Abstract
The prevalent eukaryotic RNA modification N6-methyladenosine (m6A), which is distributed in more than 50% of cases, has demonstrated significant implications in both normal development and disease progression, particularly in the context of cancer. This review aims to discuss the potential efficacy of targeting tumor cells through modulation of m6A RNA levels. Specifically, we discuss how the upregulation or downregulation of integral or specific targets is effective in treating different tumor types and patients. Additionally, we will cover the factors influencing the efficacy of m6A RNA targeting in tumor treatment. Our review will focus on the impact of targeting m6A mRNA on genes and cells and assess its potential as a therapeutic strategy for tumors. Despite the challenges involved, further research on m6A RNA in tumors and its integration with existing tumor therapy approaches is warranted.
1. Introduction
N6-methyladenosine (m6A), as the most prevalent internal modification, is catalyzed by the METTL3/METTL14/WTAP complex at the pre-RNA level before its release from chromatin, with METTL3 serving as the core enzyme, METTL14 stabilizing and positioning the complex, and WTAP assisting in proper localization and function [1,2]. m6A has been implicated as a pivotal regulator of mRNA biology in normal development or in tumorigenesis [3,4,5,6,7]. m6A is found in various nucleic acids, although the specific composition varies (see Table 1), potentially due to differences in the localization and structural compatibility between enzymes and sequences. The m6A mRNA methylation process is a dynamic and reversible modification that is regulated by three key factors. These factors include methyltransferases, known as “writers”, such as METTL3, METTL14, WTAP, RBM15, ZC3H13, and KIAA1429 (VIRMA). METTL3 serves as a core subunit with catalytic activity, while METTL14 plays a role in substrate recognition. WTAP is responsible for recruiting METTL3 and METTL14, as well as interacting with other components to form hybrids. One-third of mRNAs are m6A-modified by the METTL3-METTL14-WTAP complex, which regulates mRNA maturation and translation. Additionally, “erasers” such as FTO and ALKBH5, which share a similar core domain structure, are able to demethylate RNA. Methylated RNA is identified by various proteins for the purpose of executing distinct biological activities. These proteins, known as “readers”, encompass YTHDC1, YTHDC2, YTHDF1, YTHDF2, and HNRNPC, among others. Proteins containing the YTH domain exhibit a specific affinity for methylated mRNA, thereby governing subsequent processes of translation and degradation. In the context of potential tumor treatment strategies, a comprehensive understanding of RNA and cell fate through m6A modification is imperative. Currently, the precise effects of targeting m6A mRNA in tumors remain ambiguous, necessitating further clarification to enhance the efficacy of therapeutic approaches.

Table 1.
The core m6A catalysis enzymes of different nucleic acid types.
2. Influence of m6A on mRNA Fate
Research has demonstrated that only a subset of RRACH (R = G or A; H = A, C, or U) sites within adenosine residues are methylated by m6A [14], which might indicate the importance of the tertiary structure domain of mRNA and the selective binding sites within the m6A enzyme–mRNA complex in determining the m6A methylation levels [15]. The impact of m6A modification on the fate of m6A mRNA is influenced by various m6A readers, with the different readers binding to it being a significant factor. While a range of m6A readers have been shown to have important effects on mRNA fate, with some promoting mRNA degradation, such as YTHDF2 [16], others have been reported to have beneficial effects on mRNA translation, including mRNA structure switching (HNRNPC, HNRNPG) [17,18,19], mRNA splicing (YTHDC1) [20,21], nuclear export (YTHDC1), stability (IGFBP1/2/3, PRRC2A, FMRP) [22], and translation (YTHDF1/3, IGFBP1/2/3, YTHDC2, elF3) [23].
One critical factor influencing the selectivity of m6A readers is the presence of m6A sites, which play a significant role in determining the selection of readers and the fate of mRNA. For instance, the terminator-m6A site is more likely to be recognized by YTHDF1, leading to enhanced translation efficiency with elF3, whereas the CDS-m6A site tends to interact with YTHDC2 [24]. Additionally, the recently discovered reader eIF3, which enhances mRNA translation, has been observed to bind to a single 5′ UTR m6A site to independently initiate translation [25]. Additionally, research has indicated that YTHDF2 plays a role in the preservation of 5′ UTR-m6A mRNA translation by competitively interacting with FTO [26], rather than facilitating m6A mRNA degradation at the 3′ UTR [25]. Studies have shown that transcripts of genes with moderate expression levels are more likely to undergo m6A modification compared to genes expressed at extreme levels [13,14,27]. Thus, the localization of m6A on mRNA at the 3′ UTR site and the overall mRNA expression level in cells are significant factors influencing mRNA fate.
Furthermore, the involvement of m6A sites and the fate of m6A mRNA, as well as the expression of the m6A mRNA methylase METTL3, are becoming increasingly significant factors in mRNA translation, facilitated by the collaboration of the translation machinery. It is widely acknowledged that certain readers, specifically YTHDF1 [28] and YTHDF2 [29], have the capability to directly interact with translation initiation proteins, such as elf3, and potentially influence their expression in order to regulate m6A mRNA translation. Surprisingly, recent findings have indicated that METTL3 has the ability to localize at the transcriptional start sites of numerous actively transcribed genes, where it collaborates with CEBPZ, a CAATT-box binding protein, to facilitate translation by alleviating ribosome stalling [30]. Additionally, METTL3 was also found to enhance the translation of oncogenes EGFR and TAZ in cancer cells by directly recruiting the translation initiation factor elf3, independent of its catalytic activity or any m6A readers. The necessity of an m6A mRNA reader for METTL3-mediated regulation remains uncertain, and further research is required to determine whether this process is a normal part of cell development or a response to tumor stress.
3. Influence of m6A mRNA on Cell Fate
3.1. Influence of m6A mRNA on Normal Cell Fate
In a normal state, METTL3-mediated m6A mRNA modification has been demonstrated to play a pivotal role in terminating pluripotency and initiating differentiation in stem/progenitor cells, as well as in regulating the activation and functional competence of immune cells, such as T cells and dendritic cells (DCs). One study demonstrated that the initial 20% of m6A-labeled mRNA in embryonic stem cells (ESCs) exhibited enrichment in essential pathways related to embryonic development, proto-intestinal embryo formation, and cell cycles, underscoring the pivotal functions of m6A mRNA in cellular processes. Nevertheless, proper m6A mRNA modification appeared to have more beneficial effects on normal cell differentiation while showing less impact on normal cell survival or apoptosis. In METTL3 knockout (KO)-naïve mouse ESCs and pre-implantation epiblasts, m6A methylation on mRNA is almost entirely abolished, yet these cells remain viable. These cells fail to properly exit the naïve pluripotent state, resulting in impaired differentiation and early embryonic lethality. Mechanistically, m6A methylation facilitates the degradation of pluripotency-associated transcripts (e.g., Nanog, Klf4) by recruiting reader proteins such as Ythdf2. In METTL3 KO-naïve mESCs, the stability of these transcripts is significantly increased, while differentiation markers (e.g., Gata4, Troponin) are insufficiently expressed, thereby preventing naïve mouse ESCs from transitioning into the primed state [31]. Further investigation demonstrated that METTL3 depletion in m/hESC hinders ESC self-renewal both in vitro and in vivo, potentially due to elevated Nanog expression levels [27]. Targeting METTL3 in embryos was shown to impede the emergence of hematopoietic stem/progenitor cells (HSPCs) through the sustained activation of Notch signaling pathways [32]. Furthermore, m6A has been identified as a transcriptome flexibility marker that is essential for the differentiation of stem cells into specific lineages. Recent research has revealed its critical role in the endothelial-to-hematopoietic transition (EHT) in specifying the earliest hematopoietic stem and progenitor cells (HSPCs) [33]. In addition to its impact on early hematopoietic cells, disruptions in m6A methylation have been shown to significantly impede T cell homeostasis and differentiation through METTL3-mediated m6A mRNA catalytic activity and the regulation of the IL-7/STAT5/SOCS pathway. Naïve T cells lacking METTL3 remain in a naïve state for an extended period of 12 weeks, contributing to the resistance to colitis [34]. Furthermore, it has been demonstrated that METTL3-mediated m6A plays a role in promoting the maturation of dendritic cells without affecting their survival activity, which is achieved by the m6A-enhanced translation of CD40, CD80, and TLR4 signaling adaptor Tirap transcripts [35]. This was supported by the lack of significant differences in apoptosis observed in bone-marrow-derived dendritic cells from METTL3-WT/KO mice [35]. Additionally, m6A mRNA has been shown to significantly promote various normal biological functions such as adipogenesis, circadian rhythm, spermatogenesis [36,37,38], stem cell differentiation, precise spatiotemporal control of the cerebellum [39], and neurodevelopment [40].
3.2. Influence of m6A mRNA on Tumor Cell Fate
In tumor cells, the regulation of m6A methylation and demethylation is often significantly altered, impacting the overall m6A levels. Here, we present the expression changes of m6A methyltransferases METTL3 and METTL14, as well as demethylases ALKBH5 and FTO, across seven of the most prevalent cancers worldwide according to the TCGA database (Figure 1). This database shows frequently anomalous expressions of these enzymes, and METTL3 expression is always notably elevated in these tumor types. This suggests a potentially crucial role of abnormal m6A mRNA modification in tumor progression.
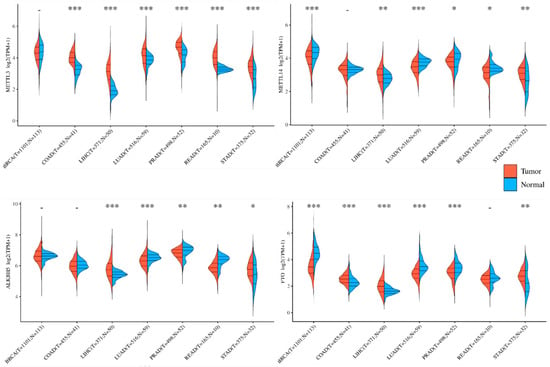
Figure 1.
Expression of m6A mRNA modifiers in five common global cancers. m6A mRNA “writers” and “erasers” indicate substantial alterations in human tumor tissues compared to normal tissues. Here, we present their expression levels in the top five most prevalent types of tumors globally, as evidenced by data from the TCGA database. Furthermore, the inconsistent tendencies of m6A mRNA modifiers observed within the same type of tumors, such as variations in METTL3 vs. METTL14 in LUAD, PRAD, and READ, as well as the contrasting changes between COAD and LIHC in METTL3 expressions, underscore the need to elucidate the factors influencing m6A mRNA modifications in tumors. Additionally, the similar patterns observed in disparate tumor types, such as LUAD and PRAD, offer a valuable opportunity to investigate their pathogenic mechanisms and determinants. BRCA: breast-invasive carcinoma; COAD: colon adenocarcinoma; LIHC: liver hepatocellular carcinoma; LUAD: lung adenocarcinoma; PRAD: prostate adenocarcinoma; READ: rectum adenocarcinoma; STAD: stomach adenocarcinoma. * p < 0.05, ** p < 0.01, *** p < 0.001, - p > 0.05.
m6A modification is pivotal in the survival of tumor cells. In contrast to normal cells, abnormal stimulation or tumor cells exhibit distinct m6A mRNA methylation patterns that may play a role in survival or apoptosis. Specifically, in the context of ultraviolet-induced DNA damage, m6A mRNA modification by METTL3/14 has been identified as a signal for the targeted recruitment of Pol κ to damaged sites, thereby promoting DNA repair and cell survival [41]. Moreover, the acceleration of apoptosis in tumor cells, specifically HepG2 cells, can be achieved by silencing METTL3 expression. This phenomenon is likely linked to the m6A-modulated p53 signaling pathway [14], which plays a critical role in cell proliferation [42]. Additionally, FTO has been shown to be significantly elevated during the formation of leukemia cells, leading to an extended survival cycle and decreased cell apoptosis by upregulating oncogene expressions [43].
m6A methylation may promote tumor cell proliferation through various pathways. In osteosarcoma (OS), the m6A level for RNA methylation and the expression level of METTL3 were upregulated in human OS tissues and OS cell lines. METTL3/m6A significantly promotes the proliferation, migration, and invasion of OS cells through the m6A-mediated stability of LEF1 mRNA and the activity of Wnt/β-catenin signaling [44]. In colorectal cancer (CRC) and bladder cancer (BC), METTL3 accelerates the cell cycle of tumor cells by directly promoting the expressions of CCNE1 [45], AF4/FMR2 family member 4 (AFF4), two key regulators of the NF-κB pathway (IKBKB and RELA), and MYC [46]. In hepatoblastoma (HB), METTL3/m6A methylation significantly affects the Wnt/β-catenin pathway by decreasing the expression and stability of CTNNB1, the coding gene of β-catenin [47]. In hepatocellular carcinoma (HCC), METTL3 has been reported to promote tumor cell proliferation through the YTHDF2-dependent silencing of SOCS2, resulting in the overactivation of the JAK/STAT pathway [48]. Moreover, KIAA1429 has also been reported to promote the proliferation of HCC cells by promoting GATA3 degradation [49].
