria. Adv. Microbiol. 2015, 5, 871. [Google Scholar] [CrossRef]
- Liang, G.; Gong, W.; Li, B.; Zuo, J.; Pan, L.; Liu, X. Analysis of Heavy Metals in Foodstuffs and an Assessment of the Health Risks to the General Public via Consumption in Beijing, China. Int J Environ Res Public Health 2019, 16, 909. [Google Scholar] [CrossRef]
- Hassani, S.; Moosavy, M.H.; Gharajalar, S.N.; Khatibi, S.A.; Hajibemani, A.; Barabadi, Z. High prevalence of antibiotic resistance in pathogenic foodborne bacteria isolated from bovine milk. Sci. Rep. 2022, 12, 3878. [Google Scholar] [CrossRef]
- Tadesse, H.A.; Gidey, N.B.; Workelule, K.; Hailu, H.; Gidey, S.; Bsrat, A.; Taddele, H. Antimicrobial Resistance Profile of E. coli Isolated from Raw Cow Milk and Fresh Fruit Juice in Mekelle, Tigray, Ethiopia. Vet. Med. Int. 2018, 2018, 8903142. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.F.; Urdaneta, A.G.; Corser, P.I.; Cagnasso, M.A.; Leal, K.V. Isolation of Gram-positive bacteria from raw milk with antimicrobial residues. Arch. Latinoam. Nutr. 2002, 52, 68–73. [Google Scholar]
- Fischer-Tenhagen, C.; Theby, V.; Krömker, V.; Heuwieser, W. Detecting Staphylococcus aureus in milk from dairy cows using sniffer dogs. J. Dairy Sci. 2018, 101, 4317–4324. [Google Scholar] [CrossRef] [PubMed]
- Earl, A.M.; Losick, R.; Kolter, R. Ecology and genomics of Bacillus subtilis. Trends Microbiol. 2008, 16, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Özer, B.; Yaman, H. Milk and milk. In Encyclopedia of Food Microbiology, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 721–727. [Google Scholar] [CrossRef]
- Singh, V.K.; Utaida, S.; Jackson, L.S.; Jayaswal, R.K.; Wilkinson, B.J.; Chamberlain, N.R. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology 2007, 153, 3162–3173. [Google Scholar] [CrossRef]
- Montanari, C.; Serrazanetti, D.I.; Felis, G.; Torriani, S.; Tabanelli, G.; Lanciotti, R.; Gardini, F. New insights in thermal resistance of staphylococcal strains belonging to the species Staphylococcus epidermidis, Staphylococcus lugdunensis and Staphylococcus aureus. Food Control 2015, 50, 605–612. [Google Scholar] [CrossRef]
- Haque, Z.F.; Sabuj, A.A.M.; Mahmud, M.M.; Pondit, A.; Islam, M.A.; Saha, S. Characterization of Staphylococcus aureus from milk and dairy products sold in some local markets of Mymensingh district of Bangladesh. J. Nutr. Food Sci. 2018, 8, 1000743. [Google Scholar] [CrossRef]
- Chen, Q.; Xie, S.; Lou, X.; Cheng, S.; Liu, X.; Zheng, W.; Zheng, Z.; Wang, H. Biofilm formation and prevalence of adhesion genes among Staphylococcus aureus isolates from different food sources. MicrobiologyOpen 2020, 9, e00946. [Google Scholar] [CrossRef]
- Amao, H.; Akimoto, T.; Komukai, Y.; Sawada, T.; Saito, M.; Takahashi, K.W. Detection of Corynebacterium kutscheri from the oral cavity of rats. Exp. Anim. 2002, 51, 99–102. [Google Scholar] [CrossRef]
- Jung, D.; Rubin, J.E. Identification of antimicrobial resistant bacteria from plant-based food products imported into Canada. Int. J. Food Microbiol. 2020, 319, 108509. [Google Scholar] [CrossRef]
- Sapkota, S.; Adhikari, S.; Pandey, A.; Khadka, S.; Adhikari, M.; Kandel, H.; Pathak, S.; Pandey, A. Multi-drug resistant extended-spectrum beta-lactamase producing E. coli and Salmonella on raw vegetable salads served at hotels and restaurants in Bharatpur, Nepal. BMC Res. Notes 2019, 12, 516. [Google Scholar] [CrossRef] [PubMed]
