The Rabep1-Mediated Endocytosis and Activation of Trypsinogen to Promote Pancreatic Stellate Cell Activation
Abstract
:1. Introduction
2. Methods and Materials
2.1. Clinical Samples
2.2. Reagents and Chemicals
Nafamostate and Bafilomycin-A1
2.3. Cell Culture and Treatment
2.4. Immunofluorescence
2.5. Datasets and Bioinformatics Analysis
2.6. GeneMANIA
2.7. Cell Transfection
2.8. Western Blot
2.9. Flow Cytometry
2.10. Statistical Methods
3. Results
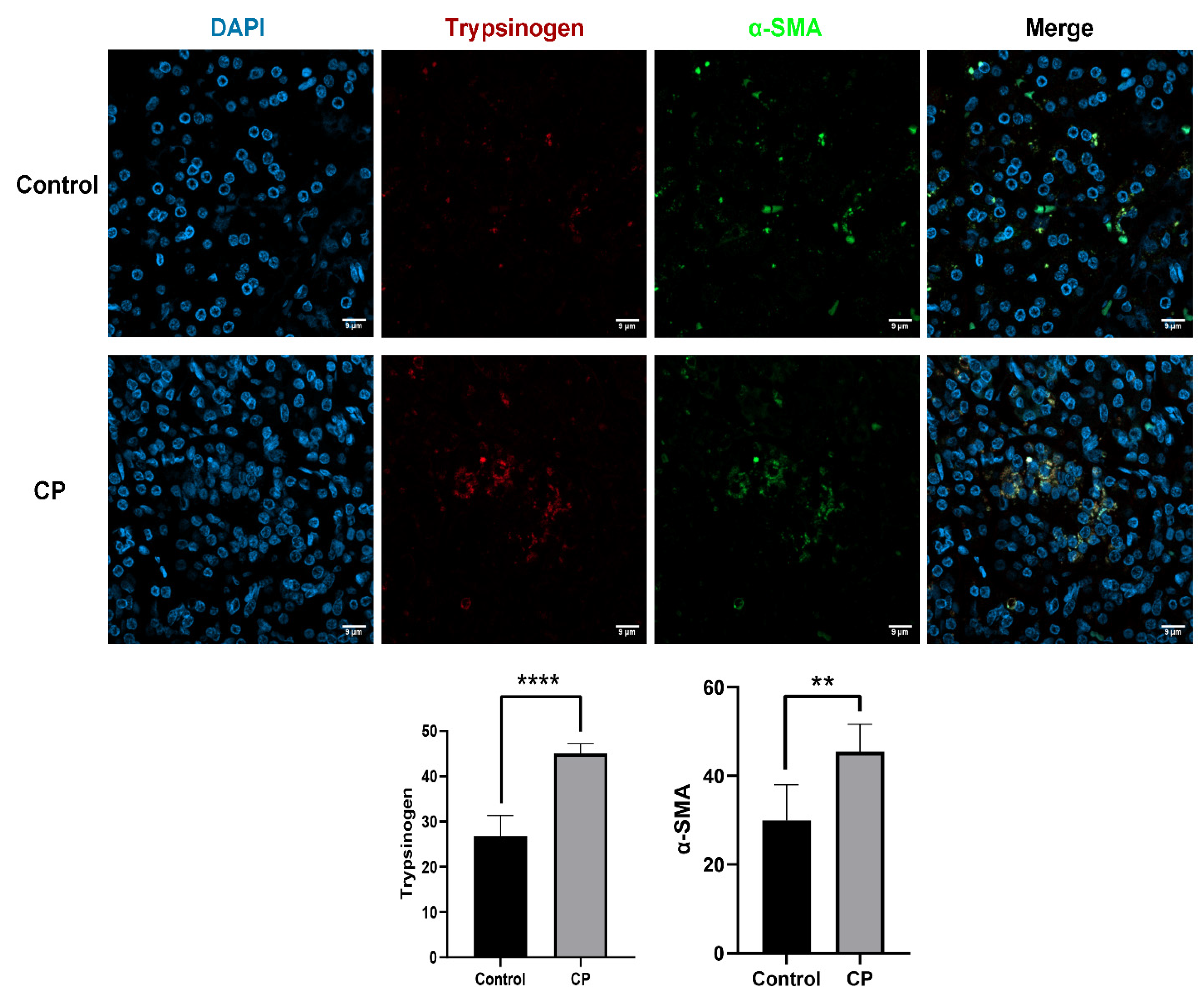
3.1. Endocytosis of Trypsinogen in PSCs Occurred in CP Samples
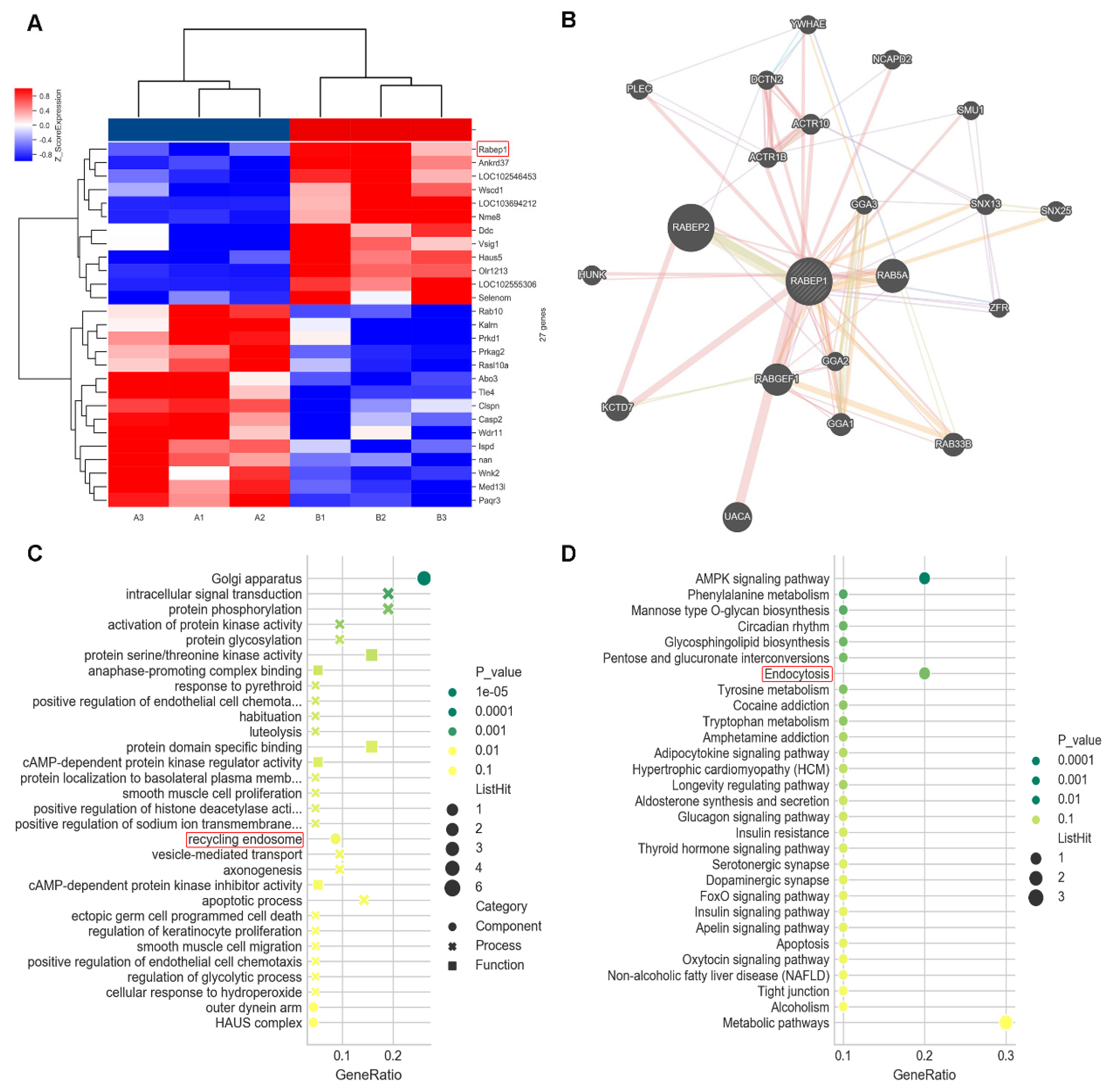
3.2. Bioinformatics Screening and Analysis in Microarray
3.3. Endocytosis Pathway Confirmed in scRNA-seq
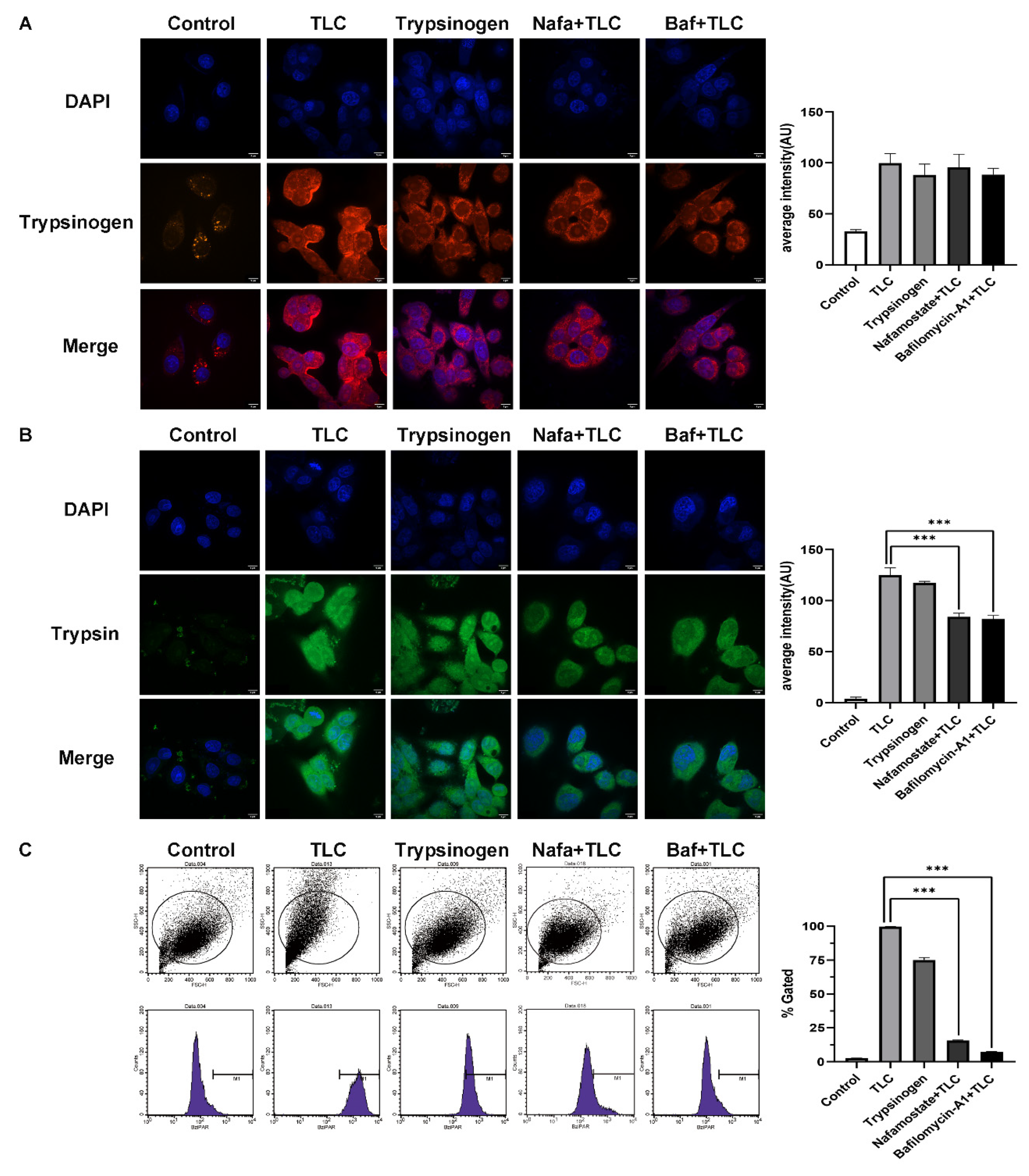
3.4. Regulation of Trypsinogen Endocytosis by Rabep1 in PSC
3.5. Detection of Trypsinogen Endocytosis and Activation in PSCs
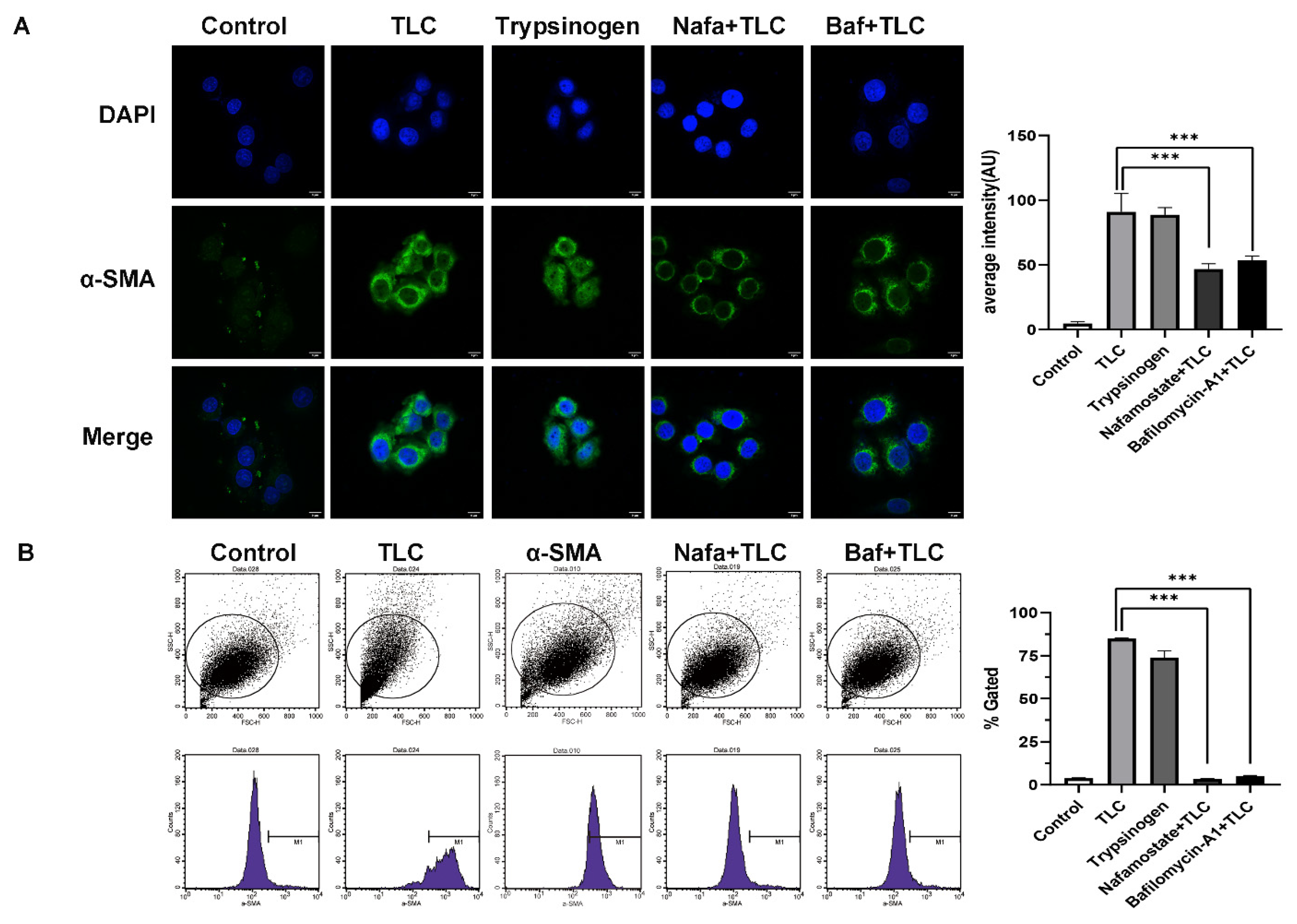
3.6. Activated Trypsinogen Provides Another Way to Activate PSCs
3.7. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kleeff, J.; Whitcomb, D.C.; Shimosegawa, T.; Esposito, I.; Markus, M.L.; Gress, T.; Julia, M.; Drewes, A.M.; Rebours, V.; Fatih Akisik, J.; et al. Chronic pancreatitis. Nat. Reviews. Dis. Primers. 2017, 3, 17060. [Google Scholar] [CrossRef] [PubMed]
- Sendler, M.; Maertin, S.; John, D.; Maria, P.; Ulrich, W.F.; Krüger, B.; Wartmann, T.; Wagh, P.; Halangk, W.; Schaschke, N.; et al. Cathepsin B Activity Initiates Apoptosis via Digestive Protease Activation in Pancreatic Acinar Cells and Experimental Pancreatitis. J. Biol. Chemistry 2016, 291, 14717–14731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apte, M.; Pirola, R.; Wilson, J. The fibrosis of chronic pancreatitis: New insights into the role of pancreatic stellate cells. Antioxid. Redox Signal. 