Besides the above basic life activities related to cell fate survival and proliferation, m6A methylation also plays critical roles in tumor invasion and metastasis. Multivariate Cox regression analysis revealed that METTL3 expression was an independent prognostic factor and effective predictor in human patients with gastric cancer (GC). Mechanistically, P300-mediated H3K27 acetylation activation in the promoter of METTL3 induced METTL3 transcription, which stimulated m6A modification and the IGF2BP3-enhanced stability of HDGF mRNA, which promoted tumor angiogenesis by activating GLUT4, ENO2 expression, and an increase in glycolysis in GC cells [50]. METTL3 can also promote the expression of transcription factor GFI-1 and α-smooth muscle actin (α-SMA) associated with EMT in GC [51]. However, in a few cases, m6A has been reported to inhibit tumor progression. In renal cancer (RCC), ATP-P2RX6 was reported to participate in the regulation of the Ca2+-mediated p-ERK1/2/MMP9 signaling pathway to enhance the migration and invasion of RCC cells, while METTL14/m6A signaling can downregulate the translation of the P2RX6 protein [52]. m6A demethylase is also suggested to play pro-tumor roles in GC, AML, and cervical cancer. In GC, ALKBH5/FTO was found to be highly expressed in tumor cells and activated Wnt and PI3K-Akt signaling to promote GC development [53]. FTO was also reported to promote tumor survival and proliferation through the inhibition of the P53/apoptotic pathway [54], and to promote E2F1 and MYC expressions [55].
The cellular state is a crucial determinant in understanding the functions of m6A mRNA. Cancer cells, characterized by heightened protein synthesis and dysregulated signaling pathways such as the p53 pathway, exhibit increased expression of mutant proteins involved in tumor cell apoptosis regulation. The removal of methylation enzymes may potentially result in the heightened sensitivity and aggressiveness of tumor cells compared to normal cells, although further evidence is required to substantiate this claim. A study revealed that the impact of METTL3 on the resting state of ESCs differed between the resting and activating states during the preparation for differentiation. Specifically, METTL3 was found to promote ESC differentiation and transition from a highly quiescent state in the resting state, while in the activating state, it primarily maintained their proliferation capacity and inhibited differentiation [56]. Similarly, in contrast to the beneficial effects of m6A in promoting normal cell differentiation, the depletion of METTL3 in AML cells resulted in a loss of self-renewal ability and an increase in differentiation capacity. This was attributed to the suppressed translation of c-MYC, BCL2, and PTEN mRNAs through m6A modification [56,57]. Additionally, the m6A-dependent negative regulation of SOCS2 has been observed in liver cancer cells [48], although a study on T cell differentiation did not find METTL3 to be responsible for the m6A modification of SOCS2. Thus, the involvement of m6A mRNA in determining cell fates may vary depending on the specific cellular state.
4. Tumor Cell Fate via m6A mRNA Targeting
4.1. Effects of Decreasing m6A
Numerous essential pathways have been documented as being directly or indirectly influenced by m6A mRNA in the progression of tumors (Figure 2); nevertheless, numerous signaling pathways exhibit inconsistent regulation or even diametrically opposed regulation across various tumor types. In this study, we delineated several potential therapeutic targets for m6A-related tumors, encompassing signaling pathways and m6A regulators that are ubiquitously present in multiple tumor types.
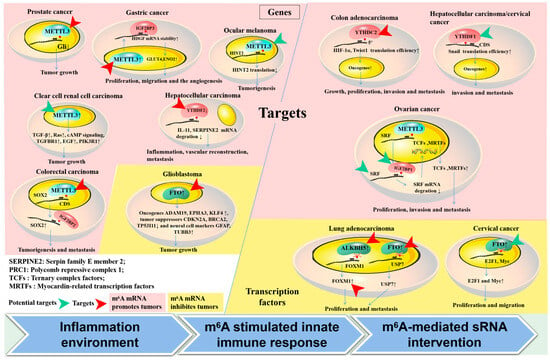
Figure 2.
Targets involved in m6A mRNA methylation in tumors. (1) The methylation of m6A mRNA has been shown to regulate gene expression through the direct modification of executive function genes (left part) and transcription factors (right part) such as HIF-1α, Snail, and FOXM1, impacting tumor status. (2) Valid targets for m6A mRNA regulators and their target genes, as demonstrated in animal models, are highlighted with red arrows. Potential targets are also suggested (indicated by a green arrow). The distinct effects of m6A mRNA modification on tumors are distinguished, both promoting (pink region) and inhibiting (yellow region). (3) Three factors are enumerated in the lower portion of the diagram that could potentially impact variations in effectiveness across various tumors following the m6A targeting of the same gene or the same m6A-associated regulators.
The Wnt/β-catenin pathway, which plays a significant role in regulating gene transcription by utilizing β-catenin as a transcription co-activator in conjunction with the LEF family, has been found to be a key factor in promoting epithelial–mesenchymal transition (EMT). A recent study on osteosarcoma (OS) revealed that the inhibition of METTL3 can disrupt Wnt signaling activity by altering the m6A levels of LEF1, resulting in the suppression of proliferation, migration, and invasion in OS cells [44]. In hepatoblastoma (HB), the knockdown of METTL3 notably decreased the expression and stability of CTNNB1 mRNA, which encodes β-catenin that binds to the TCF4/LEF1 transcription factor and activates target genes such as Jun, MYC, and CCND1, resulting in significant inhibition of HB cell proliferation and colony formation [47]. Further, the overexpression of ALKBH5 significantly inhibited the proliferation, migration, and invasion of pancreatic ductal adenocarcinoma (PDAC) cells by inhibiting the transactivation of Wnt inhibitory factor 1 (WIF-1) [58]. Hence, it is proposed that targeting Wnt signaling pathways by reducing m6A methylation may serve as an effective strategy for treating tumors.
Moreover, several key genes involved in cell proliferation and tumor cell survival have been shown to be negatively regulated by decreasing m6A mRNA in various types of tumors. For example, BCL2 and MYC, well-known oncogenes that inhibit apoptosis and promote cell proliferation, have been reported to undergo m6A-mediated translation, stabilization, or indirect promotion in bladder cancer [46,59], lung cancer [60], breast cancer [61,62], acute myeloid leukemia (AML) [63], and gastric cancer [64,65]. Moreover, decreasing m6A has demonstrated potential therapeutic efficacy through the regulation of the YAP/CCNE1, HDGF, and cAMP pathways, which are commonly associated with tumor proliferation. CCNE1, a nucleoprotein that is expressed cyclically, plays a crucial role in the mitosis of eukaryotic cells by activating CDK2 and forming a complex to traverse the G1/S phase checkpoint [66]. CCNE1 overexpression leads to chromosome instability and enhances the proliferation and metastasis of tumor cells. In CRC, the downregulation of METTL3 exhibited significantly decreased expression levels and weakened the mRNA stability of CCNE1. Notably, it has been documented that CCNE1 is positively controlled by YAP1 through the formation of a complex with TEAD4 in metformin-attenuated cancer proliferation [67]. Furthermore, the YAP signaling pathway has been shown to be subject to m6A-mediated regulation through the knockdown of METTL3, which directly enhances YAP translation by recruiting YTHDF1/3 and eIF3b to the translation initiation complex [68]. Additionally, ALKBH5 has been reported to inhibit YAP activity by regulating the miR-107/LATS2 axis in a HuR-dependent manner [68].
In addition, the activation of the HDGF-PI3K-AKT-mTOR axis could also be inhibited after decreasing m6A through the disrupted Circ-CDYL biogenesis mediated by m6A/YTHDC1 and the m6A/hnRNPA2/B1-mediated active sorting of Circ-CDYL into exosomes, ultimately attenuating tumor malignancy [69]. Consequently, targeting the m6A of the core genes essential for the survival and proliferation of tumor cells shows promise as a potential therapeutic strategy.
In addition to the aforementioned points, the potential efficacy of targeting m6A methylation as a treatment for tumors is further supported by its direct regulation of multiple transcription factors. It was discovered that decreasing m6A inhibited the expression of SRF, which controls key gene classes such as TCFs and MRTFs, through the mRNA-binding protein IGF2BP1. The depletion of Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) can accelerate the degradation of SRF directed by miRNA. This alteration was found to be particularly significant in ES-2 cells when compared to other cancer cell lines such as Huh-7 cells [70]. The downregulation of METTL3 can significantly restrain liver metastasis in gastric cancer (GC) mouse models through the disruption of the m6A reader IGF2BP3-mediated enhancement of hepatoma-derived growth factor (HDGF) transcript stability, which plays a crucial role in tumor cell proliferation, migration, and angiogenesis [50]. Another significant driver of tumor growth, HIF-1, has been shown to target approximately 50 genes and play a crucial role in various aspects of tumor progression, including proliferation, invasion, and metastasis. Recent findings indicate that alterations in the m6A modification of the HIF-1 signaling pathway are notably present in samples from patients with clear-cell renal cell carcinoma (ccRCC). Additionally, targeting YTHDC2 has been demonstrated to reduce the translation efficiency of HIF-1α, dependent on the 5′ UTR in hypoxic conditions, thereby suppressing metastasis in colon adenocarcinoma [71]. The findings indicate a significant positive correlation between YTHDC2 expression levels and tumor stage in colon cancer patients, underscoring the pivotal role of YTHDC2 in cancer progression.
Furthermore, the upregulation of cancer suppressor genes through decreasing METTL3/m6A has also been shown to effectively inhibit tumor growth. For instance, METTL3 has been reported to negatively regulate the cancer suppressor gene GIi, a crucial component of the hedgehog pathway, thereby promoting PC growth. The downregulation of METTL3 can significantly increase the expression of Gli and limit cancer growth [72]. Additionally, m6A-dependent negative regulations of TGF-β, Ras signaling pathways, and the tumor suppressor genes HINT2 and SOCS2 have also been observed in ocular melanoma, liver cancer, and other related malignancies [48,73]. Thus, decreasing m6A is potentially implicated in tumor therapy by downregulating oncogene expression and upregulating tumor suppressor genes in various key pathways, including Wnt/β-catenin, proliferation/survival via direct mRNA regulation, and transcription factors such as HIF-1, CHX8, and SRF.
