Early Prediction of Planning Adaptation Requirement Indication Due to Volumetric Alterations in Head and Neck Cancer Radiotherapy: A Machine Learning Approach
Abstract
Simple Summary
Abstract
1. Introduction
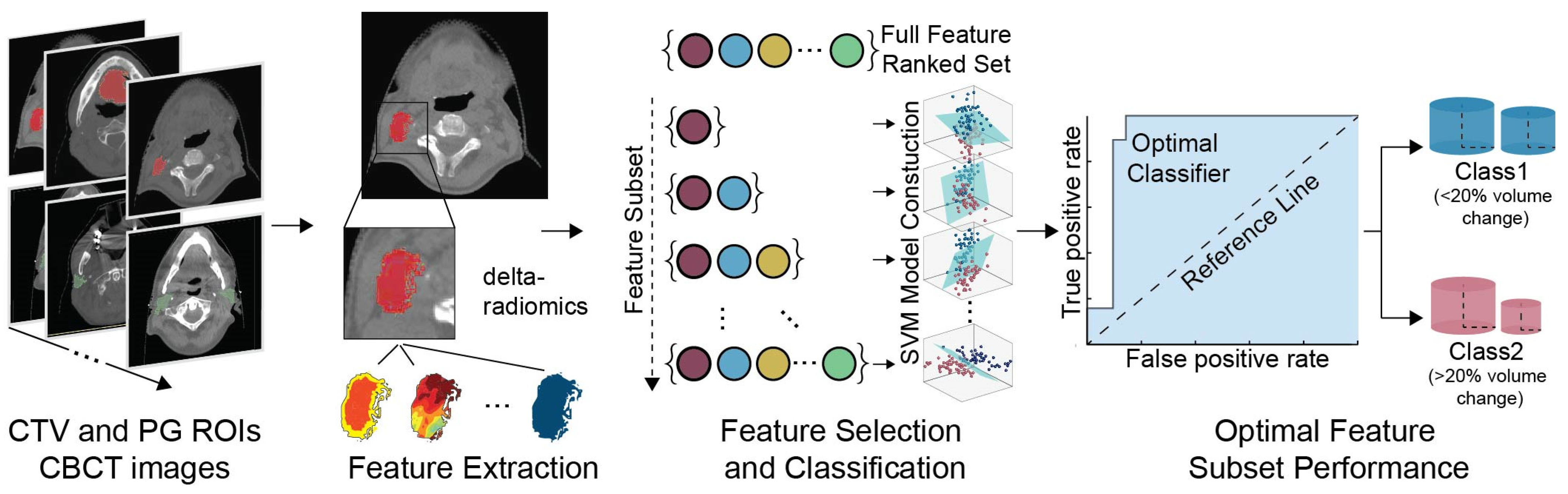
2. Materials and Methods
2.1. Patients
2.2. Data Pre-Processing and Class Designation
2.3. Feature Extraction
2.4. Feature Selection
2.5. Classification
3. Results
3.1. Classification Performance
3.2. Delta-Radiomics Features
3.3. Model Stability Evaluation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Argiris, A.; Karamouzis, M.V.; Raben, D.; Ferris, R.L. Head and Neck Cancer. Lancet 2008, 371, 1695–1709. [Google Scholar] [CrossRef]
- Kong, V.C.; Marshall, A.; Chan, H.B. Cone Beam Computed Tomography: The Challenges and Strategies in Its Application for Dose Accumulation. J. Med. Imaging Radiat. Sci. 2016, 47, 92–97. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mnejja, W.; Daoud, H.; Fourati, N.; Sahnoun, T.; Siala, W.; Farhat, L.; Daoud, J. Dosimetric Impact on Changes in Target Volumes during Intensity-Modulated Radiotherapy for Nasopharyngeal Carcinoma. Rep. Pract. Oncol. Radiother. 2020, 25, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Kaur, I.; Rawat, S.; Ahlawat, P.; Kakria, A.; Gupta, G.; Saxena, U.; Mishra, M. Dosimetric Impact of Setup Errors in Head and Neck Cancer Patients Treated by Image-Guided Radiotherapy. J. Med. Phys. 2016, 41, 144–148. [Google Scholar] [CrossRef]
- Pota, M.; Scalco, E.; Sanguineti, G.; Farneti, A.; Cattaneo, G.M.; Rizzo, G.; Esposito, M. Early Prediction of Radiotherapy-Induced Parotid Shrinkage and Toxicity Based on CT Radiomics and Fuzzy Classification. Artif. Intell. Med. 2017, 81, 41–53. [Google Scholar] [CrossRef]
- Heukelom, J.; Kantor, M.E.; Mohamed, A.S.R.; Elhalawani, H.; Kocak-Uzel, E.; Lin, T.; Yang, J.; Aristophanous, M.; Rasch, C.R.; Fuller, C.D.; et al. Differences between Planned and Delivered Dose for Head and Neck Cancer, and Their Consequences for Normal Tissue Complication Probability and Treatment Adaptation. Radiother. Oncol. 2020, 142, 100–106. [Google Scholar] [CrossRef]
- Figen, M.; Çolpan Öksüz, D.; Duman, E.; Prestwich, R.; Dyker, K.; Cardale, K.; Ramasamy, S.; Murray, P.; Şen, M. Radiotherapy for Head and Neck Cancer: Evaluation of Triggered Adaptive Replanning in Routine Practice. Front. Oncol. 2020, 10, 579917. [Google Scholar] [CrossRef]
- Iliadou, V.; Economopoulos, T.L.; Karaiskos, P.; Kouloulias, V.; Platoni, K.; Matsopoulos, G.K. Deformable Image Registration to Assist Clinical Decision for Radiotherapy Treatment Adaptation for Head and Neck Cancer Patients. Biomed. Phys. Eng. Express 2021, 7, 055012. [Google Scholar] [CrossRef]
- Karaca, S.; Kırlı, M. Adaptive Radiation Therapy for Cervical Esophageal Cancer: Dosimetric and Volumetric Analysis. J. Gastrointest. Oncol. 2019, 10, 506–512. [Google Scholar] [CrossRef]
- Green, O.L.; Henke, L.E.; Hugo, G.D. Practical Clinical Workflows for Online and Offline Adaptive Radiation Therapy. Semin. Radiat. Oncol. 2019, 29, 219–227. [Google Scholar] [CrossRef]
- Sonke, J.-J.; Aznar, M.; Rasch, C. Adaptive Radiotherapy for Anatomical Changes. Semin. Radiat. Oncol. 2019, 29, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xue, J.; Chen, T.; Qu, T.; Barbee, D.; Tam, M.; Hu, K. Adaptive Radiotherapy Based on Statistical Process Control for Oropharyngeal Cancer. J. Appl. Clin. Med. Phys. 2020, 21, 171–177. [Google Scholar] [CrossRef]
