Deep Learning Prediction of Axillary Lymph Node Metastasis in Breast Cancer Patients Using Clinical Implication-Applied Preprocessed CT Images
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. CT Imaging
2.3. Image Analysis Methods
2.3.1. Image Conversion
2.3.2. Protocol for LN Bounding Box Generation
2.3.3. Crop Strategies for Image Analysis
2.3.4. CNN Architectures
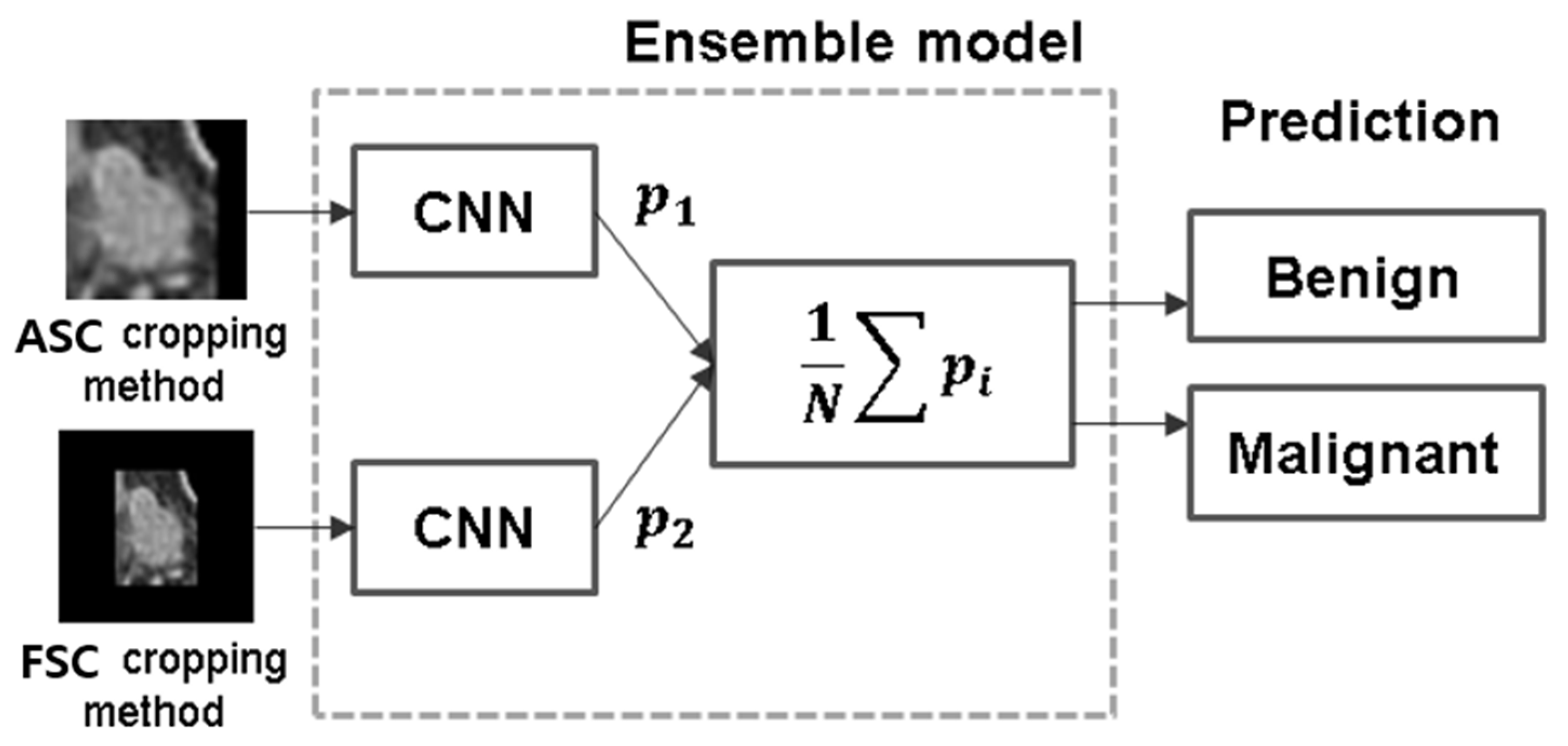
2.3.5. Ensemble Method
2.3.6. Learning the Network
3. Results
3.1. CNN Performance Based on Cropping Methods
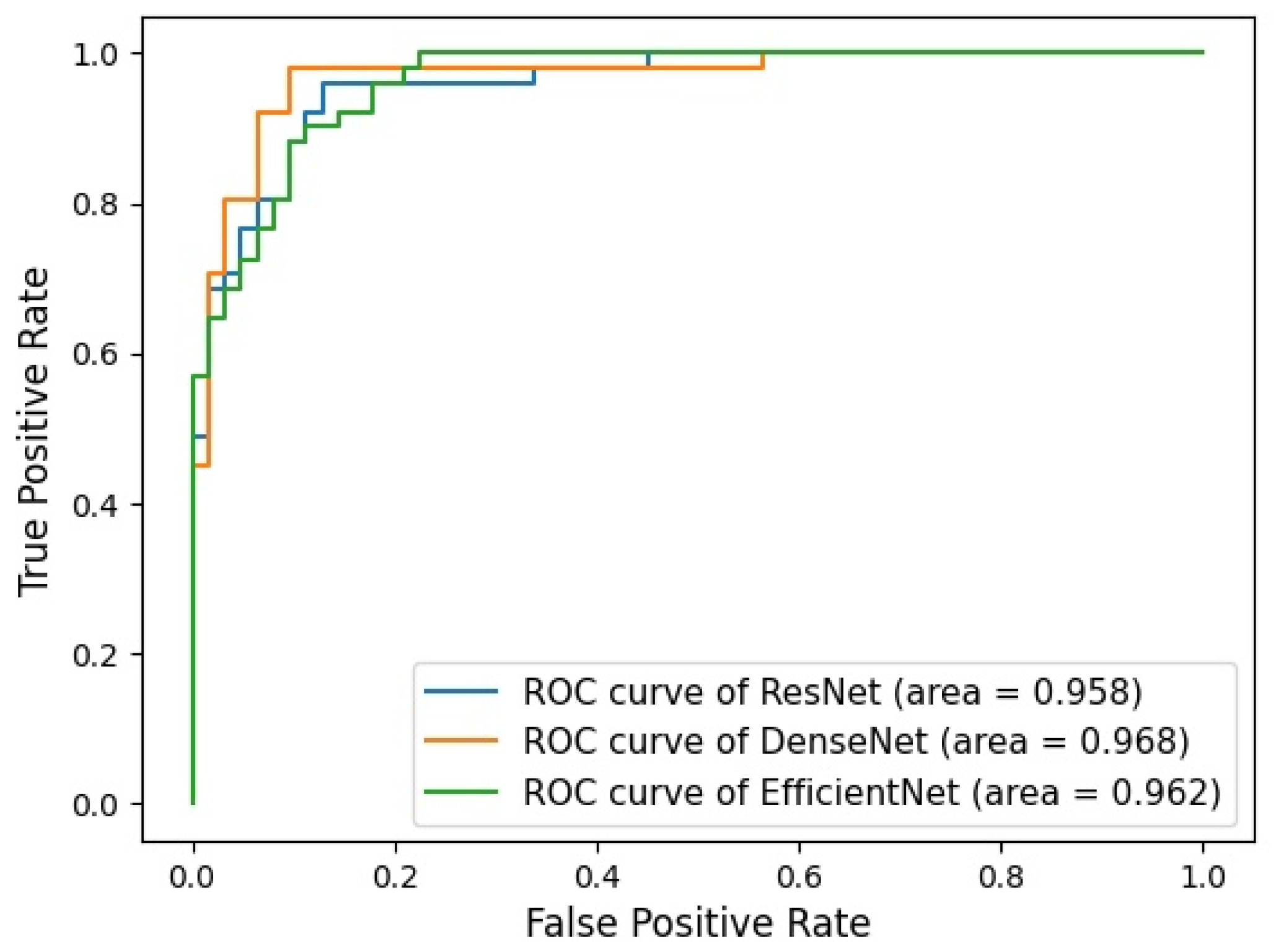
3.2. Performance of Ensemble Model
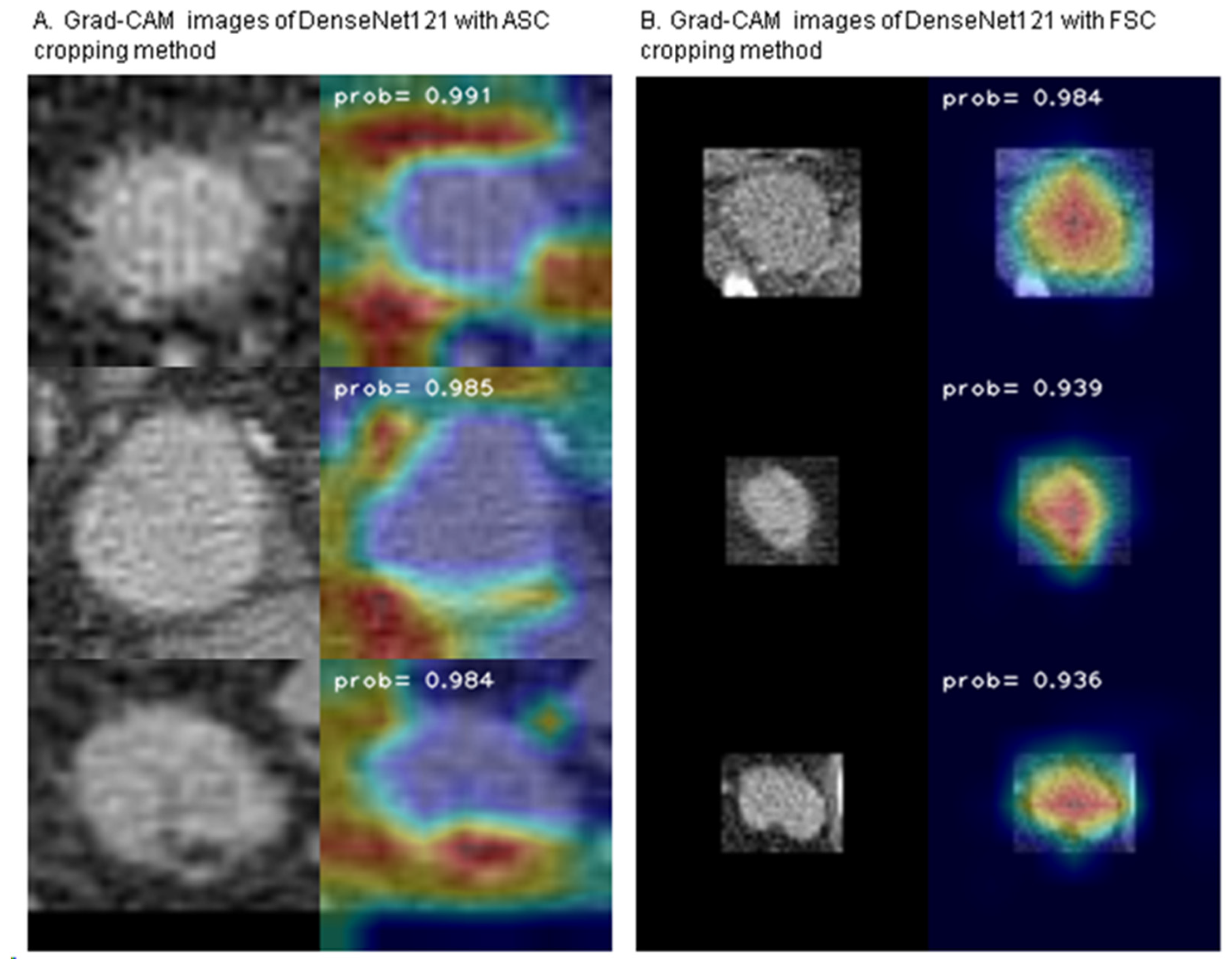
3.3. Gradient-Weighted Class Activation Mapping (Grad-CAM)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Park, K.U.; Caudle, A. Management of the Axilla in the Patient with Breast Cancer. Surg. Clin. N. Am. 2018, 98, 747–760. [Google Scholar] [CrossRef] [PubMed]
- van den Brekel, M.W.; Stel, H.V.; Castelijns, J.A.; Nauta, J.J.; van der Waal, I.; Valk, J.; Meyer, C.J.; Snow, G.B. Cervical Lymph Node Metastasis: Assessment of Radiologic Criteria. Radiology 1990, 177, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Kramer, H.; Groen, H.J.M. Current Concepts in the Mediastinal Lymph Node Staging of Nonsmall Cell Lung Cancer. Ann. Surg. 2003, 238, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Fukuya, T.; Honda, H.; Hayashi, T.; Kaneko, K.; Tateshi, Y.; Ro, T.; Maehara, Y.; Tanaka, M.; Tsuneyoshi, M.; Masuda, K. Lymph-Node Metastases: Efficacy for Detection with Helical CT in Patients with Gastric Cancer. Radiology 1995, 197, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Tiguert, R.; Gheiler, E.L.; Tefilli, M.V.; Oskanian, P.; Banerjee, M.; Grignon, D.J.; Sakr, W.; Pontes, J.E.; Wood, D.P. Lymph Node Size Does Not Correlate