4.2. Effects of Increasing m6A
While m6A methylation has been demonstrated to be carcinogenic in many tumors, the demethylation of m6A and its regulators, such as ALKBH5 and FTO, have also been shown to contribute to carcinogenesis. Additionally, increasing m6A methylation has been found to inhibit tumor progression in specific tumor types, including glioblastoma, lung cancer, cervical cancer, and endometrial tumors. In the context of glioblastoma, the inhibition of FTO rather than METTL3 or METTL14 was shown to inhibit the proliferation of glioblastoma stem cells (GSCs) and tumorigenesis through the modulation of various oncogenes (ADAM19, EPHA3, and KLF4), tumor suppressors (CDKN2A, BRCA2, and TP53I11), and neural cell markers (GFAP, TUBB3) [74]. FOXM1, a member of the Fox gene family, plays a critical role as a transcription factor in cell division, tumor progression, and distant metastasis [75]. The downregulation of ALKBH5 has been identified as a significant method for restraining the proliferation and malignancy of lung adenocarcinoma cells through decreasing the translation efficiency of FOXM1 mediated by m6A modification [76]. The deletion of ALKBH5 has also been shown to suppress the growth and invasion of lung adenocarcinoma cells. Furthermore, the downregulation of FTO in lung cancer inhibited the growth of cancer cells both in vitro and in vivo through the repression of USP7 expression in an m6A-dependent manner [77]. Similarly, the downregulation of FTO has been shown to inhibit the proliferation and migration of cervical cancer cells (HeLa and SiHa) by reducing the translation efficiency of E2F1 and MYC transcripts in an m6A-dependent manner [55].
Some evidence of an association between reduced m6A and poor cancer outcomes has also been reported. In endometrial tumors, approximately 70% of patients demonstrate decreased rather than increased m6A methylation, likely due to the presence of the METTL14 (R298P) mutation or the reduced expression of METTL3. This reduction in m6A methylation has been shown to enhance the proliferation and migration of endometrial cancer cells [78,79]. Additionally, the downregulation of PHLPP2 and upregulation of mTORC2, which respectively serve as negative and positive regulators of AKT, have been identified as key mediators in these tumor types [78]. Consequently, increasing m6A demethylation may represent a more promising therapeutic approach for these tumors.
4.3. Determiners of Targeting m6A mRNA in Tumors
As depicted in Figure 2, the presence of m6A has been shown to elicit a positive response to external stimuli through intricate regulatory mechanisms. However, it is important to note that these responses vary depending on the specific inflammatory or environmental conditions present within tumors. For example, the exposure of HEPG2 cells to UV or heat shock resulted in alterations to the 5′ UTR m6A metagene profile, whereas treatment with IFN-γ stress did not have the same effect, highlighting the unique characteristics of 5′ UTR m6A in response to stress. Additionally, environmental factors within tumors, such as hypoxia and lactic acid, have been shown to impact m6A mRNA modifications by altering the expressions of m6A regulators or impacting their functions. For instance, HIF-1α has been shown to directly stimulate the transcription of YTHDF1, thereby facilitating hepatocellular carcinoma (HCC) autophagy and malignancy by enhancing the translation of ATG2A and ATG14 [80]. In breast cancer cells, ALKBH5 has been reported to exhibit HIF-1α- and HIF-2α-dependent expression [81]. Additionally, HIF-1α has been implicated in promoting the expression of WTAP, which, in turn, mediates the expression of the glycolysis enzyme HK2 to enhance the Warburg effect in ovarian cancer [82]. Furthermore, HIF-1α has been found to promote METTL3 expression in non-small-cell lung cancer (NSCLC) under smoking-induced stimulation. Furthermore, it has been documented that HIF-1α can impede m6A-mediated mRNA regulation by exerting competitive influences [83]. Specifically, HIF-1α has the ability to enhance the expression of LncRNA STEAP3-AS1, leading to its competitive interaction with YTHDF2. This interaction serves to shield STEAP3 mRNA from YTHDF2/m6A-mediated degradation, thereby facilitating the progression of colorectal cancer [84]. Additionally, lactic acid may serve as a significant environmental factor influencing m6A modification. Studies have shown that the accumulation of lactate in the tumor microenvironment can induce the upregulation of METTL3 in tumor-infiltrating immune cells through H3K18 lactylation, a process crucial for METTL3 to bind to target RNA [85]. Furthermore, the lactylation of METTL16 has been found to promote cuproptosis via the m6A modification of FDX1 mRNA in gastric cancer. Additionally, the lactylation of histones has been implicated in oncogenesis by enhancing the expression of the m6A reader protein YTHDF2 in ocular melanoma [86]. The upregulation of lactate-induced H3K18 lactylation has also been reported to promote the transcription of YTHDF1 [87]. It is known that the degree of hypoxia/HIF-1 or lactate content within the tumor microenvironment is obviously different in different tumor types or tumor stages, which may lead to differences in the expression of m6A mRNA modification regulators and real-time changes, as well as differences in the therapeutic efficacy of targeted m6A. Therefore, all of these reports suggest that we should pay attention to not only the downstream mechanism of m6A regulation, but also the effect of targeted m6A therapy. The consideration of upstream environmental factors influencing m6A modification is essential for optimizing the effectiveness of targeted m6A therapy and adjusting dosages for personalized treatment. Additionally, investigating the specific triggers of m6A mRNA modification can enhance the understanding of pathogenesis, the dual roles potentially associated with various tumor progressions, and the development of combination immunotherapy approaches for tumors.
Additionally, the intricate regulatory mechanisms involving m6A modification play a crucial role in determining the targeting of m6A-modified mRNA in tumors. This includes factors such as disrupted mRNA binding with HuR and the impact of non-coding RNAs. On the one hand, it became evident over time that m6A modification in mRNA impedes the binding of the RNA-binding protein HuR, leading to the accelerated degradation of targeted mRNAs. For example, the upregulation of KIAA1429 has been shown to drive tumor cell proliferation and metastasis, correlating with unfavorable outcomes in hepatocellular carcinoma (HCC) patients. This upregulation can result in m6A methylation on the 3′UTR of GATA3, a transcription factor known for its role as a potent tumor suppressor involved in the growth and differentiation of malignant cells and anti-tumor immune regulation, ultimately leading to the enhanced degradation of GATA3 [88,89]. The m6A modification on the GATA3 3′UTR mechanistically initiates the dissociation of HuR from GATA3 pre-mRNA [49] in a sequential manner. Additionally, the impediment of m6A in mRNA from interacting with HuR was corroborated in a separate investigation of hepatocellular carcinoma, where WTAP-mediated m6A modification in ETS1 led to its dissociation from HuR, hastening its degradation and promoting the advancement of hepatocellular carcinoma [90]. On the other hand, it was noteworthy that the long non-coding RNA GATA3-AS, transcribed from the antisense strand of the GATA3 gene, served as a cis-acting element facilitating the preferential interaction of KIAA1429 with GATA3 pre-mRNA. This mechanism, which involves the collaboration of its homologous LncRNA and m6A regulators, aligns closely with findings in ALKBH5-mediated demethylation. Specifically, the role of LncFOXM1-AS in promoting the interaction between ALKBH5 and FOXM1 nascent transcripts, thereby contributing to the maintenance of tumorigenicity in glioblastoma stem-like cells, suggests a novel m6A regulatory pathway by non-coding RNA [91]. Hence, it is evident that diverse m6A regulation patterns are present in tumor cells and their microenvironments, necessitating the utilization of advanced technical approaches, such as organoids, to comprehensively assess the effects of targeted m6A modifications on tumors from various perspectives and mitigate the risk of obtaining inaccurate findings.
5. Contributions of m6A Non-Coding RNAs in Tumors
Non-coding RNAs, such as miRNA, LncRNA, and circRNA, are a significant group of RNA molecules that primarily regulate gene expression at the post-transcriptional level. These non-coding RNAs play a crucial role in fundamental biological processes such as cell growth, development, metabolism, and tumor progression. Recent research has indicated that the expression of non-coding RNAs is influenced by m6A modification. Furthermore, m6A has been suggested to act as a mediator between mRNA and non-coding RNAs, such as miRNA, in modulating specific cellular functions [92,93].
In normal cells, m6A modification likely enhances the processing of miRNA/LncRNA. The splicing of pri-miRNA requires the involvement of the miRNA microprocessor complex, which consists of the RNA-binding protein DGCR8 and the type III RNase DROSHA. It has been observed that m6A-modified pri-miRNAs can interact with DGCR8 and recruit DROSHA, thereby expediting the splicing of pri-miRNA. The depletion of METTL3 has been shown to decrease the binding of DGCR8 to pri-miRNAs, leading to a global reduction in mature miRNAs and the accumulation of unprocessed pri-miRNAs [94]. Furthermore, hnRNPA2B1 has the capability to recognize m6A modifications, directly interact with m6A marks on a subset of pri-miRNA transcripts, and engage with DGCR8 to facilitate the splicing process of pri-miRNAs. It has been observed that hnRNPA2B1 binds to a specific set of nuclear transcripts and induces alternative splicing effects similar to those of the m6A writer METTL3 [95]. Additionally, the long non-coding RNA X-inactive specific transcript (LncXIST) is responsible for mediating the transcriptional silencing of genes on the X chromosome and plays a crucial role in female mammalian development. In 2016, a study demonstrated that LncXIST contains a minimum of 78 m6A residues, with RBM15/RBM15B facilitating its function by facilitating the interaction between WTAP–METTL3 and LncXIST. Additionally, YTHDC1 was found to selectively recognize m6A residues on LncXIST and was essential for its function [15], highlighting the role of m6A as a regulator of transcriptional repression by long non-coding RNA.
In tumor cells, m6A modifications on miRNA/LncRNAs have been shown to promote tumor pathogenesis, as illustrated in Figure 3. For instance, in bladder cancer, the maturation of pri-miR221/222 has been found to be expedited by METTL3-mediated m6A modification through its interaction with DGCR8, resulting in decreased PTEN expression [59]. Furthermore, in hepatocellular carcinoma cells, a lipogenesis-related LncRNA known as Lnc00958 was identified as being positively regulated by m6A. This LncRNA was found to act as a sponge for miR3619-5p, leading to the upregulation of HDGF expression and ultimately promoting tumor lipogenesis and progression [96]. Additionally, the progression of liver cancer was shown to be facilitated by the m6A methyltransferase METTL14-mediated RP1-228H13.5, which targets hsa-miR-205/ZIK1 [97]. In colorectal cancer tissues, the downregulation of METTL14 resulted in reduced levels of miR-375, leading to the promotion of CRC growth and migration through the YAP1 and SP1 pathways, respectively. Based on additional research, it has been determined that the m6A-mediated regulation of the YAP pathway involves not only miRNA, but also direct interactions with LncRNA. Specifically, a negative feedback loop involving the LncRNA GAS5-YAP-YTHDF3 axis has been identified, wherein LncGAS5 promotes YAP degradation through ubiquitination and intracellular translocation, while YAP upregulates YTHDF3 expression, leading to the degradation of m6A-modified LncGAS5 and ultimately inhibiting YAP degradation, thereby contributing to the progression of colorectal cancer [98]. A recent study revealed that a novel LncRNA, RP11, plays a positive regulatory role in the migration, invasion, EMT, and liver metastasis of CRC by forming an RP11/hnRNPA2B1/mRNA complex, which is dependent on m6A-mediated upregulation [99]. Additionally, in pancreatic cancer, LncDANCR has been identified as a novel target for reader IGF2BP2 through m6A modification, leading to the stabilization of LncDANCR and the promotion of cancer proliferation and stem-like properties [100]. The m6A/HOXA10-AS/ITGA6 axis contributes to increased oxidative resistance and facilitates the malignant advancement of laryngeal squamous cell carcinoma through the modulation of the Notch and Keap1/Nrf2 pathways [101]. These studies demonstrate the significant impact of m6A modification on tumor progression via the regulation of miRNA and LncRNA expression, underscoring the promising prospects for targeted m6A RNA therapy.
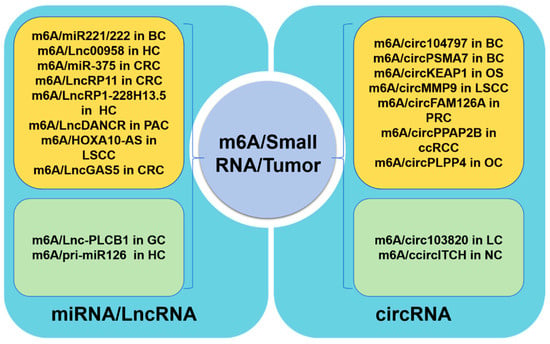
Figure 3.
