Regulatory and Interacting Partners of PDLIM7 in Thyroid Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tumor Sample Selection
2.2. miRNA and Total RNA Isolation from FFPE
2.3. RNA and Protein Isolation from Fresh Samples
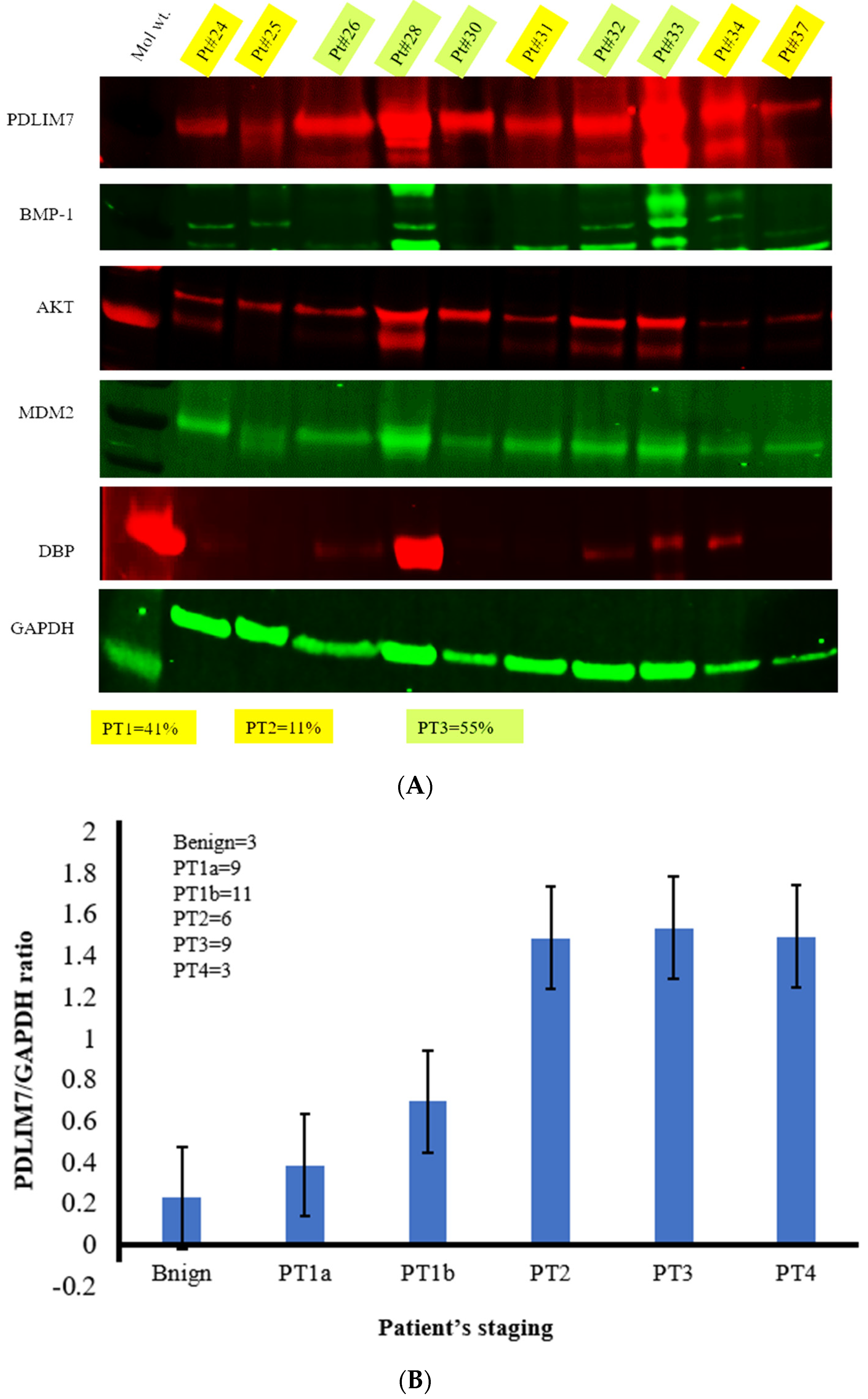
2.4. Western Blot Analysis and Quantitative Analysis by Densitometric Assay
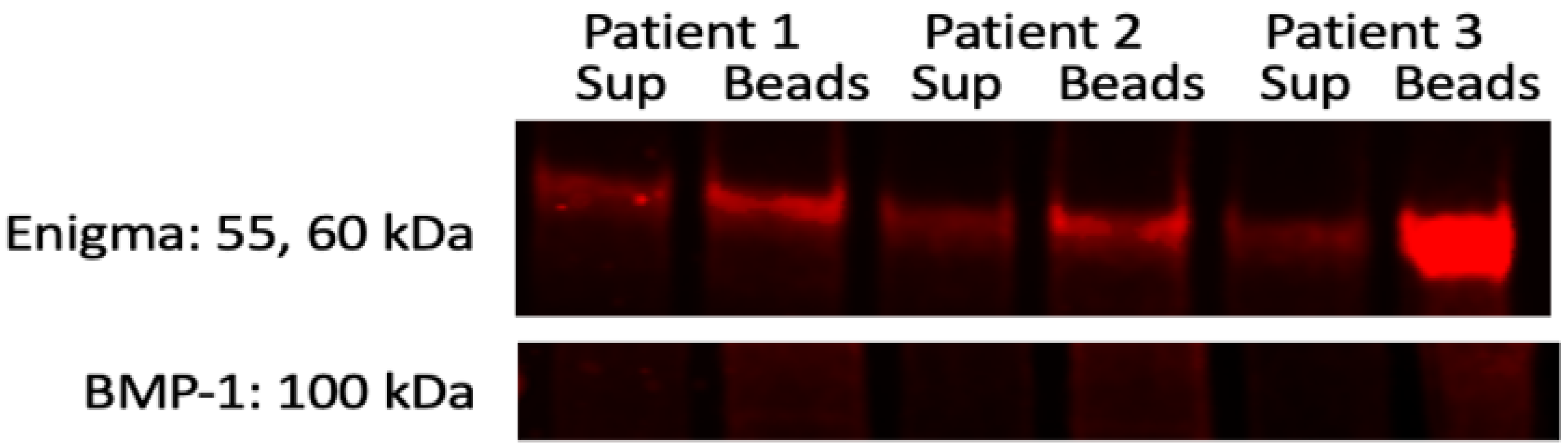
2.5. Immunoprecipitation and Reverse Immunoprecipitation
2.6. RT-qPCR (Real-Time Polymerase Chain Reaction)
2.7. Statistical Analysis
3. Results
3.1. Differential Expressions of Enigma and Its Signaling Pathways in Thyroid Cancer Tissues
3.2. Interacting Partners of Enigma in Thyroid Cancer
3.3. Quantitative Analysis of PDLIM7 and Let-7g Gene in the Same Thyroid Cancer Patient Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Firek, A.A.; Perez, M.C.; Gonda, A.; Lei, L.; Munir, I.; Simental, A.A.; Carr, F.E.; Becerra, B.J.; De Leon, M.; Khan, S. Pathologic significance of a novel oncoprotein in thyroid cancer progression. Head Neck 2017, 39, 2459–2469. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.-Y.; Durick, K.; Songyang, Z.; Cantley, L.C.; Taylor, S.S.; Gill, G.N. Specificity of LIM Domain Interactions with Receptor Tyrosine Kinases. J. Biol. Chem. 1996, 271, 15934–15941. [Google Scholar] [CrossRef] [PubMed]
- Jeleń, F.; Oleksy, A.; Smietana, K.; Otlewski, J. PDZ domains—Common players in the cell signaling. Acta Biochim. Pol. 2003, 50, 985–1017. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhou, G.; Nakamura, M.; Ozaki, T.; Mori, I.; Taniguchi, E.; Miyauchi, A.; Ito, Y.; Kakudo, K. Survival impact of psammoma body, stromal calcification, and bone formation in papillary thyroid carcinoma. Mod. Pathol. 2009, 22, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Durick, K.; Gill, G.N.; Taylor, S.S. Shc and Enigma Are Both Required for Mitogenic Signaling by Ret/ptc2. Mol. Cells Biol. 1998, 18, 2298–2308. [Google Scholar] [CrossRef]
