Dramatic Responses to High-Dose Ipilimumab Plus Temozolomide After Progression on Standard- or Low-Dose Ipilimumab in Advanced Melanoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. Whole-Exome Sequencing
2.3. RNA-Sequencing
2.4. Statistical Analysis
3. Results
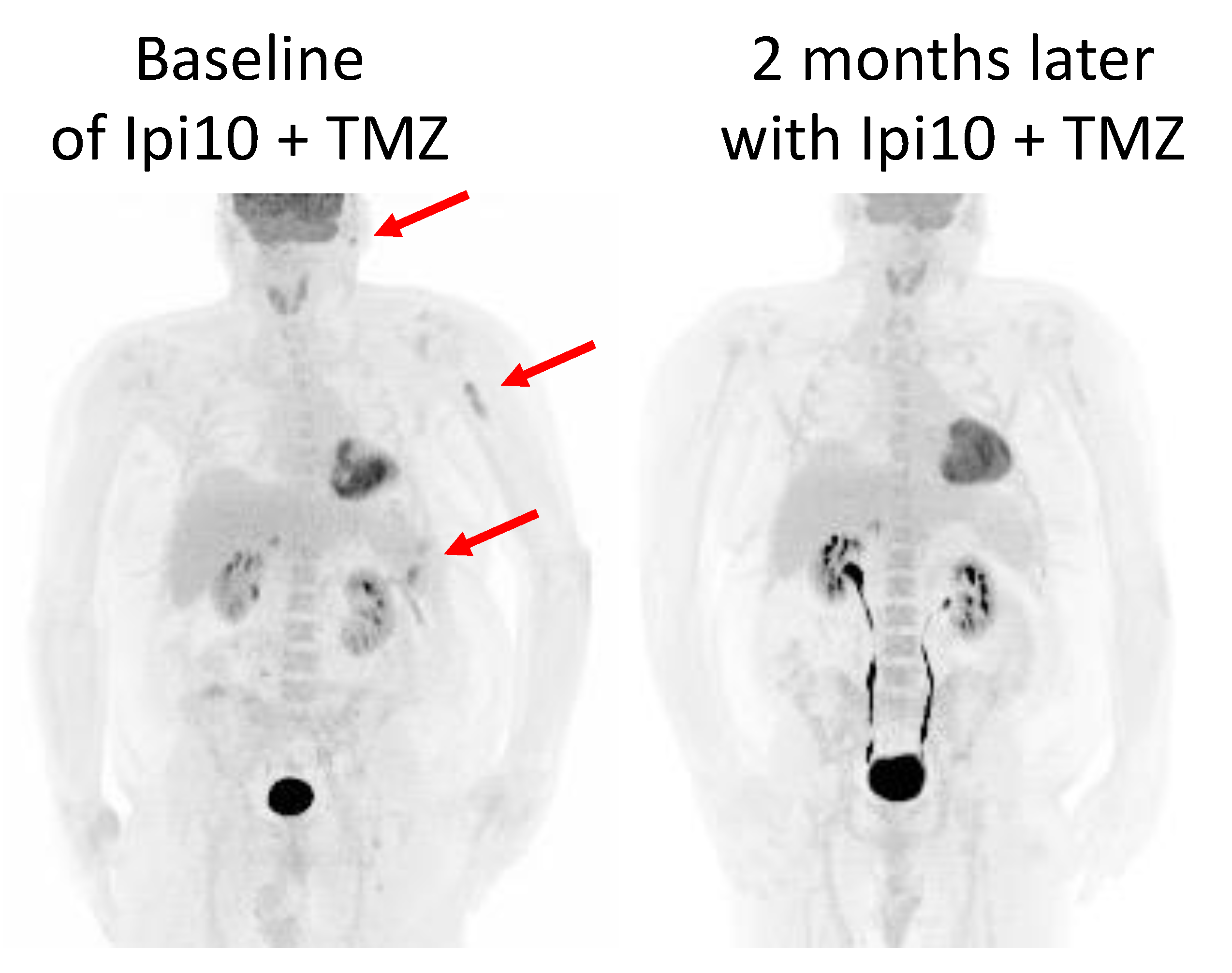
3.1. Case Study 1
3.2. Case Study 2
3.3. Cohort
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Tawbi, H.A.; Schadendorf, D.; Lipson, E.J.; Ascierto, P.A.; Matamala, L.; Castillo Gutierrez, E.; Rutkowski, P.; Gogas, H.J.; Lao, C.D.; De Menezes, J.J.; et al. Relatlimab and Nivolumab versus Nivolumab in Untreated Advanced Melanoma. N. Engl. J. Med. 2022, 386, 24–34. [Google Scholar] [CrossRef]
- Long, G.V.; Hodi, S.F.; Lipson, E.J.; Schadendorf, D.; Ascierto, P.A.; Matamala, L.; Salman, P.; Gutierrez, E.C.; Rutkowski, P.; Gogas, H.; et al. Relatlimab and nivolumab versus nivolumab in previously untreated metastatic or unresectable melanoma: Overall survival and response rates from RELATIVITY-047 (CA224-047). J. Clin. Oncol. 2022, 40. [Google Scholar] [CrossRef]
- Tawbi, H.A.; Forsyth, P.A.; Hodi, F.S.; Algazi, A.P.; Hamid, O.; Lao, C.D.; Moschos, S.J.; Atkins, M.B.; Lewis, K.; Postow, M.A.; et al. Long-term outcomes of patients with active melanoma brain metastases treated with combination nivolumab plus ipilimumab (CheckMate 204): Final results of an open-label, multicentre, phase 2 study. Lancet Oncol. 2021, 22, 1692–1704. [Google Scholar] [CrossRef] [PubMed]
- Lebbe, C.; Meyer, N.; Mortier, L.; Marquez-Rodas, I.; Robert, C.; Rutkowski, P.; Menzies, A.M.; Eigentler, T.; Ascierto, P.A.; Smylie, M.; et al. Evaluation of Two Dosing Regimens for Nivolumab in Combination With Ipilimumab in Patients With Advanced Melanoma: Results From the Phase IIIb/IV CheckMate 511 Trial. J. Clin. Oncol. 2019, 37, 867–875. [Google Scholar] [CrossRef]
- Ascierto, P.A.; Del Vecchio, M.; Robert, C.; Mackiewicz, A.; Chiarion-Sileni, V.; Arance, A.; Lebbe, C.; Bastholt, L.; Hamid, O.; Rutkowski, P.; et al. Ipilimumab 10 mg/kg versus ipilimumab 3 mg/kg in patients with unresectable or metastatic melanoma: A randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol. 2017, 18, 611–622. [Google Scholar] [CrossRef]
- Lai-Kwon, J.E.; Jacques, S.; Carlino, M.S.; Benannoune, N.; Robert, C.; Allayous, C.; Baroudjian, B.; Lebbe, C.; Zimmer, L.; Eroglu, Z.; et al. Efficacy of ipilimumab 3mg/kg following progression on low dose ipilimumab in metastatic melanoma. J. Clin. Oncol. 2022. [Google Scholar] [CrossRef]