- Tanooka, M.; Doi, H.; Ishida, T.; Kitajima, K.; Wakayama, T.; Sakai, T.; Inoue, H.; Kotoura, N.; Kosaka, K.; Tarutani, K.; et al. Usability of Deformable Image Registration for Adaptive Radiotherapy in Head and Neck Cancer and an Automatic Prediction of Replanning. Int. J. Med. Phys. Clin. Eng. Radiat. Oncol. 2017, 6, 10–20. [Google Scholar] [CrossRef][Green Version]
- Kavanaugh, J.; Roach, M.; Ji, Z.; Fontenot, J.; Hugo, G.D. A Method for Predictive Modeling of Tumor Regression for Lung Adaptive Radiotherapy. Med. Phys. 2021, 48, 2083–2094. [Google Scholar] [CrossRef] [PubMed]
- Ramella, S.; Fiore, M.; Greco, C.; Cordelli, E.; Sicilia, R.; Merone, M.; Molfese, E.; Miele, M.; Cornacchione, P.; Ippolito, E.; et al. A Radiomic Approach for Adaptive Radiotherapy in Non-Small Cell Lung Cancer Patients. PLoS ONE 2018, 13, e0207455. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Mahon, R.N.; Mukhopadhyay, N.D.; Hugo, G.D.; Weiss, E. Changes in Radiomic Features During Radiation Therapy as Predictors for Outcome in Locally Advanced Non-Small Cell Lung Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, S71. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, Y.; Wang, Y.; Jiang, S.; Fan, R.; Zhang, H.; Jiang, W. Application of Radiomics and Machine Learning in Head and Neck Cancers. Int. J. Biol. Sci. 2021, 17, 475–486. [Google Scholar] [CrossRef]
- Giraud, P.; Giraud, P.; Gasnier, A.; El Ayachy, R.; Kreps, S.; Foy, J.-P.; Durdux, C.; Huguet, F.; Burgun, A.; Bibault, J.-E. Radiomics and Machine Learning for Radiotherapy in Head and Neck Cancers. Front. Oncol. 2019, 9, 174. [Google Scholar] [CrossRef]
- Guha, A.; Connor, S.; Anjari, M.; Naik, H.; Siddiqui, M.; Cook, G.; Goh, V. Radiomic Analysis for Response Assessment in Advanced Head and Neck Cancers, a Distant Dream or an Inevitable Reality? A Systematic Review of the Current Level of Evidence. Br. J. Radiol. 2020, 93, 20190496. [Google Scholar] [CrossRef]
- Zhou, N.; Chu, C.; Dou, X.; Chen, W.; He, J.; Yan, J.; Zhou, Z.; Yang, X. Early Evaluation of Radiation-Induced Parotid Damage in Patients with Nasopharyngeal Carcinoma by T2 Mapping and MDIXON Quant Imaging: Initial Findings. Radiat. Oncol. 2018, 13, 22. [Google Scholar] [CrossRef]
- Cui, S.; Tseng, H.; Pakela, J.; Ten Haken, R.K.; El Naqa, I. Introduction to Machine and Deep Learning for Medical Physicists. Med. Phys. 2020, 47, e127–e147. [Google Scholar] [CrossRef] [PubMed]
- Huynh, E.; Hosny, A.; Guthier, C.; Bitterman, D.S.; Petit, S.F.; Haas-Kogan, D.A.; Kann, B.; Aerts, H.J.W.L.; Mak, R.H. Artificial Intelligence in Radiation Oncology. Nat. Rev. Clin. Oncol. 2020, 17, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.-D.; Su, X.-H.; Wang, Y.-F.; Shi, G.-F.; Han, C.; Zhang, N. Predicting the Effects of Radiotherapy Based on Diffusion Kurtosis Imaging in a Xenograft Mouse Model of Esophageal Carcinoma. Exp. Ther. Med. 2021, 21, 327. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, H.; Huang, S.; Chen, X.; Zhou, H.; Chang, H.; Xia, Y.; Wang, G.; Yang, X. Early Prediction of Acute Xerostomia during Radiation Therapy for Nasopharyngeal Cancer Based on Delta Radiomics from CT Images. Quant. Imaging Med. Surg. 2019, 9, 1288–1302. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, K.; Lee, S.H.; Cheng, Z.; Lakshminarayanan, P.; Peng, L.; Han, P.; McNutt, T.R.; Quon, H.; Lee, J. Predicting Acute Radiation Induced Xerostomia in Head and Neck Cancer Using MR and CT Radiomics of Parotid and Submandibular Glands. Radiat. Oncol. 2019, 14, 131. [Google Scholar] [CrossRef]