2011, 15, 2711–2722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omary, M.B.; Lugea, A.; Lowe, A.W.; Pandol, S.J. The pancreatic stellate cell: A star on the rise in pancreatic diseases. J. Clin. Investigation 2007, 117, 50–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bynigeri, R.R.; Jakkampudi, A.; Jangala, R.; Subramanyam, C.; Sasikala, M.; Rao, G.V.; Nageshwar, D.R.; Rupjyoti, T. Pancreatic stellate cell: Pandora’s box for pancreatic disease biology. World J. Gastroenterol. 2017, 23, 382–405. [Google Scholar] [CrossRef]
- Sendler, M.; Weiss, F.U.; Golchert, J.; Homuth, G.; den Brandt, C.; Mahajan, U.M.; Partecke, L.; Döring, P.; Gukovsky, I.; Gukovskaya, A.S.G.; et al. Cathepsin B-Mediated Activation of Trypsinogen in Endocytosing Macrophages Increases Severity of Pancreatitis in Mice. Gastroenterology 2018, 154, 704–718.e710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef]
- Stenmark, H.; Vitale, G.; Ullrich, O.; Zerial, M. Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell 1995, 83, 423–432. [Google Scholar] [CrossRef] [Green Version]
- Zeigerer, A.; Gilleron, J.; Bogorad, R.L.; Marsico, G.; Nonaka, H.; Seifert, S.; Zerial, M. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature 2012, 485, 465–470. [Google Scholar] [CrossRef]