- Banissi, C.; Ghiringhelli, F.; Chen, L.; Carpentier, A.F. Treg depletion with a low-dose metronomic temozolomide regimen in a rat glioma model. Cancer Immunol. Immunother. 2009, 58, 1627–1634. [Google Scholar] [CrossRef]
- Ridolfi, L.; Petrini, M.; Granato, A.M.; Gentilcore, G.; Simeone, E.; Ascierto, P.A.; Pancisi, E.; Ancarani, V.; Fiammenghi, L.; Guidoboni, M.; et al. Low-dose temozolomide before dendritic-cell vaccination reduces (specifically) CD4+CD25++Foxp3+ regulatory T-cells in advanced melanoma patients. J. Transl. Med. 2013, 11, 135. [Google Scholar] [CrossRef]
- Patel, S.P.; Kim, D.W.; Bassett, R.L.; Cain, S.; Washington, E.; Hwu, W.J.; Kim, K.B.; Papadopoulos, N.E.; Homsi, J.; Hwu, P.; et al. A phase II study of ipilimumab plus temozolomide in patients with metastatic melanoma. Cancer Immunol. Immunother. 2017, 66, 1359–1366. [Google Scholar] [CrossRef]
- Robert, C.; Schachter, J.; Long, G.V.; Arance, A.; Grob, J.J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2015, 372, 2521–2532. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Kallberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 2001, 11, 863–874. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Suda, K.; Kim, J.; Murakami, I.; Rozeboom, L.; Shimoji, M.; Shimizu, S.; Rivard, C.J.; Mitsudomi, T.; Tan, A.C.; Hirsch, F.R. Innate Genetic Evolution of Lung Cancers and Spatial Heterogeneity: Analysis of Treatment-Naive Lesions. J. Thorac. Oncol. 2018, 13, 1496–1507. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic. Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martinez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Freshwater, T.; Kondic, A.; Ahamadi, M.; Li, C.H.; de Greef, R.; de Alwis, D.; Stone, J.A. Evaluation of dosing strategy for pembrolizumab for oncology indications. J. Immunother. Cancer 2017, 5, 43. [Google Scholar] [CrossRef]
- Munhoz, R.R.; Postow, M.A. Clinical Development of PD-1 in Advanced Melanoma. Cancer J. 2018, 24, 7–14. [Google Scholar] [CrossRef]
- Feng, Y.; Roy, A.; Masson, E.; Chen, T.T.; Humphrey, R.; Weber, J.S. Exposure-response relationships of the efficacy and safety of ipilimumab in patients with advanced melanoma. Clin. Cancer Res. 2013, 19, 3977–3986. [Google Scholar] [CrossRef]
- Wolchok, J.D.; Neyns, B.; Linette, G.; Negrier, S.; Lutzky, J.; Thomas, L.; Waterfield, W.; Schadendorf, D.; Smylie, M.; Guthrie, T., Jr.; et al. Ipilimumab monotherapy in patients with pretreated advanced melanoma: A randomised, double-blind, multicentre, phase 2, dose-ranging study. Lancet Oncol. 2010, 11, 155–164. [Google Scholar] [CrossRef]