with the Presence of Prostate Cancer Metastasis. Urology 1999, 53, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Yuen, S.; Yamada, K.; Goto, M.; Sawai, K.; Nishimura, T. CT-Based Evaluation of Axillary Sentinel Lymph Node Status in Breast Cancer: Value of Added Contrast-Enhanced Study. Acta Radiol. 2004, 45, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Shien, T.; Akashi-Tanaka, S.; Yoshida, M.; Hojo, T.; Iwamoto, E.; Miyakawa, K.; Kinoshita, T. Evaluation of Axillary Status in Patients with Breast Cancer Using Thin-Section CT. Int. J. Clin. Oncol. 2008, 13, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Vrdoljak, J.; Krešo, A.; Kumrić, M.; Martinović, D.; Cvitković, I.; Grahovac, M.; Vickov, J.; Bukić, J.; Božic, J. The Role of AI in Breast Cancer Lymph Node Classification: A Comprehensive Review. Cancers 2023, 15, 2400. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ni, S.; Yang, C.; Sun, W.; Huang, D.; Su, H.; Shu, J.; Qin, N. Axillary Lymph Node Metastasis Prediction by Contrast-Enhanced Computed Tomography Images for Breast Cancer Patients Based on Deep Learning. Comput. Biol. Med. 2021, 136, 104715. [Google Scholar] [CrossRef]
- Yang, X.; Wu, L.; Ye, W.; Zhao, K.; Wang, Y.; Liu, W.; Li, J.; Li, H.; Liu, Z.; Liang, C. Deep Learning Signature Based on Staging CT for Preoperative Prediction of Sentinel Lymph Node Metastasis in Breast Cancer. Acad. Radiol. 2020, 27, 1226–1233. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019. [Google Scholar]
- Liu, Y.; Yao, X. Ensemble Learning via Negative Correlation. Neural Netw. 1999, 12, 1399–1404. [Google Scholar] [CrossRef]