- Yang, X.; Yan, F.; He, Z.; Liu, S.; Cheng, Y.; Wei, K.; Xiang, S. ITSN2L Interacts with and Negatively Regulates RABEP1. Int. J. Mol. Sciences 2015, 16, 28242–28254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Homma, S.; Hayashi, K.; Yoshida, K.; Sagawa, Y.; Kamata, Y.; Ito, M. Nafamostat mesilate, a serine protease inhibitor, suppresses interferon-gamma-induced up-regulation of programmed cell death ligand 1 in human cancer cells. Int. Immunopharmacol. 2018, 54, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.S.; Huh, K.R.; Kim, Y.J.; Kim Kyoung Oh, M.D.; Park Cheol Hee, M.D.; Hahn Taeho, M.D.; Park Sang Hoon, M.D.; Kim Jong Hyeok, M.D.; Park Choong Kee, M.D.; Kwon Young-Jun, M.D.; et al. Nafamostat mesilate for prevention of post-endoscopic retrograde cholangiopancreatography pancreatitis: A prospective, randomized, double-blind, controlled trial. Pancreas 2011, 40, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Akshintala, V.S.; Hutfless, S.M.; Colantuoni, E.; Kim, K.J.; Khashab, M.A.; Li, T.; Elmunzer, B.J.; Puhan, M.A.; Sinha, A.; Kamal, A.; et al. Systematic review with network meta-analysis: Pharmacological prophylaxis against post-ERCP pancreatitis. Aliment. Pharm. Ther. 2013, 38, 1325–1337. [Google Scholar] [CrossRef] [Green Version]
- Gammella, E.; Lomoriello, I.S.; Conte, A.; Stefano, F.; Alessandra, A.; Maura, P.; Sara, S.; Gaetano, C.; Stefania, R. Unconventional endocytosis and trafficking of transferrin receptor induced by iron. Mol. Biol. Cell. 2021, 32, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Mauthe, M.; Orhon, I.; Rocchi, C.; Zhou, X.; Luhr, M.; Hijlkema, K.; Coppes, R.P.; Engedal, N.; Mari, M.; Reggiori, F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy 2018, 14, 1435–1455. [Google Scholar] [CrossRef] [PubMed]