Roles of m6A methylation on small RNA in tumor cells. The regulation of tumor progression in tumor cells by m6A RNA is partially mediated through the modulation of miRNA/LncRNA (the left part) or circRNA (the right part). Specifically, the yellow region signifies a positive regulation of tumor progression by m6A, while the green region denotes a negative regulation of tumor progression. BC: bladder cancer; HC: hepatocellular carcinoma; CRC: colorectal cancer; PAC: pancreatic cancer; LSCC: laryngeal squamous cell carcinoma; GC: gastric cancer; OS: osteosarcoma; PRC: prostate cancer; ccRCC: clear-cell renal cell carcinoma; OC: ovarian cancer; LC: lung cancer; NC: nasopharyngeal carcinoma.
circRNA, functioning as a molecular sponge for microRNA (miRNA), modulates gene expression within signaling pathways, thereby influencing tumor cell proliferation, migration, invasion, apoptosis, and drug resistance. This characteristic renders circRNA a promising target for therapeutic interventions in cancer. The regulation of circular RNAs through m6A modification has recently been extensively proposed [102], with evidence suggesting that the upregulation of m6A-mediated circRNA expression is linked to the progression of various types of tumors including bladder cancer, clear-cell renal cell carcinoma (ccRCC), osteosarcoma, laryngeal squamous cell carcinoma, nasopharyngeal carcinoma, prostate cancer, and ovarian cancer [103]. For instance, research has demonstrated that elevated levels of m6A in circ_104797 [104] and circPSMA7 [105] play a crucial role in maintaining cisplatin resistance and advancing bladder cancer progression. Additionally, the m6A modification on circPPAP2B modulates the interaction between HNRNPC and the splicing factors PTBP1 and HNPNPK, thereby regulating pre-mRNA alternative splicing in ccRCC [106]. In osteosarcoma, the decreased expression of m6A-mediated circKEAP1 promotes the proliferation and invasion of tumor cells. In the context of lung cancer, the suppression of hsa_circRNA_103820 expression by IGF2BP3/m6A has been observed to augment the migratory and invasive properties of cancer cells [107]. Additionally, the stabilization of CircMMP9 by IGF2BP2/m6A has been shown to expedite the progression of laryngeal squamous cell carcinoma by facilitating TRIM59 transcription [108]. Furthermore, the downregulation of circITCH mediated by HNRNPC has been implicated in the pathogenesis of nasopharyngeal carcinoma by impeding the sequestration of miR-224-3p [109]. Lastly, the m6A modification of circFAM126A has been demonstrated to enhance transcriptional stability and promote the progression of prostate cancer [110]. Moreover, a recent study identified a new interaction between circular RNA (circRNA) and m6A reader proteins, demonstrating a regulatory function in m6A modifications. Specifically, circRARS has been shown to cooperate with IGF2BP3 to increase m6A recognition, thereby promoting the advancement of renal cell carcinoma [111].
Hence, the ability of non-coding RNA to modulate the expression of numerous genes—potentially hundreds—represents a distinctive benefit of non-coding RNA in the context of anti-tumor therapy. It is increasingly evident that m6A modification is pivotal in driving tumor advancement by controlling the expression of a significant array of non-coding RNAs. Manipulating the expression of non-coding RNAs via m6A modification to impact multiple downstream pathways, thereby reshaping and redefining the tumor microenvironment, emerges as a highly promising avenue for further research. Further investigation into the impact of m6A modifications on non-coding RNAs in tumor-related processes has the potential to inform novel therapeutic approaches targeting m6A in tumor treatment strategies.
6. Perspectives on m6A RNA Targeting for Tumor Treatment
6.1. Strategies for Targeting m6A mRNA in Tumor Therapy
According to reports, the silencing of METTL3 in HepG2 cells resulted in a significant enrichment of the p53 signaling pathway, with 22 out of 23 genes showing differentially expressed splice variants, 18 of which were found to be m6A-methylated. Table 2 demonstrates the significant regulation of m6A mRNA methylation in tumor formation. Here, we propose four strategies for targeting m6A mRNA in tumor therapy (Figure 4). The first strategy involves targeting m6A regulators, such as FTO or YTHDF2, which have been identified as potential adjuvant therapeutic targets in AMLs. These targets have shown promise in effectively regulating the proliferation and life cycle of leukemic cells, as well as enhancing ATRA-induced differentiation [43,112]. Numerous studies have demonstrated the potential efficacy of targeting m6A regulators, specifically METTL3, for improving the treatment of gemcitabine in pancreatic cancer [113]. Additionally, R-2HG has been shown to possess anti-tumor activity through its targeting of the FTO/m6A/MYC/CEBPA signaling pathway [114]. However, due to the complex regulatory mechanisms and dual roles of m6A mRNA regulators in certain tumors, the therapeutic effectiveness of strategies aimed at inhibiting or promoting methylase expressions may be constrained or unpredictable. Hence, the integration of this approach with siRNA-based biomedical engineering holds promise for enhancing therapeutic outcomes and may lead to the exploration of more precise methods for tumor treatment as potential avenues for further research. In the second approach, the modification of m6A sites through RCas9 technology [115] may be utilized to precisely alter the m6A sites of specific targets. The implementation of such procedures is reliant on advancements in tumor-targeting technology.

Table 2.
Roles of m6A methylation on mRNA in tumor cells.
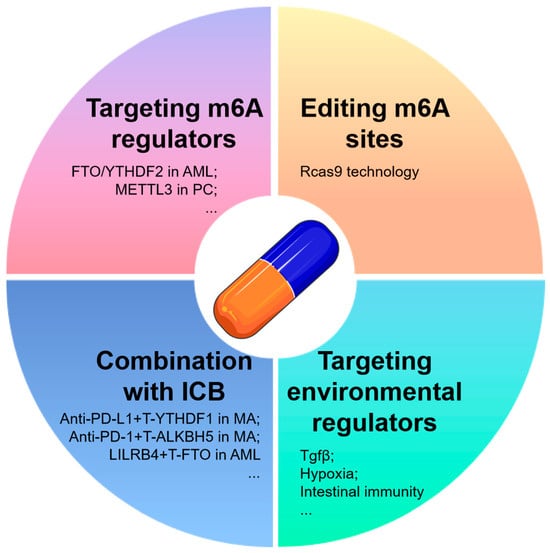
Figure 4.
Therapy strategies targeting m6A RNA in tumors. Here, we present four tumor treatment strategies focused on targeting m6A RNA: targeted m6A RNA regulators, the utilization of Cas9 or other advanced technologies for the direct manipulation of gene m6A targets, combination therapy with traditional tumor treatments or tumor immune checkpoint (ICB) inhibitors to enhance therapeutic efficacy, and the targeted modulation of m6A-related tumor microenvironmental factors. PC, pancreatic cancer; ICB, immune checkpoint blockade; T-, targeting; MA, melanoma.
Disrupting the mechanisms that link environmental factors to m6A alterations could potentially have comparable effects across various cancer types, including, but not limited to, TGF-β, intestinal immunity, heat shock [118], lactate acid, and hypoxia [76]. Research shows that m6A is a dynamic event directly modulated by extracellular signals such as TGFβ. In hPSCs, SMAD2/3 binds with METTL3/METTL14/WTAP in a TGFβ-dependent Activin/Nodal signaling manner, promoting the m6A modification of pluripotency genes (e.g., NANOG, Nodal, LEFTY1) near the termination codon, inducing their degradation and prompting the cells to exit pluripotency [119]. Additionally, TGF-β promotes METTL3 liquid–liquid phase separation, reducing ITIH1 mRNA stability and advancing HCC progression [120]. As a secreted protein, ITIH1 acts as a ligand for the integrin α5β1, antagonizing fibronectin, inhibiting the focal adhesion kinase pathway, and suppressing HCC progression. In preclinical models (mouse, patient-derived organoids, patient-derived xenografts), purified recombinant ITIH1 (r-ITIH1) protein targets HCC and synergizes with TGF-β inhibitors in therapy [120]. ALKBH5 promotes FOXA1 protein expression by inhibiting m6A modification in its CDS region, while TGF-β1 reduces ALKBH5’s binding to the FOXA1 CDS, increasing m6A modification and suppressing FOXA1 expression, thereby promoting GBC EMT and metastasis [121]. Thus, TGFβ, as a key regulator of m6A modification and tumor metastasis, may potentially enhance m6A-targeted therapies when inhibited.
Research shows that butyrate, a classic intestinal microbial metabolite, downregulates METTL3 and cyclin E1 expression to inhibit CRC development, highlighting a potential strategy for intervening in m6A modification by regulating gut microbial metabolism in colorectal cancer [45]. As described in Section 4.3, hypoxia-triggered HIF-1α in the tumor microenvironment promotes tumor progression by enhancing the expression of m6A regulators (e.g., YTHDF1, WTAP, METTL3) and altering their functions (e.g., competitively inhibiting YTHDF2 recognition). Lactate also promotes tumor progression by regulating the expression and function of m6A factors such as METTL3, YTHDF2, and METTL16. Therefore, these m6A-regulated factors may serve as novel targets for cancer diagnosis and the development of therapeutic interventions. Investigating the specific triggers of m6A mRNA modification can enhance the understanding of pathogenesis, the dual roles potentially associated with various tumor progressions, and the development of combination immunotherapy approaches for tumors.
In the field of clinical oncology, monotherapy using chemical agents is often ineffective due to the emergence of drug resistance. The study of cancer biology is intricately linked to epigenomic regulation. In recent years, there has been significant progress in the development of therapies targeting epigenetic mechanisms, exemplified by the use of DNA methyltransferase inhibitors such as azacytidine and decitabine, as well as histone deacetylase inhibitors. The integration of conventional tumor therapy with novel epigenetic medications represents a promising avenue for the development of anticancer drugs in the future. The involvement of m6A mRNA regulators in the resistance to tumor radiotherapy, kinase inhibitors (KIs), 5-fluorouracil (5-FU), gemcitabine, anthracyclines, and cisplatin has demonstrated significant clinical utility [122,123]. Furthermore, the combination of m6A mRNA regulation with immune checkpoint blockade (ICB) has emerged as a viable strategy for the treatment of tumors. According to reports, the deletion of YTHDF1 in conjunction with PD-L1 checkpoint blockade has been shown to significantly improve the therapeutic effectiveness in melanoma mouse models [124]. Additionally, ALKBH5, but not FTO, has been implicated in having a detrimental impact on the response to anti-PD-1 therapy in both melanoma mouse models and patients [125]. The removal of ALKBH5 in melanoma cell lines may impact lactate levels and the presence of immunosuppressive cells in the tumor microenvironment, such as regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs), through m6A-mediated splicing regulation. This alteration could significantly improve the efficacy of the melanoma response to GVAX/anti-PD-1 treatment, which has also demonstrated effectiveness in a colorectal cancer model during anti-PD-1 therapy [125]. Furthermore, the attenuation of leukemia by suppressing immune checkpoint genes, particularly LILRB4, has been proposed to exhibit enhanced efficacy with the assistance of genetic depletion or the inhibition of FTO. This effect is attributed to the restriction of stem cell self-renewal and the reprogramming of the immune response [126]. Additionally, decreased expression of ALKBH5, METTL3, HNRNPC, and KIAA1429 in lung squamous cell carcinoma (LUSC) has been identified as a potential biomarker for increased sensitivity to immunotherapy and chemotherapy [127], suggesting potential targets for tumor treatment. Hence, anticipating the potential efficacy of combining immunotherapy with epigenetic therapy is warranted. Additionally, the consideration of combining multiple drugs targeting either similar or different types of epigenetic modifications for neoadjuvant therapy should be taken into account, despite the potential limitations observed in certain combinations such as the reduced effectiveness seen with the combination of DNMTi and HDAC inhibitors in myelodysplastic syndrome. The identification of new therapeutic targets and ongoing clinical trials are essential for the development of more effective therapy combinations in the evolving landscape of epigenetic molecular mechanisms.