- Tanaka, S.; Kadoya, N.; Sugai, Y.; Umeda, M.; Ishizawa, M.; Katsuta, Y.; Ito, K.; Takeda, K.; Jingu, K. A Deep Learning-Based Radiomics Approach to Predict Head and Neck Tumor Regression for Adaptive Radiotherapy. Sci. Rep. 2022, 12, 8899. [Google Scholar] [CrossRef]
- Alves, N.; Dias, J.; Ventura, T.; Mateus, J.; Capela, M.; Khouri, L.; do Carmo Lopes, M. Predicting the Need for Adaptive Radiotherapy in Head and Neck Patients from CT-Based Radiomics and Pre-Treatment Data. In Computational Science and Its Applications—ICCSA 2021; Gervasi, O., Murgante, B., Misra, S., Garau, C., Blečić, I., Taniar, D., Apduhan, B.O., Rocha, A.M.A.C., Tarantino, E., Torre, C.M., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2021; Volume 12953, pp. 429–444. ISBN 978-3-030-86975-5. [Google Scholar]
- Gehani, A.; Sen, S.; Chatterjee, S.; Mukhopadhyay, S. Imaging Features of Postradiotherapy Changes in Head and Neck Cancers. Indian J. Radiol. Imaging 2021, 31, 661–669. [Google Scholar] [CrossRef]
- Yang, X.; Tridandapani, S.; Beitler, J.J.; Yu, D.S.; Yoshida, E.J.; Curran, W.J.; Liu, T. Ultrasound GLCM Texture Analysis of Radiation-Induced Parotid-Gland Injury in Head-and-Neck Cancer Radiotherapy: An in Vivo Study of Late Toxicity: Ultrasound Assessment of Post-RT Parotid Gland. Med. Phys. 2012, 39, 5732–5739. [Google Scholar] [CrossRef]
- Panek, R.; Welsh, L.; Baker, L.C.J.; Schmidt, M.A.; Wong, K.H.; Riddell, A.M.; Koh, D.-M.; Dunlop, A.; Mcquaid, D.; d’Arcy, J.A.; et al. Noninvasive Imaging of Cycling Hypoxia in Head and Neck Cancer Using Intrinsic Susceptibility MRI. Clin. Cancer Res. 2017, 23, 4233–4241. [Google Scholar] [CrossRef]
- Kikinis, R.; Pieper, S.D.; Vosburgh, K.G. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In Intraoperative Imaging and Image-Guided Therapy; Jolesz, F.A., Ed.; Springer: New York, NY, USA, 2014; pp. 277–289. ISBN 978-1-4614-7656-6. [Google Scholar]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Kakkos, I.; Dimitrakopoulos, G.N.; Sun, Y.; Yuan, J.; Matsopoulos, G.K.; Bezerianos, A.; Sun, Y. EEG Fingerprints of Task-Independent Mental Workload Discrimination. IEEE J. Biomed. Health Inform. 2021, 25, 3824–3833. [Google Scholar] [CrossRef] [PubMed]
- Kakkos, I.; Ventouras, E.M.; Asvestas, P.A.; Karanasiou, I.S.; Matsopoulos, G.K. A Condition-Independent Framework for the Classification of Error-Related Brain Activity. Med. Biol. Eng. Comput. 2020, 58, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Golland, P.; Liang, F.; Mukherjee, S.; Panchenko, D. Permutation Tests for Classification. In Proceedings of the Learning Theory; Auer, P., Meir, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 501–515. [Google Scholar]
- Avgousti, R.; Antypas, C.; Armpilia, C.; Simopoulou, F.; Liakouli, Z.; Karaiskos, P.; Kouloulias, V.; Kyrodimos, E.; Moulopoulos, L.A.; Zygogianni, A. Adaptive Radiation Therapy: When, How and What Are the Benefits That Literature Provides? Cancer Radiothérapie 2022, 26, 622–636. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhu, S. Visual Interpretability for Deep Learning: A Survey. Front. Inf. Technol. Electron. Eng. 2018, 19, 27–39. [Google Scholar] [CrossRef]