- Moon, W.K.; Lee, Y.-W.; Ke, H.-H.; Lee, S.H.; Huang, C.-S.; Chang, R.-F. Computer-Aided Diagnosis of Breast Ultrasound Images Using Ensemble Learning from Convolutional Neural Networks. Comput. Methods Programs Biomed. 2020, 190, 105361. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A Large-Scale Hierarchical Image Database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Perkins, N.J.; Schisterman, E.F. The Inconsistency of “Optimal” Cutpoints Obtained Using Two Criteria Based on the Receiver Operating Characteristic Curve. Am. J. Epidemiol. 2006, 163, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, H.; Li, J.; Chen, J.; Meng, F.; Li, H.; Han, L.; Zhou, S.; Yu, T. Preoperative Prediction of Axillary Lymph Node Metastasis in Breast Cancer Using CNN Based on Multiparametric MRI. J. Magn. Reson. Imaging 2022, 56, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, M.; Ren, W.; Sun, J.; Wang, K.; Xi, X.; Zhang, G. Predicting of Axillary Lymph Node Metastasis in Invasive Breast Cancer Using Multiparametric MRI Dataset Based on CNN Model. Front. Oncol. 2022, 12, 1069733. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Mutasa, S.; Liu, M.Z.; Nemer, J.; Sun, M.; Siddique, M.; Desperito, E.; Jambawalikar, S.; Ha, R.S. Deep Learning Prediction of Axillary Lymph Node Status Using Ultrasound Images. Comput. Biol. Med. 2022, 143, 105250. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Shi, Y.; Yin, P.; Liu, F.; Fang, Y.; Li, X.; Zhang, Q.; Zhang, Z. A Machine Learning Model Based on Ultrasound Image Features to Assess the Risk of Sentinel Lymph Node Metastasis in Breast Cancer Patients: Applications of Scikit-Learn and SHAP. Front. Oncol. 2022, 12, 944569. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Yao, Z.; Huang, Y.; Yu, Y.; Wang, Y.; Liu, Y.; Mao, R.; Li, F.; Xiao, Y.; Wang, Y.; et al. Deep Learning Radiomics Can Predict Axillary Lymph Node Status in Early-Stage Breast Cancer. Nat. Commun. 2020, 11, 1236. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Ren, C.; Liu, G.; Shui, R.; Zhang, Y.; Li, J.; Shao, Z. Development of High-Resolution Dedicated PET-Based Radiomics Machine Learning Model to Predict Axillary Lymph Node Status in Early-Stage Breast Cancer. Cancers 2022, 14, 950. [Google Scholar] [CrossRef] [PubMed]



| Whole Dataset | Training Set | Tuning Dataset | Test Dataset | |||||
|---|---|---|---|---|---|---|---|---|
| Image N | Patient N | Image N | Patient N | Image N | Patient N | Image N | Patient N | |
| Overall | 1127 | 523 | 890 | 417 | 113 | 53 | 124 | 53 |
| Malignant | 538 | 303 | 422 | 241 | 53 | 31 | 63 | 31 |
| Benign | 589 | 220 | 468 | 176 | 60 | 22 | 61 | 22 |
| Accuracy | AUROC | Sensitivity | Specificity | PPV | NPV | F1 Score | p-Value * | |
|---|---|---|---|---|---|---|---|---|
| ResNet 152 [12] | 0.83 ± 0.039 | 0.929 ± 0.021 | 0.874 ± 0.068 | 0.878 ± 0.024 | 0.868 ± 0.02 | 0.885 ± 0.062 | 0.869 ± 0.028 | 0.292 |
| DenseNet 121 [13] | 0.87 ± 0.043 | 0.939 ± 0.026 | 0.900 ± 0.043 | 0.883 ± 0.037 | 0.878 ± 0.033 | 0.904 ± 0.045 | 0.889 ± 0.038 | |
| EfficientNet B7 [14] | 0.862 ± 0.019 | 0.927 ± 0.020 | 0.874 ± 0.075 | 0.884 ± 0.052 | 0.876 ± 0.052 | 0.888 ± 0.064 | 0.87 ± 0.013 | 0.274 |
| Accuracy | AUROC | Sensitivity | Specificity | PPV | NPV | F1 Score | p-Value * | |
|---|---|---|---|---|---|---|---|---|
| ResNet 152 [12] | 0.851 ± 0.024 | 0.929 ± 0.023 | 0.858 ± 0.024 | 0.9 ± 0.041 | 0.891 ± 0.034 | 0.872 ± 0.033 | 0.874 ± 0.025 | 0.171 |
| DenseNet 121 [13] | 0.875 ± 0.038 | 0.934 ± 0.03 | 0.921 ± 0.059 | 0.844 ± 0.042 | 0.847 ± 0.03 | 0.921 ± 0.063 | 0.881 ± 0.034 | |
| EfficientNet B7 [14] | 0.814 ± 0.038 | 0.933 ± 0.024 | 0.893 ± 0.034 | 0.857 ± 0.039 | 0.853 ± 0.025 | 0.896 ± 0.037 | 0.872 ± 0.021 | 0.118 |
| Accuracy | AUROC | Sensitivity | Specificity | PPV | NPV | F1 Score | |