An additional approach to selectively targeting m6A involves the utilization of RNA drugs, which have demonstrated efficacy in diverse therapeutic contexts. The successful creation and widespread administration of the COVID-19 mRNA vaccine has enhanced confidence in the practicality of RNA drugs for clinical applications. While RNA drugs show potential for exploration by emerging researchers, it is important to note that RNA can also trigger an innate immune response similar to mammalian DNA [128] through the activation of the TLR family [129]. The extent of this impact is closely linked to the specific type and quantity of RNA modifications present [130]. The activation of Toll-like receptors 7 and 8 by RNA-containing m6A and s2U modifications and Toll-like receptor 3 by RNA-containing m5C, m5U, or Ψ modifications has been documented. Unmodified RNA has the ability to activate all human Toll-like receptors. Therefore, aside from the challenges associated with delivering RNA drugs to specific anatomical sites and the endogenous cellular mechanisms involved in protein modifications derived from RNA drugs, the RNA-induced innate immune response is recognized as a key factor in determining tumor outcomes. The presence of m6A modification may prove to be advantageous in this context. Research has indicated that the presence of m6A in RNA can diminish or abolish the activation of human DCs triggered by RNA. This apparent inverse correlation between RNA modification and immune activation may elucidate the mechanism by which RNA from necrotic cells can stimulate DCs to secrete IFN-α, while RNA from apoptotic cells cannot, underscoring the importance of RNA degradation during apoptosis [131]. The presence of m6A modifications in viral mRNAs, with up to eight modifications per 1.8 kb segment of influenza RNA, has been shown to effectively inhibit the ability of RNA to activate dendritic cells. Consequently, the potential benefits of incorporating m6A modifications or other RNA modifications to enhance drug stability should be carefully considered when developing RNA-based therapies for tumors.
In the realm of comprehending the mechanisms of m6A RNA, artificial intelligence shows promise in facilitating the prediction of RNA structures and RNA–protein interactions. Nevertheless, the efficacy of machine learning algorithms is contingent upon the availability of comprehensive training data sets, which are presently more prevalent for proteins than for RNA. Moreover, artificial intelligence technology can be harnessed to pinpoint potential therapeutic targets and develop customized coding RNA drugs aimed at specific proteins. Additionally, endeavors can be undertaken to forecast m6A RNA splicing patterns based on genomic sequences. Several predictive models have been developed to forecast patterns of gene expression by analyzing chromatin modification [132]. While some predictive patterns for cancer prognosis based on m6A mRNA levels or regulators have been proposed, there is potential for the discovery of more reliable models to predict gene expression patterns using m6A mRNA modification. This study posits that the validity of results and conclusions derived from animal models must be extrapolated to human patients. The potential implications of this extrapolation include advancements in the manipulation and regulation of gene expression levels, as well as the study and treatment of cancers associated with specific gene expressions, potentially leading to personalized therapeutic approaches.
6.2. Clinical Trials and Drug Development for Tumor Therapy Targeting m6A
Currently, clinical trials for drugs targeting m6A regulators in cancer therapy are still in the early stages, and no anticancer drugs targeting m6A have been approved by the FDA. However, several drugs targeting m6A regulators have been developed and are undergoing clinical research for cancer treatment. It was reported that multiple AML cell lines and PDX models exhibited sensitivity to METTL3 inhibition by STC-15. METTL3 inhibition resulted in the dose-dependent downregulation of the anti-apoptotic protein BCL-2. A matrix combination experiment demonstrated strong synergy between STC-15 and venetoclax, a BCL-2 inhibitor, in MOLM-13 cells and an AML PDX model, significantly extending animal survival. At present, a highly selective METTL3 inhibitor, STC-15, has been developed as a specific inhibitor of m6A methylase METTL3 for stage I clinical use in malignant tumors whose indications include lung/hepatic/bone/colorectal/ skin/mammary gland cancer and malignant melanoma (NCT05584111, Table 3).

Table 3.
Clinical trials for tumor therapy targeting m6A.
Two small-molecule compounds (CS1 and CS2; CS1 also means Bisantrene) which specifically target FTO and inhibit its m6A demethylase activity have been identified through in silico screening and validation. CS1 and CS2 bind to the catalytic pocket of FTO, disrupting its interaction with m6A-modified RNAs. Both compounds significantly reduced the viability and growth of human AML cells, with IC50 values in the low nanomolar range, at least 10-fold more potent than previous FTO inhibitors (e.g., FB23-2 and MO-I-500). Mechanistically, CS1 and CS2 promote anti-leukemic effects by inhibiting FTO activity, activating apoptosis signaling, and suppressing MYC pathways [126]. Targets of one clinical trial involving CS1 for the treatment of relapsed or refractory AML by combination therapy with fludarabine and clofarabine have entered Phase II (NCT04989335). In addition, the combined vaccine of IGFBP2, HER2, and IGF-1R and a small-molecule inhibitor, AST-201, have also entered clinical trials for AML and breast cancer therapy, respectively. Some other drugs have also entered the preclinical stage, such as a METTL3 inhibitor, next-generation CS1 targeting FTO, and EP-102 targeting METTL3 in malignant tumors. We list some emerging drugs targeting m6A that have not yet entered clinical trials in Table 4.

Table 4.
Drugs not yet in clinical trials for tumor therapy targeting m6A.
6.3. Challenges in the Development of Drugs Targeting m6A for Tumor Therapy
Targeting m6A with small-molecule inhibitors, RNA drugs, and other therapeutic approaches holds great promise in modulating gene expression and controlling various biological processes. Targeting m6A (N6-methyladenosine) in cancer therapy presents several challenges that need to be addressed for its effective clinical application.
First, these therapies may cause off-target effects, which refer to unintended interactions with molecules other than the intended targets, potentially leading to adverse biological responses and affecting drug efficacy. Small-molecule inhibitors that target m6A-associated enzymes, such as METTL3, METTL14, and FTO, may interact with other enzymes or molecular pathways with similar structures, resulting in the unintended modulation of RNA modification processes or other metabolic pathways, thus inducing off-target effects. RNA drugs, such as mRNA, siRNA, or antisense oligonucleotides (ASOs), may bind to non-target RNAs, disrupting cellular function and gene expression balance. Moreover, external factors such as TGFβ in the tumor microenvironment may also interfere with m6A regulation, further complicating the effects of these drugs.
To improve the selectivity and targeting of m6A-based drugs, significant advancements must be made in drug design and molecular optimization. By refining the molecular structure and increasing the affinity of drugs for specific m6A-related enzymes or RNA sequences, the possibility of unintended interactions with non-target molecules can be minimized. Utilizing computational drug design tools, such as computer-aided drug design (CADD), could help predict the binding affinity of drug candidates for both target and off-target molecules, guiding the development of more specific therapies. Additionally, RNA drugs can be designed to precisely recognize and regulate specific mRNA targets, reducing the likelihood of interactions with non-target RNAs. Integrating gene-editing technologies, such as CRISPR/Cas, can also offer a more precise approach to m6A modification, further improving therapeutic specificity.
Optimizing drug delivery systems is another promising avenue for reducing off-target effects. Techniques such as nanoparticle carriers, liposomes, and antibody–drug conjugates (ADCs) can enhance the accumulation of drugs in target tissues or cells, minimizing their impact on non-target areas. By incorporating receptor-targeted and cell-penetrating strategies, drugs can be directed specifically to the affected regions, improving therapeutic outcomes while reducing side effects. Moreover, advances in precision medicine and personalized therapies, supported by genomic, transcriptomic, and single-cell technologies, could enable tailored treatments based on individual genetic profiles. By identifying patient-specific variations in m6A regulation, personalized therapeutic approaches can be developed, optimizing drug efficacy and minimizing off-target effects. Combining multi-omics data for the systemic evaluation of off-target effects and utilizing high-throughput screening methods will be crucial for assessing the safety of m6A-targeting drugs.
The complexity of m6A’s role across different tumor types complicates its use as a therapeutic target as its effects may vary, either inhibiting or promoting cancer progression depending on the context. The regulation of m6A involves multiple enzymes, such as methyltransferases (e.g., METTL3) and demethylases (e.g., FTO, ALKBH5), whose interplay adds another layer of complexity to its functional understanding and therapeutic targeting. Moreover, individual variability in genetic backgrounds, tumor types, and m6A expression levels requires personalized therapeutic approaches, as differences in m6A modifications between tumor and normal tissues may lead to unintended toxicity. The lack of specific and effective drugs for targeting m6A remains a major hurdle, as existing inhibitors often lack selectivity and may interfere with critical biological processes. Additionally, the dynamic nature of m6A modifications, influenced by various factors such as the tumor microenvironment and disease progression, makes it challenging to predict therapeutic outcomes. The potential side effects and toxicity associated with modulating m6A, such as immune suppression or liver damage, further complicate its clinical application. Finally, while m6A’s potential has been demonstrated in laboratory studies, the lack of substantial clinical trial data on its impact in cancer therapy makes its translation into clinical practice difficult. Overall, while targeting m6A holds promise, overcoming these challenges will require further research and the development of more specific, efficient, and safe therapeutic strategies.
6.4. Causes of Heterogeneity of m6A Modification and Future Research Directions
N6-methyladenosine (m6A) methylation has been shown to play a crucial role in cancer biology, influencing various processes such as gene expression regulation, tumor progression, and metastasis. However, its role in different tumors, and even within the same tumor type, can be contradictory, which complicates the development of m6A-targeted cancer therapies. Some key reasons for these contradictions and suggestions for future research directions are as follows: 1. Tumor heterogeneity. Tumors are often heterogeneous, consisting of subpopulations of cells with distinct molecular and genetic profiles. These variations can result in different responses to m6A modifications. In some cancer cells, m6A may promote tumor growth by enhancing the gene expression of oncogenes, while in other cells, it may suppress tumor progression by regulating tumor suppressors. Therefore, it is important to conduct single-cell RNA sequencing studies to better understand how m6A modification affects different subpopulations within a tumor. This will help identify specific tumor cell types that are more responsive to m6A modulation, enabling personalized therapies. 2. Differential expression of m6A regulators. The expression levels of m6A methyltransferases (such as METTL3 and METTL14), demethylases (such as FTO and ALKBH5), and reader proteins (such as YTHDF1 and YTHDF2) vary between different tumors and even between tumor stages. High or low levels of these regulators can lead to opposing effects on m6A-mediated RNA modifications, resulting in pro-tumorigenic or anti-tumorigenic outcomes. Therefore, future studies should focus on profiling m6A regulators in various tumors to determine how their expression correlates with clinical outcomes. This will aid in identifying key regulators whose manipulation may have therapeutic potential for specific tumor types. Additionally, developing targeted inhibitors or activators of these regulators could provide more precise therapeutic interventions. 3. Microenvironmental influence. The tumor microenvironment (TME), which includes immune cells, fibroblasts, extracellular matrix components, and various signaling molecules, can significantly influence m6A modifications. For example, hypoxic conditions in tumors have been shown to alter the expression of m6A regulators and contribute to altered gene expression, promoting tumor survival and metastasis. However, the TME can also induce m6A-mediated regulation of immune responses, affecting the tumor’s immune evasion strategies. Therefore, to better understand how the TME affects m6A dynamics, researchers should examine the interplay between tumor cells and immune cells in the microenvironment. Moreover, studies should investigate the potential of combining m6A-targeted therapies with immunotherapy to improve therapeutic outcomes by reprogramming immune responses. 4. The dual role of m6A in tumor suppression and promotion. m6A modification has been reported to both inhibit and promote cancer progression depending on the context. For instance, m6A methylation can regulate the stability of tumor suppressor genes and oncogenes, with different tumor types showing opposing effects. In some cancers, m6A enhances the stability of oncogenic mRNAs, promoting cell proliferation, while in others, it may inhibit tumor growth by destabilizing pro-cancerous transcripts. Therefore, in-depth functional studies are needed to explore the specific mechanisms through which m6A affects tumor suppression and promotion in different cancer types. Investigating how m6A regulators interact with other key molecular pathways, such as those involved in cell cycle regulation and apoptosis, will help identify therapeutic targets that can modulate the dual role of m6A in tumor progression. 5. Post-transcriptional modifications and feedback loops. m6A affects RNA splicing, translation, and decay, and it may engage in complex feedback loops that influence tumor behavior. For example, the m6A-mediated regulation of certain RNA transcripts may lead to changes in the expression of m6A regulators themselves, further modulating tumorigenic processes. These feedback mechanisms can result in varying effects in different tumor contexts. Therefore, to uncover the full extent of m6A’s impact, it will be important to study the interactions between m6A-modified transcripts and their regulators. Comprehensive multi-omics analyses, including transcriptomics, proteomics, and epigenomics, could provide insights into the feedback loops involving m6A and their role in tumor biology.