- Mahmud, M.; Kaiser, M.S.; Hussain, A.; Vassanelli, S. Applications of Deep Learning and Reinforcement Learning to Biological Data. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 2063–2079. [Google Scholar] [CrossRef]
- Glastonbury, C.M.; Parker, E.E.; Hoang, J.K. The Postradiation Neck: Evaluating Response to Treatment and Recognizing Complications. Am. J. Roentgenol. 2010, 195, W164–W171. [Google Scholar] [CrossRef]
- Saito, N.; Nadgir, R.N.; Nakahira, M.; Takahashi, M.; Uchino, A.; Kimura, F.; Truong, M.T.; Sakai, O. Posttreatment CT and MR Imaging in Head and Neck Cancer: What the Radiologist Needs to Know. Radiographics 2012, 32, 1261–1282. [Google Scholar] [CrossRef]
- Haufe, S.; Meinecke, F.; Görgen, K.; Dähne, S.; Haynes, J.-D.; Blankertz, B.; Bießmann, F. On the Interpretation of Weight Vectors of Linear Models in Multivariate Neuroimaging. Neuroimage 2014, 87, 96–110. [Google Scholar] [CrossRef]
- Tolosi, L.; Lengauer, T. Classification with Correlated Features: Unreliability of Feature Ranking and Solutions. Bioinformatics 2011, 27, 1986–1994. [Google Scholar] [CrossRef]
- Luo, H.-S.; Chen, Y.-Y.; Huang, W.-Z.; Wu, S.-X.; Huang, S.-F.; Xu, H.-Y.; Xue, R.-L.; Du, Z.-S.; Li, X.-Y.; Lin, L.-X.; et al. Development and Validation of a Radiomics-Based Model to Predict Local Progression-Free Survival after Chemo-Radiotherapy in Patients with Esophageal Squamous Cell Cancer. Radiat. Oncol. 2021, 16, 201. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.F.; Liu, Y.; Parmar, C.; Li, Q.; Liu, S.; Qu, F.; Ye, Z.; Gillies, R.J.; Aerts, H.J.W.L. Associations between Radiologist-Defined Semantic and Automatically Computed Radiomic Features in Non-Small Cell Lung Cancer. Sci. Rep. 2017, 7, 3519. [Google Scholar] [CrossRef] [PubMed]
- Soufi, M.; Arimura, H.; Nakamoto, T.; Hirose, T.; Ohga, S.; Umezu, Y.; Honda, H.; Sasaki, T. Exploration of Temporal Stability and Prognostic Power of Radiomic Features Based on Electronic Portal Imaging Device Images. Phys. Med. 2018, 46, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Bogowicz, M.; Riesterer, O.; Ikenberg, K.; Stieb, S.; Moch, H.; Studer, G.; Guckenberger, M.; Tanadini-Lang, S. Computed Tomography Radiomics Predicts HPV Status and Local Tumor Control After Definitive Radiochemotherapy in Head and Neck Squamous Cell Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, 921–928. [Google Scholar] [CrossRef]
- McNitt-Gray, M.F.; Hart, E.M.; Wyckoff, N.; Sayre, J.W.; Goldin, J.G.; Aberle, D.R. A Pattern Classification Approach to Characterizing Solitary Pulmonary Nodules Imaged on High Resolution CT: Preliminary Results. Med. Phys. 1999, 26, 880–888. [Google Scholar] [CrossRef]
- Coroller, T.P.; Agrawal, V.; Huynh, E.; Narayan, V.; Lee, S.W.; Mak, R.H.; Aerts, H.J.W.L. Radiomic-Based Pathological Response Prediction from Primary Tumors and Lymph Nodes in NSCLC. J. Thorac. Oncol. 2017, 12, 467–476. [Google Scholar] [CrossRef]
- Yu, T.; Lam, S.; To, L.; Tse, K.; Cheng, N.; Fan, Y.; Lo, C.; Or, K.; Chan, M.; Hui, K.; et al. Pretreatment Prediction of Adaptive Radiation Therapy Eligibility Using MRI-Based Radiomics for Advanced Nasopharyngeal Carcinoma Patients. Front. Oncol. 2019, 9, 1050. [Google Scholar] [CrossRef]