- Mauvezin, C.; Neufeld, T.P. Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy 2015, 11, 1437–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Wen, L.; Zhang, R.; Wei, Z.; Shi, N.; Xiong, Q.; Xia, Q.; Xing, Z.; Zeng, Z.; Niu, H.; et al. Dihydrodiosgenin protects against experimental acute pancreatitis and associated lung injury through mitochondrial protection and PI3Kγ/Akt inhibition. Br. J. Pharmacol. 2018, 175, 1621–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zha, Z.Q.; You, S.; Hu, Y.H.; Zhang, F.; Chen, Y.W.; Wang, J. Asn57 N-glycosylation promotes the degradation of hemicellulose by β-1,3-1,4-glucanase from Rhizopus homothallicus. Environ. Sci. Pollut. Res. Int. 2022. Advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Christian, T.L.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Sherwood, M.W.; Prior, I.A.; Voronina, S.G.; Barrow, S.L.; Woodsmith, J.D.; Gerasimenko, O.V.; Petersen, O.H.; Tepikin, A.V. Activation of trypsinogen in large endocytic vacuoles of pancreatic acinar cells. Proc. Natl. Acad. Sci. USA 2007, 104, 5674–5679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, B.; Wu, L.; Lu, M.; Gao, B.; Qiao, X.; Sun, B.; Xue, D.; Zhang, W. Differentially expressed kinase genes associated with trypsinogen activation in rat pancreatic acinar cells treated with taurolithocholic acid 3-sulfate. Mol. Med. Rep. 2013, 7, 1591–1596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, S.; Nakata, K.; Ohuchida, K.; Takesue, S.; Nakayama, H.; Abe, T.; Koikawa, K.; Okumura, T.; Sada, M.; Horioka, K.; et al. Autophagy Is Required for Activation of Pancreatic Stellate Cells, Associated With Pancreatic Cancer Progression and Promotes Growth of Pancreatic Tumors in Mice. Gastroenterology 2017, 152, 1492–1506.e1424. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Wang, G.; Hu, J.S.; Zhang, G.; Chen, H.; Yue, Y.; Yi-Long, L.; Xin-Jian, L.; Feng-Yu, T.; Shang-Ha, P. RB1CC1-enhanced autophagy facilitates PSCs activation and pancreatic fibrogenesis in chronic pancreatitis. Cell Death Dis. 2018, 9, 952. [Google Scholar] [CrossRef] [PubMed]
- Mace, T.A.; Ameen, Z.; Collins, A.; Wojcik, S.; Mair, M.; Young, G.S.; Fuchs, J.R.; Eubank, T.D.; Frankel, W.L.; Bekaii-Saab, T.; et al. Pancreatic cancer-associated stellate cells promote differentiation of myeloid-derived suppressor cells in a STAT3-dependent manner. Cancer Res. 2013, 73, 3007–3018. [Google Scholar] [CrossRef] [Green Version]
- Jin, G.; Hong, W.; Guo, Y.; Yongheng, B.; Bicheng, C. Molecular Mechanism of Pancreatic Stellate Cells Activation in Chronic Pancreatitis and Pancreatic Cancer. J. Cancer 2020, 11, 1505–1515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, X.; Wan, J.; Zhang, G.; Song, L.; Gui, F.; Zhang, Y.; Li, Y.; Guo, J.; Dawra, R.K.; Saluja, A.K.; et al. Elevated intracellular trypsin exacerbates acute pancreatitis and chronic pancreatitis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 316, G816–g825. [Google Scholar] [CrossRef] [PubMed]
- Geisz, A.; Sahin-Tóth, M. A preclinical model of chronic pancreatitis driven by trypsinogen autoactivation. Nat. Commun. 2018, 9, 5033. [Google Scholar] [CrossRef] [Green Version]