- van Hoek, A.H.; Veenman, C.; van Overbeek, W.M.; Lynch, G.; de Roda Husman, A.M.; Blaak, H. Prevalence and characterization of ESBL- and AmpC-producing Enterobacteriaceae on retail vegetables. Int. J. Food Microbiol. 2015, 204, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Wu, Q.; Zhang, S.; Zhang, J.; Yang, G.; Wang, H.; Huang, J.; Chen, M.; Xue, L.; Wang, J. Antibiotic-Resistant Extended Spectrum ß-Lactamase- and Plasmid-Mediated AmpC-Producing Enterobacteriaceae Isolated from Retail Food Products and the Pearl River in Guangzhou, China. Front. Microbiol. 2017, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef]
- Di Domenico, E.G.; Farulla, I.; Prignano, G.; Gallo, M.T.; Vespaziani, M.; Cavallo, I.; Sperduti, I.; Pontone, M.; Bordignon, V.; Cilli, L.; et al. Biofilm is a Major Virulence Determinant in Bacterial Colonization of Chronic Skin Ulcers Independently from the Multidrug Resistant Phenotype. Int. J. Mol. Sci. 2017, 18, 1077. [Google Scholar] [CrossRef]
- Radovanovic, R.S.; Savic, N.R.; Ranin, L.; Smitran, A.; Opavski, N.V.; Tepavcevic, A.M.; Ranin, J.; Gajic, I. Biofilm Production and Antimicrobial Resistance of Clinical and Food Isolates of Pseudomonas spp. Curr. Microbiol. 2020, 77, 4045–4052. [Google Scholar] [CrossRef]
- Marek, A.; Pyzik, E.; Stępień-Pyśniak, D.; Dec, M.; Jarosz, Ł.S.; Nowaczek, A.; Sulikowska, M. Biofilm-Formation Ability and the Presence of Adhesion Genes in Coagulase-Negative Staphylococci Isolates from Chicken Broilers. Animals 2021, 11, 728. [Google Scholar] [CrossRef]
- Senobar Tahaei, S.A.; Stájer, A.; Barrak, I.; Ostorházi, E.; Szabó, D.; Gajdács, M. Correlation Between Biofilm-Formation and the Antibiotic Resistant Phenotype in Staphylococcus aureus Isolates: A Laboratory-Based Study in Hungary and a Review of the Literature. Infect. Drug Resist. 2021, 14, 1155–1168. [Google Scholar] [CrossRef]
- Pavlickova, S.; Klancnik, A.; Dolezalova, M.; Mozina, S.S.; Holko, I. Antibiotic resistance, virulence factors and biofilm formation ability in Escherichia coli strains isolated from chicken meat and wildlife in the Czech Republic. J. Environ. Sci. Health B 2017, 52, 570–576. [Google Scholar] [CrossRef]
- Karmakar, R.; Bindiya, S.; Hariprasad, P. Convergent evolution in bacteria from multiple origins under antibiotic and heavy metal stress, and endophytic conditions of host plant. Sci. Total Environ. 2019, 650, 858–867. [Google Scholar] [CrossRef]
- Khan, M.U.; Malik, R.N.; Muhammad, S.; Ullah, F.; Qadir, A. Health risk assessment of consumption of heavy metals in market food crops from Sialkot and Gujranwala Districts, Pakistan. Hum. Ecol. Risk Assess. Int. J. 2015, 21, 327–337. [Google Scholar] [CrossRef]
- Akhtar, S. Food safety challenges—A Pakistan’s perspective. Crit. Rev. Food Sci. Nutr. 2015, 55, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, M.; Ahmad, G.; Anjum, F.; Asghar, A.; Sajid, M. Heavy metal contents and their daily intake in vegetables under peri-urban farming system of Multan, Pakistan. Pak. J. Agric. Sci. 2014, 51, 1025–1031. [Google Scholar]
- Iftikhar, B.; Arif, S.; Siddiqui, S.; Khattak, R. Assessment of Toxic Metals in Dairy Milk and Animal Feed in Peshawar, Pakistan. Br. Biotechnol. J. 2014, 4, 883–893. [Google Scholar] [CrossRef]
- Harrison, J.J.; Ceri, H.; Turner, R.J. Multimetal resistance and tolerance in microbial biofilms. Nat. Rev. Microbiol. 2007, 5, 928–938. [Google Scholar] [CrossRef]

| Type of Food Sample | Number (%) of Bacterial Isolates (n = 357) | CFU/mL (log) Mean ± SD | Number (%) of Gram-Positive Isolates (n = 328) | Number (%) of Gram-Negative Isolates (n = 29) | |
|---|---|---|---|---|---|
| Dairy food samples (n = 100) | Liquid (n = 44) | 180 (50.4%) | 2.9 ± 0.9 | 180 (54.9%) | 0 (0%) |
| Solid (n = 20) | |||||
| Semisolid (n = 18) | |||||
| Powdered (n = 18) | |||||
| Non-dairy food samples (n = 100) | Liquid (n = 20) | 177 (49.6%) | 5.1 ± 0.3 | 148 (45.1%) | 29 (100%) |
| Solid (n = 43) | |||||
| Semisolid (n = 11) | |||||
| Powdered (n = 26) | |||||