- Kavanagh, B.; O’Brien, S.; Lee, D.; Hou, Y.; Weinberg, V.; Rini, B.; Allison, J.P.; Small, E.J.; Fong, L. CTLA4 blockade expands FoxP3+ regulatory and activated effector CD4+ T cells in a dose-dependent fashion. Blood 2008, 112, 1175–1183. [Google Scholar] [CrossRef]
- Goleva, E.; Lyubchenko, T.; Kraehenbuehl, L.; Lacouture, M.E.; Leung, D.Y.M.; Kern, J.A. Our current understanding of checkpoint inhibitor therapy in cancer immunotherapy. Ann. Allergy Asthma Immunol. 2021, 126, 630–638. [Google Scholar] [CrossRef]
- Hodi, F.S.; Wolchok, J.D.; Schadendorf, D.; Larkin, J.; Long, G.V.; Qian, X.; Saci, A.; Young, T.C.; Srinivasan, S.; Chang, H.; et al. TMB and Inflammatory Gene Expression Associated with Clinical Outcomes following Immunotherapy in Advanced Melanoma. Cancer Immunol. Res. 2021, 9, 1202–1213. [Google Scholar] [CrossRef]
- Zhu, W.; Zhou, L.; Qian, J.Q.; Qiu, T.Z.; Shu, Y.Q.; Liu, P. Temozolomide for treatment of brain metastases: A review of 21 clinical trials. World J. Clin. Oncol. 2014, 5, 19–27. [Google Scholar] [CrossRef]



| Pt ID | Gender | Age at C1D1 | Ipilimumab mg/kg | Temodar 200 mg/m2 D1–D4 | Melanoma Subtype | BRAF | Prior Therapies | No of Cycles | Best Response | PFS | OS | Alive | Next Lines of Therapy | irAE | Grade | HD Steroids? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10A | F | 30 | 10 | Y | Nodular | V600E | neoadj pembro, D/T/Pembro, Ipi1+nivo3 | 4 | PR | 219 | 537 | Y | ipi3/nivo1 | Hepatitis | 3 | Y |
| 10B | M | 52 | 10 | Y | Superficial Spreading | V600E | Adj nivo, D/T/pembro, Ipi3/Nivo1, E/B | 4 | PR | 114 | 114 | Y | None (Continued response) | Rash | 3 | Y |
| 10C | F | 48 | 10 | Y | Acral | WT | Ipi3+nivo1, Palbociclib+nivo | 5 | Mixed response | 175 | 267 | N | lenvatinib + pembro | Diarrhea | 2 | Y |
| 10D | M | 35 | 10 | Y | Non-Acral Cutaneous | V600E | Ipi3, Pembro, Ipi3+Nivo1, D/T/P | 3 | PD | 96 | 96 | N | None | PE, SOB | 3 | Y (for brain mets) |
| 10E | F | 67 | 10 | Y | Acral | WT | Neoadj Nivo, Ipi3+nivo1, microbiome+nivo, lenvatinib+pembro | 1 | PD | 195 | 195 | N | None | Diarrhea | 3 | Y |
| 10F | M | 66 | 10 | Y | Nodular | V600E | Adj nivo, D/T, E/B, Ipi3/Nivo1 | 1 | PD | 27 | 27 | N | None | No | 0 | N |
| 3A | M | 47 | 3 | Y | Non-Acral Cutaneous | V600E | Ipi3, IL-2, Dabra, pembro, D/T | 3 | PD | 75 | 124 | N | Carbo | No | 0 | N |