- Masamune, A.; Kikuta, K.; Satoh, M.; Noriaki, S.; Tooru, S. Protease-activated receptor-2-mediated proliferation and collagen production of rat pancreatic stellate cells. J. Pharmacol. Exp. Ther. 2005, 312, 651–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaksonen, M.; Roux, A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell. Biol. 2018, 19, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Hinze, C.; Boucrot, E. Endocytosis in proliferating, quiescent and terminally differentiated cells. J. Cell Sci. 2018, 131, jcs216804. [Google Scholar] [CrossRef] [Green Version]
- Dilsizoglu Senol, A.; Tagliafierro, L.; Gorisse-Hussonnois, L.; Rebeillard, F.; Huguet, L.; Geny, D.; Allinquant, B. Protein interacting with Amyloid Precursor Protein tail-1 (PAT1) is involved in early endocytosis. Cell Mol. Life Sci. 2019, 76, 4995–5009. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Roche, O.; Yan, M.S.; Finak, G.; Evans, A.J.; Metcalf, J.L.; Hast, B.E.; Hanna, S.C.; Wondergem, B.; Furge, K.A. Regulation of endocytosis via the oxygen-sensing pathway. Nat. Med. 2009, 15, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Dolai, S.; Takahashi, T.; Qin, T.; Liang, T.; Xie, L.; Kang, F.; Miao, Y.; Xie, H.; Kang, Y.; Manuel, J. Pancreas-specific SNAP23 depletion prevents pancreatitis by attenuating pathological basolateral exocytosis and formation of trypsin-activating autolysosomes. Autophagy 2020, 1–14, 3068–3081. [Google Scholar] [CrossRef] [PubMed]