| Isolated Bacteria | Number (%) of Isolates from Dairy Food Products (n = 180) | Number (%) of Isolates from Non-Dairy Food Products (n = 177) | |
|---|---|---|---|
| Gram-positive isolates (n = 328) | Corynebacterium kutscheri (n = 74) | - | 74 (41.8%) |
| Staphylococcus aureus (n = 56) | 30 (16.7%) | 26 (14.7%) | |
| Lactobacillus spp. (n = 73) | 45 (25%) | 28 (15.8%) | |
| Corynebacterium xerosis (n = 45) | 33 (18.4%) | 12 (6.8%) | |
| Bacillus subtilis (n = 44) | 44 (24.4%) | - | |
| Staphylococcus epidermidis (n = 26) | 18 (10%) | 8 (4.5%) | |
| Micrococcus spp. (n = 10) | 10 (5.5) | - | |
| Gram-negative isolates (n = 29) | Shigella spp. (n = 10) | - | 10 (5.6%) |
| Proteus mirabilis (n = 5) | - | 5 (2.8%) | |
| Pseudomonas aeruginosa (n = 4) | - | 4 (2.2%) | |
| Klebsiella pneumoniae (n = 2) | - | 2 (1.1%) | |
| Enterobacter cloacae (n = 2) | - | 2 (1.1%) | |
| Escherichia coli (n = 2) | - | 2 (1.1%) | |
| Yersinia enterocolitica (n = 2) | - | 2 (1.1%) | |
| Salmonella Enteritidis (n = 1) | - | 1 (0.5%) | |
| Salmonella Typhimurium (n = 1) | - | 1 (0.5%) | |
| Biofilm Type | Isolates from Dairy Food Products (n = 180) | Isolates from Non-Dairy Food Products (n = 177) | |
| Biofilm non-producers (n = 77) | 50 (27.8%) | 27 (15.7%) | |
| Biofilm producers (n = 280; 78.4%) | Weak (n = 161; 57.5%) | 81 (45%) | 80 (44.9%) |
| Moderate (n = 99; 35.4%) | 43 (23.9%) | 56 (31.5%) | |
| Strong (n = 20; 7.1%) | 6 (3.3%) | 14 (7.9%) | |
| Antibiotic | Total Number of Resistant Isolates | Number of (%) Biofilm Producers (n = 280) | Number (%) of Biofilm Non-Producers (n = 77) | p-Value |
|---|---|---|---|---|
| Ampicillin | 281 | 207 (73.7%) | 74 (26.3%) | 0.002 |
| Cefotaxime | 157 | 135 (86%) | 22 (14%) | 0.001 |
| Cefuroxime | 154 | 117 (76%) | 37 (24%) | 0.26 |
| Ceftazidime | 143 | 87 (60.8%) | 56 (39.2%) | 0.01 |
| Ceftriaxone | 136 | 98 (72.1%) | 38 (27.9%) | 0.01 |
| Amikacin | 125 | 64 (51.2%) | 61 (48.8%) | <0.001 |
| Gentamicin | 114 | 66 (57.9%) | 48 (42.1%) | 0.001 |
| Co-trimoxazole | 107 | 71 (66.4%) | 36 (33.6%) | 0.001 |
| Aztreonam | 103 | 61 (59.2%) | 42 (40.8%) | <0.001 |
| Levofloxacin | 73 | 71 (97.3%) | 2 (2.7%) | <0.001 |
| Cefepime | 69 | 65 (94.2%) | 4 (5.8%) | 0.01 |
| Ciprofloxacin | 68 | 62 (91.2%) | 6 (8.8%) | 0.01 |
| Cefoxitin | 67 | 50 (74.6%) | 17 (25.4%) | 0.27 |
| Imipenem | 63 | 56 (88.9%) | 7 (11.1%) | 0.05 |
| Meropenem | 62 | 53 (85.5%) | 9 (14.5%) | 0.23 |
| Piperacillin-tazobactam | 50 | 46 (92%) | 4 (8%) | 0.02 |
| Tigecycline | 28 | 22 (78.6%) | 6 (21.4%) | 0.67 |
| Colistin | 11 | 9 (81.8%) | 2 (18.2%) | 0.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ejaz, H.; Junaid, K.; Yasmeen, H.; Naseer, A.; Alam, H.; Younas, S.; Qamar, M.U.; Abdalla, A.E.; Abosalif, K.O.A.; Ahmad, N.; et al. Multiple Antimicrobial Resistance and Heavy Metal Tolerance of Biofilm-Producing Bacteria Isolated from Dairy and Non-Dairy Food Products. Foods 2022, 11, 2728. https://doi.org/10.3390/foods11182728
Ejaz H, Junaid K, Yasmeen H, Naseer A, Alam H, Younas S, Qamar MU, Abdalla AE, Abosalif KOA, Ahmad N, et al. Multiple Antimicrobial Resistance and Heavy Metal Tolerance of Biofilm-Producing Bacteria Isolated from Dairy and Non-Dairy Food Products. Foods. 2022; 11(18):2728. https://doi.org/10.3390/foods11182728
Chicago/Turabian StyleEjaz, Hasan, Kashaf Junaid, Humaira Yasmeen, Amina Naseer, Hafsa Alam, Sonia Younas, Muhammad Usman Qamar, Abualgasim E. Abdalla, Khalid O. A. Abosalif, Naveed Ahmad, and et al. 2022. "Multiple Antimicrobial Resistance and Heavy Metal Tolerance of Biofilm-Producing Bacteria Isolated from Dairy and Non-Dairy Food Products" Foods 11, no. 18: 2728. https://doi.org/10.3390/foods11182728