- Ganeshan, B.; Miles, K.A. Quantifying Tumour Heterogeneity with CT. Cancer Imaging 2013, 13, 140–149. [Google Scholar] [CrossRef]
- Kuno, H.; Qureshi, M.M.; Chapman, M.N.; Li, B.; Andreu-Arasa, V.C.; Onoue, K.; Truong, M.T.; Sakai, O. CT Texture Analysis Potentially Predicts Local Failure in Head and Neck Squamous Cell Carcinoma Treated with Chemoradiotherapy. AJNR Am. J. Neuroradiol. 2017, 38, 2334–2340. [Google Scholar] [CrossRef]
- Mroz, E.A.; Rocco, J.W. Intra-Tumor Heterogeneity in Head and Neck Cancer and Its Clinical Implications. World J. Otorhinolaryngol. Head Neck Surg. 2016, 2, 60–67. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in Medical Imaging—“How-to” Guide and Critical Reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Chiesa-Estomba, C.M.; Echaniz, O.; Larruscain, E.; Gonzalez-Garcia, J.A.; Sistiaga-Suarez, J.A.; Graña, M. Radiomics and Texture Analysis in Laryngeal Cancer. Looking for New Frontiers in Precision Medicine through Imaging Analysis. Cancers 2019, 11, 1409. [Google Scholar] [CrossRef] [PubMed]


| Feature Family | Number of Features | Description |
|---|---|---|
| Shape | 12 | Descriptors of the two/three-dimensional size and shape of the ROI. Gray level intensity distribution in the ROI does not affect these feature values. |
| First Order | 17 | Describes the distribution of grey values within the image region. |
| GLDM | 14 | Quantifies gray level dependencies in an image. A gray level dependency is the number of connected voxels within a specific distance that are dependent on the center voxel. |
| GLCM | 24 | Represents the frequency that gray level value pairs with the same distance in the image appear within an ROI. |
| GLRLM | 16 | Quantifies gray level runs. Run is the length in the number of pixels, of consecutive pixels that have the same gray value. |
| GLSZM | 16 | Quantifies gray level zones in an image. A gray level zone is the number of connected voxels that share the same gray level value. |
| NGTDM | 5 | Quantifies the difference between a voxel’s gray value and the average gray value of its neighbor voxels within a specific distance. |
| Accuracy | Sensitivity | Specificity | F1-Score | Area Under the Curve |
|---|---|---|---|---|
| 0.90 ** | 0.95 | 0.86 | 0.90 | 0.91 |
| Feature | Feature Family | Equation | Ranking | Definition |
|---|---|---|---|---|
| Gray Level Non Uniformity | GLDM | (15) | Quantifies the gray level intensity values similarity in the image. A lower GLN value implies a greater similarity in intensity values | |
| Small Dependence Low Gray Level Emphasis | GLDM | Lower gray-level values imply a joint distribution of small dependence. | ||
| Difference Variance | GLCM | (8) | Measures the heterogeneity. Higher weights on differing intensity level pairs deviate more from the mean | |
| Correlation | GLCM | (4) | Quantifies the linear dependence of gray level values to their respective voxels in the GLCM | |
| Cluster Prominence | GLCM | (16) | Quantifies the skewness and asymmetry of the GLCM. Lower values imply lower asymmetry about the mean. | |
| Interquartile Range | FIRST ORDER | (19) | Difference between percentiles of the image array | |
| Energy | FIRST ORDER | (3) | Measures the magnitude of voxel values in an image. Larger values show a greater sum of the squares of these values | |