The contradictory effects of m6A in different tumor types, as well as its complex regulation and interaction with various tumor-related pathways, pose significant challenges for its therapeutic targeting. To address these issues, future research should focus on understanding the context-dependent roles of m6A, identifying key regulators, and exploring combinatory strategies with other therapeutic modalities. This will pave the way for the development of more effective and personalized cancer treatments targeting m6A.
Author Contributions
Z.M. drafted the manuscript. M.L. drafted and revised the manuscript. S.W. designed the study and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Jiangsu Provincial Medical Key Discipline Cultivation Unit (Grant No. JSDW202241), the Research Project of the Jiangsu Commission of Health (Grant No. K2024017), and the Postgraduate Research & Practice Innovation Program of Jiangsu Province (Grant No. KYCX22_3710).
Conflicts of Interest
There are no potential conflicts of interest.
Abbreviations
| Abbreviation | Full Form/Description | Abbreviation | Full Form/Description |
| ALKBH5 | AlkB homolog 5 | NSCLC | Non-small-cell lung cancer |
| α-SMA | α-smooth muscle actin | OS | Osteosarcoma |
| AML | Acute myeloid leukemia | PC | Prostate cancer |
| BC | Bladder cancer | P2RX6 | Purinergic receptor P2X, ligand-gated ion channel 6 |
| BRCA | Breast cancer gene | PI3K-Akt | Phosphoinositide 3-kinase-Akt |
| CBX8 | Chromobox 8 | PDAC | Pancreatic ductal adenocarcinoma |
| CCNE1 | Cyclin E1 | PRAD | Prostate adenocarcinoma |
| CDK2 | Cyclin-dependent kinase 2 | PRC1 | Polycomb repressive complex 1 |
| COAD | Colon adenocarcinoma | RBM15 | RNA-binding motif protein 15 |
| CRC | Colorectal cancer | READ | Rectum adenocarcinoma |
| DCs | Dendritic cells | RCC | Renal cancer |
| EMT | Epithelial–mesenchymal transition | SRF | Serum response factor |
| ESCs | Embryonic stem cells | STAD | Stomach adenocarcinoma |
| FMRP | Fragile X mental retardation protein | TLR | Toll-like receptor |
| FTO | Fat mass and obesity-associated protein | Tregs | Regulatory T cells |
| GATA3 | GATA-binding protein 3 | USP7 | Ubiquitin-specific peptidase 7 |
| GC | Gastric cancer | WIF-1 | Wnt inhibitory factor 1 |
| GSCs | Glioblastoma stem cells | WTAP | Wilms tumor 1-associating protein |
| HB | Hepatoblastoma | Wnt | Wingless-related integration site |
| HCC | Hepatocellular carcinoma | YAP | Yes-associated protein |
| HDGF | Hepatoma-derived growth factor | YAP1 | Yes-associated protein 1 |
| HSPCs | Hematopoietic stem and progenitor cells | YTHDC1 | YTH domain containing 1 |
| HIF-1 | Hypoxia-inducible factor 1 | YTHDC2 | YTH domain containing 2 |
| HIF-1α | Hypoxia-inducible factor 1 alpha | YTHDF1 | YTH N6-methyladenosine RNA binding protein 1 |
| ICB | Immune checkpoint blockade | YTHDF2 | YTH N6-methyladenosine RNA binding protein 2 |
| IGF2BP1 | Insulin-like growth factor 2 mRNA binding protein 1 | YTHDF3 | YTH N6-methyladenosine RNA binding protein 3 |
| IGF2BP2 | Insulin-like growth factor 2 mRNA binding protein 2 | ZC3H13 | Zinc finger CCCH-type containing 13 |
| IGF2BP3 | Insulin-like growth factor 2 mRNA binding protein 3 | ccRCC | Clear-cell renal cell carcinoma |
| JAK/STAT | Janus kinase/signal transducer and activator of transcription | circRNA | Circular RNA |
| KIAA1429 | KIAA1429 gene | eIF3 | Eukaryotic translation initiation factor 3 |
| LGR5 | Leucine-rich repeat-containing G-protein coupled receptor 5 | m6A | N6-methyladenosine |
| LIHC | Liver hepatocellular carcinoma | mRNA | Messenger RNA |
| LUAD | Lung adenocarcinoma | mTOR | Mechanistic target of rapamycin |
| LUSC | Lung squamous cell carcinoma | miRNA | MicroRNA |
| LncRNA | Long non-coding RNA | ||
| LncXIST | Long non-coding RNA X-inactive specific transcript | ||
| MDSCs | Myeloid-derived suppressor cells | ||
| METTL14 | Methyltransferase-like protein 14 | ||
| METTL3 | Methyltransferase-like protein 3 | ||
| MYC | Myelocytomatosis oncogene | ||
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
References
- Ke, S.; Pandya-Jones, A.; Saito, Y.; Fak, J.J.; Vagbo, C.B.; Geula, S.; Hanna, J.H.; Black, D.L.; Darnell, J.E., Jr.; Darnell, R.B. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017, 31, 990–1006. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- He, C. Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol. 2010, 6, 863–865. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [PubMed]
- Machnicka, M.A.; Milanowska, K.; Osman Oglou, O.; Purta, E.; Kurkowska, M.; Olchowik, A.; Januszewski, W.; Kalinowski, S.; Dunin-Horkawicz, S.; Rother, K.M.; et al. MODOMICS: A database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013, 41, D262–D267. [Google Scholar] [CrossRef] [PubMed]
- Rottman, F.; Shatkin, A.J.; Perry, R.P. Sequences containing methylated nucleotides at the 5’ termini of messenger RNAs: Possible implications for processing. Cell 1974, 3, 197–199. [Google Scholar] [CrossRef]
- Wei, C.M.; Gershowitz, A.; Moss, B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell 1975, 4, 379–386. [Google Scholar] [CrossRef]
- Xiao, C.L.; Zhu, S.; He, M.; Chen, D.; Zhang, Q.; Chen, Y.; Yu, G.; Liu, J.; Xie, S.Q.; Luo, F.; et al. N6-Methyladenine DNA Modification in the Human Genome. Mol. Cell 2018, 71, 306–318.e307. [Google Scholar] [CrossRef]
- Ren, W.; Lu, J.; Huang, M.; Gao, L.; Li, D.; Wang, G.G.; Song, J. Structure and regulation of ZCCHC4 in mA-methylation of 28S rRNA. Nat. Commun. 2019, 10, 5042. [Google Scholar] [CrossRef]
- van Tran, N.; Ernst, F.G.M.; Hawley, B.R.; Zorbas, C.; Ulryck, N.; Hackert, P.; Bohnsack, K.E.; Bohnsack, M.T.; Jaffrey, S.R.; Graille, M.; et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019, 47, 7719–7733. [Google Scholar] [CrossRef]
- Pendleton, K.E.; Chen, B.; Liu, K.; Hunter, O.V.; Xie, Y.; Tu, B.P.; Conrad, N.K. The U6 snRNA m6A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 2017, 169, 824–835.e14. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vagbo, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Patil, D.P.; Chen, C.-K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Liu, N.; Zhou, K.I.; Parisien, M.; Dai, Q.; Diatchenko, L.; Pan, T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017, 45, 6051–6063. [Google Scholar] [CrossRef]
- Zhou, K.I.; Shi, H.; Lyu, R.; Wylder, A.C.; Matuszek, Z.; Pan, J.N.; He, C.; Parisien, M.; Pan, T. Regulation of Co-transcriptional Pre-mRNA Splicing by m6A through the Low-Complexity Protein hnRNPG. Mol. Cell 2019, 76, 70–81.e79. [Google Scholar] [CrossRef]
- Ozkurede, U.; Kala, R.; Johnson, C.; Shen, Z.; Miller, R.A.; Garcia, G.G. Cap-independent mRNA translation is upregulated in long-lived endocrine mutant mice. J. Mol. Endocrinol. 2019, 63, 123–138. [Google Scholar] [CrossRef]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.J.; Shi, H.; Zhu, A.C.; Lu, Z.; Miller, N.; Edens, B.M.; Ma, Y.C.; He, C. The RNA-binding protein FMRP facilitates the nuclear export of N6-methyladenosine-containing mRNAs. J. Biol. Chem. 2019, 294, 19889–19895. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Dong, L.; Liu, X.M.; Guo, J.; Ma, H.; Shen, B.; Qian, S.B. m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019, 10, 5332. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5’ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.-B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m6A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef]
- Orouji, E.; Peitsch, W.K.; Orouji, A.; Houben, R.; Utikal, J. Oncogenic Role of an Epigenetic Reader of m6A RNA Modification: YTHDF1 in Merkel Cell Carcinoma. Cancers 2020, 12, 202. [Google Scholar] [CrossRef]
- Barbieri, I.; Tzelepis, K.; Pandolfini, L.; Shi, J.; Millán-Zambrano, G.; Robson, S.C.; Aspris, D.; Migliori, V.; Bannister, A.J.; Han, N.; et al. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature 2017, 552, 126. [Google Scholar] [CrossRef]
- Geula, S.; Moshitch-Moshkovitz, S.; Dominissini, D.; Mansour, A.A.; Kol, N.; Salmon-Divon, M.; Hershkovitz, V.; Peer, E.; Mor, N.; Manor, Y.S.; et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 2015, 347, 1002–1006. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, Y.; Sun, B.; Wang, L.; Yang, Y.; Ma, D.; Lv, J.; Heng, J.; Ding, Y.; Xue, Y.; et al. m6A modulates haematopoietic stem and progenitor cell specification. Nature 2017, 549, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhang, Y.; Gao, S.; Zhang, C.; Chen, Y.; Li, W.; Yang, Y.G.; Zhou, Q.; Liu, F. Endothelial-specific m6A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res. 2018, 28, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Cirovic, B.; Schönheit, J.; Kowenz-Leutz, E.; Ivanovska, J.; Klement, C.; Pronina, N.; Bégay, V.; Leutz, A. C/EBP-Induced Transdifferentiation Reveals Granulocyte-Macrophage Precursor-like Plasticity of B Cells. Stem Cell Rep. 2017, 8, 346–359. [Google Scholar] [CrossRef]
- Wang, H.; Hu, X.; Huang, M.; Liu, J.; Gu, Y.; Ma, L.; Zhou, Q.; Cao, X. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat. Commun. 2019, 10, 1898. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Lin, Z.; Hsu, P.J.; Xing, X.; Fang, J.; Lu, Z.; Zou, Q.; Zhang, K.-J.; Zhang, X.; Zhou, Y.; Zhang, T.; et al. Mettl3-/Mettl14-mediated mRNA N-methyladenosine modulates murine spermatogenesis. Cell Res. 2017, 27, 1216–1230. [Google Scholar] [CrossRef]
- Xu, K.; Yang, Y.; Feng, G.H.; Sun, B.F.; Chen, J.Q.; Li, Y.F.; Chen, Y.S.; Zhang, X.X.; Wang, C.X.; Jiang, L.Y.; et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017, 27, 1100–1114. [Google Scholar] [CrossRef]
- Ma, C.; Chang, M.; Lv, H.; Zhang, Z.-W.; Zhang, W.; He, X.; Wu, G.; Zhao, S.; Zhang, Y.; Wang, D.; et al. RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018, 19, 68. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Y.-C.; Huang, C.; Shen, H.; Sun, B.; Cheng, X.; Zhang, Y.-J.; Yang, Y.-G.; Shu, Q.; Yang, Y.; et al. m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genom. Proteom. Bioinform. 2019, 17, 154–168. [Google Scholar] [CrossRef]