| 3B | M | 54 | 3 | Y | Nodular | V600K | Ipi3, D/T, neoadj TVEC, adj nivo, D/T/P | 2 | PD | 47 | 548 | Y | Carbo/Taxol, lenvatinib+pembro, rela+nivo | Eye muscle inflammation | 3 | Y |
| 3C | M | 69 | 3 | Y | Acral | WT | TVEC+pembro, Ipi3+Cavatak, Ipi3+Nivo1 | 1 | PD | 26 | 26 | N | None | No | 0 | N |
| 3D | M | 45 | 3 | Y | Non-Acral Cutaneous | V600E | Ipi3+nivo1, nivo, D/T, pembro, D/T/P | 1 | PD | 55 | 55 | N | None | No | 0 | N |
| 3E | F | 21 | 3 | Y | Nodular | V600E | Adj D/T, Neoadj pembro, Ipi3+Nivo1, TVEC+Pembro, temodar 75mg/kg daily | 1 | PD | 39 | 39 | N | None | No | 0 | N |
| 3F | M | 52 | 3 | Y | Choroidal | WT | Tram+AKT inh | 1 | PD | 41 | 194 | N | Pembro, Disulfiram/Zinc | No | 0 | N |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Williamson, J.; Fadlullah, M.Z.H.; Kovacsovics-Bankowski, M.; Gibson, B.; Swami, U.; Erickson-Wayman, A.; Jamison, D.; Sageser, D.; Jeter, J.; Bowles, T.L.; et al. Dramatic Responses to High-Dose Ipilimumab Plus Temozolomide After Progression on Standard- or Low-Dose Ipilimumab in Advanced Melanoma. Curr. Oncol. 2025, 32, 144. https://doi.org/10.3390/curroncol32030144
Williamson J, Fadlullah MZH, Kovacsovics-Bankowski M, Gibson B, Swami U, Erickson-Wayman A, Jamison D, Sageser D, Jeter J, Bowles TL, et al. Dramatic Responses to High-Dose Ipilimumab Plus Temozolomide After Progression on Standard- or Low-Dose Ipilimumab in Advanced Melanoma. Current Oncology. 2025; 32(3):144. https://doi.org/10.3390/curroncol32030144
Chicago/Turabian StyleWilliamson, Julie, Muhammad Zaki Hidayatullah Fadlullah, Magdalena Kovacsovics-Bankowski, Berit Gibson, Umang Swami, Alyssa Erickson-Wayman, Debra Jamison, Dan Sageser, Joanne Jeter, Tawnya L. Bowles, and et al. 2025. "Dramatic Responses to High-Dose Ipilimumab Plus Temozolomide After Progression on Standard- or Low-Dose Ipilimumab in Advanced Melanoma" Current Oncology 32, no. 3: 144. https://doi.org/10.3390/curroncol32030144
APA StyleWilliamson, J., Fadlullah, M. Z. H., Kovacsovics-Bankowski, M., Gibson, B., Swami, U., Erickson-Wayman, A., Jamison, D., Sageser, D., Jeter, J., Bowles, T. L., Cannon, D. M., Haaland, B., Schroeder, J. D., Nix, D. A., Atkinson, A., Hyngstrom, J., McPherson, J., Tan, A.-C., & Hu-Lieskovan, S. (2025). Dramatic Responses to High-Dose Ipilimumab Plus Temozolomide After Progression on Standard- or Low-Dose Ipilimumab in Advanced Melanoma. Current Oncology, 32(3), 144. https://doi.org/10.3390/curroncol32030144