| Total Energy | FIRST ORDER | (7) | Is the value of energy feature scales by the volume of the voxel in cubic mm | |
| Kurtosis | FIRST ORDER | (18) | Measures the ROI’s distributions of values peakedness. The mass of the distribution is concentrated towards the tail(s) for higher kurtosis values. | |
| Short Run Low Gray Level Emphasis | GLRLM | (17) | Measures the joint distribution for shorter run lengths with smaller gray level values | |
| Low Gray Level Run Emphasis | GLRLM | (11) | Measures the distribution of low gray level values. Higher values indicate greater concentration of low gray level values in the image | |
| Gray Level Non Uniformity | GLSZM | (2) | Measures the distribution of large area size zones. Greater value indicative larger size zones and more coarse textures | |
| Long Run Low Gray Level Emphasis | GLRLM | (14) | Quantifies the joint distribution of shorter run lengths with higher gray level values |
| Feature | Feature Family | Equation | Ranking | Definition |
|---|---|---|---|---|
| Gray Level Non Uniformity | GLDM | (10) | Same as (Gray Level Non Uniformity) | |
| Low Gray Level Emphasis | GLDM | (12) | Measures the distribution of low gray level values. Higher values indicate greater concentration of low gray level values in the image | |
| Maximum Probability | GLCM | (6) | Measures the occurrences of the most predominant pair of neighboring intensity values | |
| Correlation | GLCM | (1) | Same as (Correlation) | |
| Maximum | FIRST ORDER | (9) | The maximum gray level intensity within the ROI | |
| Gray Level Non Uniformity | GLSZM | (5) | Same as (Gray Level Non Uniformity) |
| Week/CBCT | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| 1/2 | 0.90 ** | 0.95 | 0.86 |
| 2/3 | 0.85 ** | 0.86 | 0.84 |
| 3/4 | 0.75 * | 0.76 | 0.74 |
| 4/5 | 0.72 * | 0.73 | 0.72 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iliadou, V.; Kakkos, I.; Karaiskos, P.; Kouloulias, V.; Platoni, K.; Zygogianni, A.; Matsopoulos, G.K. Early Prediction of Planning Adaptation Requirement Indication Due to Volumetric Alterations in Head and Neck Cancer Radiotherapy: A Machine Learning Approach. Cancers 2022, 14, 3573. https://doi.org/10.3390/cancers14153573
Iliadou V, Kakkos I, Karaiskos P, Kouloulias V, Platoni K, Zygogianni A, Matsopoulos GK. Early Prediction of Planning Adaptation Requirement Indication Due to Volumetric Alterations in Head and Neck Cancer Radiotherapy: A Machine Learning Approach. Cancers. 2022; 14(15):3573. https://doi.org/10.3390/cancers14153573
Chicago/Turabian StyleIliadou, Vasiliki, Ioannis Kakkos, Pantelis Karaiskos, Vassilis Kouloulias, Kalliopi Platoni, Anna Zygogianni, and George K. Matsopoulos. 2022. "Early Prediction of Planning Adaptation Requirement Indication Due to Volumetric Alterations in Head and Neck Cancer Radiotherapy: A Machine Learning Approach" Cancers 14, no. 15: 3573. https://doi.org/10.3390/cancers14153573
APA StyleIliadou, V., Kakkos, I., Karaiskos, P., Kouloulias, V., Platoni, K., Zygogianni, A., & Matsopoulos, G. K. (2022). Early Prediction of Planning Adaptation Requirement Indication Due to Volumetric Alterations in Head and Neck Cancer Radiotherapy: A Machine Learning Approach. Cancers, 14(15), 3573. https://doi.org/10.3390/cancers14153573