- Xiang, Y.; Laurent, B.; Hsu, C.-H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Gao, Y.; Lv, D.; Wang, C.; Wang, D.; Li, Q. METTL3 mediated m6A modification plays an oncogenic role in cutaneous squamous cell carcinoma by regulating DeltaNp63. Biochem. Biophys. Res. Commun. 2019, 515, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Miao, W.; Chen, J.; Jia, L.; Ma, J.; Song, D. The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem. Biophys. Res. Commun. 2019, 516, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Si, Y.; Xu, J.; Lin, Y.; Wang, J.-Z.; Cao, M.; Sun, S.; Ding, Q.; Zhu, L.; Wei, J.-F. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J. Cell. Mol. Med. 2020, 24, 3521–3533. [Google Scholar] [CrossRef]
- Cheng, M.; Sheng, L.; Gao, Q.; Xiong, Q.; Zhang, H.; Wu, M.; Liang, Y.; Zhu, F.; Zhang, Y.; Zhang, X.; et al. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene 2019, 38, 3667–3680. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Sun, G.; Wu, Q.; Ma, J.; Zhang, X.; Huang, N.; Bian, Z.; Gu, S.; Xu, M.; et al. m6A mRNA methylation regulates CTNNB1 to promote the proliferation of hepatoblastoma. Mol. Cancer 2019, 18, 188. [Google Scholar] [CrossRef]
- Chen, M.; Wei, L.; Law, C.-T.; Tsang, F.H.-C.; Shen, J.; Cheng, C.L.-H.; Tsang, L.-H.; Ho, D.W.-H.; Chiu, D.K.-C.; Lee, J.M.-F.; et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018, 67, 2254–2270. [Google Scholar] [CrossRef]
- Lan, T.; Li, H.; Zhang, D.; Xu, L.; Liu, H.; Hao, X.; Yan, X.; Liao, H.; Chen, X.; Xie, K.; et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer 2019, 18, 186. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, C.; Ding, Q.; Zhao, Y.; Wang, Z.; Chen, J.; Jiang, Z.; Zhang, Y.; Xu, G.; Zhang, J.; et al. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020, 69, 1193–1205. [Google Scholar] [CrossRef]
- Liu, T.; Yang, S.; Sui, J.; Xu, S.-Y.; Cheng, Y.-P.; Shen, B.; Zhang, Y.; Zhang, X.-M.; Yin, L.-H.; Pu, Y.-P.; et al. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J. Cell. Physiol. 2020, 235, 548–562. [Google Scholar] [CrossRef]
- Gong, D.; Zhang, J.; Chen, Y.; Xu, Y.; Ma, J.; Hu, G.; Huang, Y.; Zheng, J.; Zhai, W.; Xue, W. The m6A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca2+ influx modulating ERK1/2 phosphorylation and MMP9 signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, M.; Ge, S.; Huang, W.; Lin, X.; Gao, J.; Gong, J.; Shen, L. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019, 8, 4766–4781. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Su, R.; Sheng, Y.; Dong, L.; Dong, Z.; Xu, H.; Ni, T.; Zhang, Z.S.; Zhang, T.; Li, C.; et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 2019, 35, 677–691.E10. [Google Scholar] [CrossRef]
- Zou, D.; Dong, L.; Li, C.; Yin, Z.; Rao, S.; Zhou, Q. The m6A eraser FTO facilitates proliferation and migration of human cervical cancer cells. Cancer Cell Int. 2019, 19, 321. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M.; et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef]
- Tang, B.; Yang, Y.; Kang, M.; Wang, Y.; Wang, Y.; Bi, Y.; He, S.; Shimamoto, F. m6A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol. Cancer 2020, 19, 3. [Google Scholar] [CrossRef]
- Han, J.; Wang, J.-Z.; Yang, X.; Yu, H.; Zhou, R.; Lu, H.-C.; Yuan, W.-B.; Lu, J.-C.; Zhou, Z.-J.; Lu, Q.; et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 2019, 18, 110. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, S.; Zhao, T.; Dang, C. METTL3-mediated m6A modification of Bcl-2 mRNA promotes non-small cell lung cancer progression. Oncol. Rep. 2021, 46, 163. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Qian, F.; Zhu, Y.; He, G.; Yang, J.; Wu, X.; Zhang, H.; Yu, X.; Liu, X. Deubiquitinase USP19 modulates apoptotic calcium release and endoplasmic reticulum stress by deubiquitinating BAG6 in triple negative breast cancer. Clin. Transl. Med. 2023, 13, e1398. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, B.; Shi, J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene 2020, 722, 144076. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Xie, X.; Han, G.; Zhang, T.; Li, Y.; Li, Y.; Yin, R.; Wang, Q.; Zhang, T.; Wang, P.; et al. YBX1 is required for maintaining myeloid leukemia cell survival by regulating BCL2 stability in an m6A-dependent manner. Blood 2021, 138, 71–85. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhou, B.; Hu, X.; Ying, S.; Zhou, Q.; Xu, W.; Feng, L.; Hou, T.; Wang, X.; Zhu, L.; et al. LncRNA LINC00942 promotes chemoresistance in gastric cancer by suppressing MSI2 degradation to enhance c-Myc mRNA stability. Clin. Transl. Med. 2022, 12, e703. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Jiang, X.; Li, D.; Jiang, X. HBXIP promotes gastric cancer via METTL3-mediated MYC mRNA m6A modification. Aging 2020, 12, 24967–24982. [Google Scholar] [CrossRef]
- Aziz, K.; Limzerwala, J.F.; Sturmlechner, I.; Hurley, E.; Zhang, C.; Jeganathan, K.B.; Nelson, G.; Bronk, S.; Fierro Velasco, R.O.; van Deursen, E.J.; et al. Ccne1 Overexpression Causes Chromosome Instability in Liver Cells and Liver Tumor Development in Mice. Gastroenterology 2019, 157, 210–226.e212. [Google Scholar] [CrossRef]
- Wu, Y.; Zheng, Q.; Li, Y.; Wang, G.; Gao, S.; Zhang, X.; Yan, X.; Zhang, X.; Xie, J.; Wang, Y.; et al. Metformin targets a YAP1-TEAD4 complex via AMPKalpha to regulate CCNE1/2 in bladder cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 376. [Google Scholar] [CrossRef]
- Jin, D.; Guo, J.; Wu, Y.; Yang, L.; Wang, X.; Du, J.; Dai, J.; Chen, W.; Gong, K.; Miao, S.; et al. m6A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol. Cancer 2020, 19, 40. [Google Scholar] [CrossRef]
- Wei, Y.; Fu, J.; Zhang, H.; Ling, Y.; Tang, X.; Liu, S.; Yu, M.; Liu, F.; Zhuang, G.; Qian, H.; et al. N6-methyladenosine modification promotes hepatocarcinogenesis through circ-CDYL-enriched and EpCAM-positive liver tumor-initiating exosomes. iScience 2023, 26, 108022. [Google Scholar] [CrossRef]
- Müller, S.; Glaß, M.; Singh, A.K.; Haase, J.; Bley, N.; Fuchs, T.; Lederer, M.; Dahl, A.; Huang, H.; Chen, J.; et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019, 47, 375–390. [Google Scholar] [CrossRef]
- Tanabe, A.; Tanikawa, K.; Tsunetomi, M.; Takai, K.; Ikeda, H.; Konno, J.; Torigoe, T.; Maeda, H.; Kutomi, G.; Okita, K.; et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1alpha mRNA is translated. Cancer Lett. 2016, 376, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Yang, F.; Zhan, H.; Situ, J.; Li, W.; Mao, Y.; Luo, Y. RNA m6A Methyltransferase METTL3 Promotes The Growth of Prostate Cancer By Regulating Hedgehog Pathway. OncoTargets Ther. 2019, 12, 9143–9152. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, C.; Sun, Y.; He, X.; Xue, D. m6A RNA modification modulates gene expression and cancer-related pathways in clear cell renal cell carcinoma. Epigenomics 2019, 12, 87–99. [Google Scholar] [CrossRef]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.-G.; et al. m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef] [PubMed]
- Marceau, A.H.; Brison, C.M.; Nerli, S.; Arsenault, H.E.; McShan, A.C.; Chen, E.; Lee, H.W.; Benanti, J.A.; Sgourakis, N.G.; Rubin, S.M. An order-to-disorder structural switch activates the FoxM1 transcription factor. elife 2019, 8, e46131. [Google Scholar] [CrossRef]
- Chao, Y.; Shang, J.; Ji, W. ALKBH5-m6A-FOXM1 signaling axis promotes proliferation and invasion of lung adenocarcinoma cells under intermittent hypoxia. Biochem. Biophys. Res. Commun. 2020, 521, 499–506. [Google Scholar] [CrossRef]
- Li, J.; Han, Y.; Zhang, H.; Qian, Z.; Jia, W.; Gao, Y.; Zheng, H.; Li, B. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem. Biophys. Res. Commun. 2019, 512, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Eckert, M.A.; Harada, B.T.; Liu, S.-M.; Lu, Z.; Yu, K.; Tienda, S.M.; Chryplewicz, A.; Zhu, A.C.; Yang, Y.; et al. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018, 20, 1074–1083. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.; Kong, B.; Song, C.; Cong, J.; Hou, J.; Wang, S. Reduced m6A mRNA methylation is correlated with the progression of human cervical cancer. Oncotarget 2017, 8, 98918–98930. [Google Scholar] [CrossRef]
- Li, Q.; Ni, Y.; Zhang, L.; Jiang, R.; Xu, J.; Yang, H.; Hu, Y.; Qiu, J.; Pu, L.; Tang, J.; et al. HIF-1α-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct. Target. Ther. 2021, 6, 76. [Google Scholar] [CrossRef]
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Y.; Zhang, Y.; Wang, Y.; Luo, Y.; Ding, H.; Li, P.; Ni, G. HIF-1α Regulated WTAP Overexpression Promoting the Warburg Effect of Ovarian Cancer by m6A-Dependent Manner. J. Immunol. Res. 2022, 2022, 6130806. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cheng, C.; He, B.; Du, X.; Liu, J.; Xia, H.; Wang, P.; Wu, M.; Wu, H.; Liu, Q. Cigarette smoking, by accelerating the cell cycle, promotes the progression of non-small cell lung cancer through an HIF-1α-METTL3-m6A/CDK2AP2 axis. J. Hazard. Mater. 2023, 455, 131556. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, J.; Huang, Z.; Jin, P.; Peng, L.; Luo, M.; Zhang, Z.; Chen, Y.; Xie, N.; Gao, W.; et al. Hypoxia-induced lncRNA STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal cancer progression by preventing m6A-mediated degradation of STEAP3 mRNA. Mol. Cancer 2022, 21, 168. [Google Scholar] [CrossRef]
- Xiong, J.; He, J.; Zhu, J.; Pan, J.; Liao, W.; Ye, H.; Wang, H.; Song, Y.; Du, Y.; Cui, B.; et al. Lactylation-driven METTL3-mediated RNA m6A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell 2022, 82, 1660–1677.E10. [Google Scholar] [CrossRef]
- Yu, J.; Chai, P.; Xie, M.; Ge, S.; Ruan, J.; Fan, X.; Jia, R. Histone lactylation drives oncogenesis by facilitating m6A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 2021, 22, 85. [Google Scholar] [CrossRef]
- Wang, P.; Xie, D.; Xiao, T.; Cheng, C.; Wang, D.; Sun, J.; Wu, M.; Yang, Y.; Zhang, A.; Liu, Q. H3K18 lactylation promotes the progression of arsenite-related idiopathic pulmonary fibrosis via YTHDF1/m6A/NREP. J. Hazard. Mater. 2024, 461, 132582. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.Y. GATA3: A master of many trades in immune regulation. Trends Immunol. 2014, 35, 233–242. [Google Scholar] [CrossRef]