|---|---|---|---|---|---|---|---|
| ResNet 152 | 0.912 (0.859–0.964) | 0.958 (0.952–0.960) | 0.961 (0.868–0.988) | 0.871 (0.765–0.933) | 0.860 (0.746–0.927) | 0.964 (0.879–0.989) | 0.907 (0.9–0.914) |
| DenseNet 121 | 0.938 (0.894–0.982) | 0.968 (0.965–0.971) | 0.980 (0.897–0.995) | 0.903 (0.804–0.954) | 0.893 (0.785–0.949) | 0.982 (0.908–0.996) | 0.935 (0.930–0.940) |
| EfficientNet B7 | 0.894 (0.837–0.951) | 0.962 (0.960–0.966) | 0.902 (0.790–0.956) | 0.887 (0.784–0.944) | 0.868 (0.751–0.934) | 0.917 (0.819–0.963) | 0.885 (0.879–0.892) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, T.Y.; Kwon, L.M.; Hyeon, J.; Cho, B.-J.; Kim, B.J. Deep Learning Prediction of Axillary Lymph Node Metastasis in Breast Cancer Patients Using Clinical Implication-Applied Preprocessed CT Images. Curr. Oncol. 2024, 31, 2278-2288. https://doi.org/10.3390/curroncol31040169
Park TY, Kwon LM, Hyeon J, Cho B-J, Kim BJ. Deep Learning Prediction of Axillary Lymph Node Metastasis in Breast Cancer Patients Using Clinical Implication-Applied Preprocessed CT Images. Current Oncology. 2024; 31(4):2278-2288. https://doi.org/10.3390/curroncol31040169
Chicago/Turabian StylePark, Tae Yong, Lyo Min Kwon, Jini Hyeon, Bum-Joo Cho, and Bum Jun Kim. 2024. "Deep Learning Prediction of Axillary Lymph Node Metastasis in Breast Cancer Patients Using Clinical Implication-Applied Preprocessed CT Images" Current Oncology 31, no. 4: 2278-2288. https://doi.org/10.3390/curroncol31040169
APA StylePark, T. Y., Kwon, L. M., Hyeon, J., Cho, B.-J., & Kim, B. J. (2024). Deep Learning Prediction of Axillary Lymph Node Metastasis in Breast Cancer Patients Using Clinical Implication-Applied Preprocessed CT Images. Current Oncology, 31(4), 2278-2288. https://doi.org/10.3390/curroncol31040169