- Haeberle, L.; Steiger, K.; Schlitter, A.M.; Safi, S.A.; Knoefel, W.T.; Erkan, M.; Esposito, I. Stromal heterogeneity in pancreatic cancer and chronic pancreatitis. Pancreatology 2018, 18, 536–549. [Google Scholar] [CrossRef] [PubMed]


| PIN | Primer Name | Sequence (5′ to 3′) | Length | MW (g/mol) | Tm °C | GC % | Purification |
|---|---|---|---|---|---|---|---|
| A2276696 | Rabep1 (Norway rat) siRNA-1962 | CAGAAGAGCUGGUGAGGUUTT | 21 | 6789.21 | 52.4 | 47.6 | HPLC |
| A2276697 | AACCUCACCAGCUCUUCUGTT | 21 | 6525.99 | 52.4 | 47.6 | HPLC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, W.; Luo, D.; Lv, Z.; Yang, Y.; Wang, L.; Ma, B.; Xue, D.; Hao, C.; Zhang, Y. The Rabep1-Mediated Endocytosis and Activation of Trypsinogen to Promote Pancreatic Stellate Cell Activation. Biomolecules 2022, 12, 1063. https://doi.org/10.3390/biom12081063
Yao W, Luo D, Lv Z, Yang Y, Wang L, Ma B, Xue D, Hao C, Zhang Y. The Rabep1-Mediated Endocytosis and Activation of Trypsinogen to Promote Pancreatic Stellate Cell Activation. Biomolecules. 2022; 12(8):1063. https://doi.org/10.3390/biom12081063
Chicago/Turabian StyleYao, Wenchao, Dankun Luo, Zhenyi Lv, Yang Yang, Liyi Wang, Biao Ma, Dongbo Xue, Chenjun Hao, and Yingmei Zhang. 2022. "The Rabep1-Mediated Endocytosis and Activation of Trypsinogen to Promote Pancreatic Stellate Cell Activation" Biomolecules 12, no. 8: 1063. https://doi.org/10.3390/biom12081063
APA StyleYao, W., Luo, D., Lv, Z., Yang, Y., Wang, L., Ma, B., Xue, D., Hao, C., & Zhang, Y. (2022). The Rabep1-Mediated Endocytosis and Activation of Trypsinogen to Promote Pancreatic Stellate Cell Activation. Biomolecules, 12(8), 1063. https://doi.org/10.3390/biom12081063