- Du, F.; Yuan, P.; Wang, T.; Zhao, J.; Zhao, Z.; Luo, Y.; Xu, B. The Significance and Therapeutic Potential of GATA3 Expression and Mutation in Breast Cancer: A Systematic Review. Med. Res. Rev. 2015, 35, 1300–1315. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, C.; Chen, J.; Chen, D.; Yang, B.; He, B.; Hu, W.; Zhang, Y.; Liu, H.; Dai, L.; et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer 2019, 18, 127. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bogler, O.; et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606.e596. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; He, C. Fate by RNA methylation: m6A steers stem cell pluripotency. Genome Biol. 2015, 16, 43. [Google Scholar] [CrossRef] [PubMed]
- Roignant, J.Y.; Soller, M. m6A in mRNA: An Ancient Mechanism for Fine-Tuning Gene Expression. Trends Genet. TIG 2017, 33, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Alarcon, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef]
- Zuo, X.; Chen, Z.; Gao, W.; Zhang, Y.; Wang, J.; Wang, J.; Cao, M.; Cai, J.; Wu, J.; Wang, X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020, 13, 5. [Google Scholar] [CrossRef]
- Xu, J.; Liu, C.; Qu, K.; Zhang, J.; Liu, S.; Meng, F.; Wan, Y. m6A methyltransferase METTL14-mediated RP1-228H13.5 promotes the occurrence of liver cancer by targeting hsa-miR-205/ZIK1. Oncol. Rep. 2024, 51, 59. [Google Scholar] [CrossRef]
- Ni, W.; Yao, S.; Zhou, Y.; Liu, Y.; Huang, P.; Zhou, A.; Liu, J.; Che, L.; Li, J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol. Cancer 2019, 18, 143. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, X.; Chen, Z.; Tian, L.; Jiang, G.; Chen, F.; Li, J.; An, P.; Lu, L.; Luo, N.; et al. m6A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol. Cancer 2019, 18, 87. [Google Scholar] [CrossRef]
- Hu, X.; Peng, W.-X.; Zhou, H.; Jiang, J.; Zhou, X.; Huang, D.; Mo, Y.-Y.; Yang, L. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2019, 27, 1782–1794. [Google Scholar] [CrossRef]
- Zhao, K.; Chen, L.; Xie, Y.; Ren, N.; Li, J.; Zhai, X.; Zheng, S.; Liu, K.; Wang, C.; Qiu, Q.; et al. m6A/HOXA10-AS/ITGA6 axis aggravates oxidative resistance and malignant progression of laryngeal squamous cell carcinoma through regulating Notch and Keap1/Nrf2 pathways. Cancer Lett. 2024, 587, 216735. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, C.; Chen, C.; Guo, Y.; Yuan, W.; Yin, D.; Liu, J.; Sun, Z. The role of N6-methyladenosine (m6A) modification in the regulation of circRNAs. Mol. Cancer 2020, 19, 105. [Google Scholar] [CrossRef]
- Li, H.; Lin, R.; Zhang, Y.; Zhu, Y.; Huang, S.; Lan, J.; Lu, N.; Xie, C.; He, S.; Zhang, W. N6-methyladenosine-modified circPLPP4 sustains cisplatin resistance in ovarian cancer cells via PIK3R1 upregulation. Mol. Cancer 2024, 23, 5. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhou, J.; Zhang, X.; Kang, X.; Liu, S.; Song, M.; Chang, C.; Lin, Y.; Wang, Y. N6-methyladenosine-modified circ_104797 sustains cisplatin resistance in bladder cancer through acting as RNA sponges. Cell Mol. Biol. Lett. 2024, 29, 28. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Ma, X.; Ying, Y.; Liu, Z.; Tang, Y.; Shu, X.; Sun, J.; Wu, Y.; Lu, D.; Wang, X.; et al. N6-methyladenosine-modified CircPSMA7 enhances bladder cancer malignancy through the miR-128-3p/MAPK1 axis. Cancer Lett. 2024, 585, 216613. [Google Scholar] [CrossRef]
- Zheng, Z.; Zeng, X.; Zhu, Y.; Leng, M.; Zhang, Z.; Wang, Q.; Liu, X.; Zeng, S.; Xiao, Y.; Hu, C.; et al. CircPPAP2B controls metastasis of clear cell renal cell carcinoma via HNRNPC-dependent alternative splicing and targeting the miR-182-5p/CYP1B1 axis. Mol. Cancer 2024, 23, 4. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yao, L.; Su, Y.; Tian, L. IGF2BP3 loss inhibits cell progression by upregulating has_circRNA_103820, and hsa_circRNA_103820-encoded peptide inhibits cell progression by inactivating the AKT pathway in lung cancer. Chem. Biol. Drug Des. 2024, 103, e14473. [Google Scholar] [CrossRef]
- Li, J.; Cao, H.; Yang, J.; Wang, B. IGF2BP2-m6A-circMMP9 axis recruits ETS1 to promote TRIM59 transcription in laryngeal squamous cell carcinoma. Sci. Rep. 2024, 14, 3014. [Google Scholar] [CrossRef]
- Zhou, Q.; Song, W.; Li, X.; Lin, J.; Zhu, C.; Cao, L.; Li, W.; Lin, S. N6-Methyladenosine reader HNRNPC-mediated downregulation of circITCH prevents miR-224-3p sequestering and contributes to tumorigenesis in nasopharyngeal carcinoma. Environ. Toxicol. 2024, 39, 2893–2907. [Google Scholar] [CrossRef]
- Luo, L.; Li, P.; Xie, Q.; Wu, Y.; Qin, F.; Liao, D.; Zeng, K.; Wang, K. n6-methyladenosine-modified circular RNA family with sequence similarity 126, member A affects cholesterol synthesis and malignant progression of prostate cancer cells by targeting microRNA-505-3p to mediate calnexin. J. Cancer 2024, 15, 966–980. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, K.; Shou, Y.; Li, S.; Wang, J.; Zhang, Q.; Huang, Z.; Xu, J.; Li, M.; Liu, D.; et al. circRARS synergises with IGF2BP3 to regulate RNA methylation recognition to promote tumour progression in renal cell carcinoma. Clin. Transl. Med. 2023, 13, e1512. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.; Morgan, M.; Campos, J.; Spencer, G.J.; Shmakova, A.; Ivanova, I.; Mapperley, C.; Lawson, H.; Wotherspoon, D.A.; Sepulveda, C.; et al. Targeting the RNA m6A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell 2019, 25, 137–148.e136. [Google Scholar] [CrossRef]
- Taketo, K.; Konno, M.; Asai, A.; Koseki, J.; Toratani, M.; Satoh, T.; Doki, Y.; Mori, M.; Ishii, H.; Ogawa, K. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol. 2018, 52, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Dong, L.; Li, C.; Nachtergaele, S.; Wunderlich, M.; Qing, Y.; Deng, X.; Wang, Y.; Weng, X.; Hu, C.; et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/mA/MYC/CEBPA Signaling. Cell 2018, 172, 90–105.E23. [Google Scholar] [CrossRef] [PubMed]
- Rau, K.; Roesner, L.; Rentmeister, A. Sequence-specific m6A demethylation in RNA by FTO fused to RCas9. RNA 2019, 25, 1311–1323. [Google Scholar] [CrossRef]
- Liu, Q.; Li, M.; Jiang, L.; Jiang, R.; Fu, B. METTL3 promotes experimental osteoarthritis development by regulating inflammatory response and apoptosis in chondrocyte. Biochem. Biophys. Res. Commun. 2019, 516, 22–27. [Google Scholar] [CrossRef]
- Jia, R.; Chai, P.; Wang, S.; Sun, B.; Xu, Y.; Yang, Y.; Ge, S.; Jia, R.; Yang, Y.G.; Fan, X. m6A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol. Cancer 2019, 18, 161. [Google Scholar] [CrossRef]
- Hou, J.; Zhang, H.; Liu, J.; Zhao, Z.; Wang, J.; Lu, Z.; Hu, B.; Zhou, J.; Zhao, Z.; Feng, M.; et al. YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer 2019, 18, 163. [Google Scholar] [CrossRef]
- Bertero, A.; Brown, S.; Madrigal, P.; Osnato, A.; Ortmann, D.; Yiangou, L.; Kadiwala, J.; Hubner, N.C.; de Los Mozos, I.R.; Sadee, C.; et al. The SMAD2/3 interactome reveals that TGFbeta controls m6A mRNA methylation in pluripotency. Nature 2018, 555, 256–259. [Google Scholar] [CrossRef]
- Liao, Z.; Zhang, H.; Liu, F.; Wang, W.; Liu, Y.; Su, C.; Zhu, H.; Chen, X.; Zhang, B.; Zhang, Z. m6A-Dependent ITIH1 Regulated by TGF-β Acts as a Target for Hepatocellular Carcinoma Progression. Adv. Sci. 2024, 11, e2401013. [Google Scholar] [CrossRef]
- Wu, Z.; Ke, Q.; Jiang, L.; Hong, H.; Pan, W.; Chen, W.; Abudukeremu, X.; She, F.; Chen, Y. TGF-β1 facilitates gallbladder carcinoma metastasis by regulating FOXA1 translation efficiency through m6A modification. Cell Death Dis. 2024, 15, 422. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Yu, B.; Tao, D.; Xu, X.; Xu, Y.; Wang, J.; Jiao, Y.; Wang, L. The role of m6A methylation in therapy resistance in cancer. Mol. Cancer 2023, 22, 91. [Google Scholar] [CrossRef]
- Lin, H.; Wang, Y.; Wang, P.; Long, F.; Wang, T. Mutual regulation between N6-methyladenosine (m6A) modification and circular RNAs in cancer: Impacts on therapeutic resistance. Mol. Cancer 2022, 21, 148. [Google Scholar] [CrossRef]
- Han, D.; Liu, J.; Chen, C.; Dong, L.; Liu, Y.; Chang, R.; Huang, X.; Liu, Y.; Wang, J.; Dougherty, U.; et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019, 566, 270–274. [Google Scholar] [CrossRef]
- Li, N.; Kang, Y.; Wang, L.; Huff, S.; Tang, R.; Hui, H.; Agrawal, K.; Gonzalez, G.M.; Wang, Y.; Patel, S.P.; et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. USA 2020, 117, 20159–20170. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, Y.; Gao, M.; Han, L.; Wunderlich, M.; Deng, X.; Li, H.; Huang, Y.; Gao, L.; et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 2020, 38, 79–96.E11. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhang, H.; Chen, J.; Lin, L.; Chen, Y. Immune signature of T follicular helper cells predicts clinical prognostic and therapeutic impact in lung squamous cell carcinoma. Int. Immunopharmacol. 2020, 81, 105932. [Google Scholar] [CrossRef] [PubMed]
- Hemmi, H.; Takeuchi, O.; Kawai, T.; Kaisho, T.; Sato, S.; Sanjo, H.; Matsumoto, M.; Hoshino, K.; Wagner, H.; Takeda, K.; et al. A Toll-like receptor recognizes bacterial DNA. Nature 2000, 408, 740–745. [Google Scholar] [CrossRef]
- Ronnblom, L.; Eloranta, M.L.; Alm, G.V. Role of natural interferon-alpha producing cells (plasmacytoid dendritic cells) in autoimmunity. Autoimmunity 2003, 36, 463–472. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef]
- Karikó, K.; Ni, H.; Capodici, J.; Lamphier, M.; Weissman, D. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 2004, 279, 12542–12550. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Lluch, S.; Blanco, E.; Tilgner, H.; Curado, J.; Ruiz-Romero, M.; Corominas, M.; Guigó, R. Absence of canonical marks of active chromatin in developmentally regulated genes. Nat. Genet. 2015, 47, 1158–1167. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Smith, A.R.; Qian, Z.; Zheng, G. Patent landscape of small molecule inhibitors of METTL3 (2020-present). Expert. Opin. Ther. Pat. 2024, 35, 305–320. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).