- Kuroda, S.; Tokunaga, C.; Kiyohara, Y.; Higuchi, O.; Konishi, H.; Mizuno, K.; Gill, G.N.; Kikkawa, U. Protein-Protein Interaction of Zinc Finger LIM Domains with Protein Kinase C. J. Biol. Chem. 1996, 271, 31029–31032. [Google Scholar] [CrossRef]
- Guo, Z.S.; Qu, Z. PDLIM2: Signaling pathways and functions in cancer suppression and host immunity. Biochim. Biophys. Acta (BBA) Rev. Cancer 2021, 1876, 188630. [Google Scholar] [CrossRef]
- Iwakuma; Lozano, G. MDM2, an introduction. Mol. Cancer Res. 2003, 1, 993–1000. [Google Scholar]
- Jung, C.-R.; Lim, J.H.; Choi, Y.; Kim, D.-G.; Kang, K.J.; Noh, S.-M.; Im, D.-S. Enigma negatively regulates p53 through MDM2 and promotes tumor cell survival in mice. J. Clin. Investig. 2010, 120, 4493–4506. [Google Scholar] [CrossRef]
- Porta, C.; Paglino, C.; Mosca, A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Front. Oncol. 2014, 4, 64. [Google Scholar] [CrossRef]
- Kim, Y.J.; Hwang, H.-J.; Kang, J.G.; Kim, C.S.; Ihm, S.-H.; Choi, M.G.; Lee, S.J. Enigma Plays Roles in Survival of Thyroid Carcinoma Cells through PI3K/AKT Signaling and Survivin. Anticancer. Res. 2018, 38, 3515–3525. [Google Scholar] [CrossRef] [PubMed]
- Cocolos, A.-M.; Muresan, A.; Caragheorgheopol, A.; Ghemigian, M.; Ioachim, D.; Poiana, C. Vitamin D Status and VDR Polymorphisms as Prognostic Factors in Differentiated Thyroid Carcinoma. Vivo 2022, 36, 2434–2441. [Google Scholar] [CrossRef] [PubMed]
- Mull, B.; Davis, R.; Munir, I.; Perez, M.C.; Simental, A.A.; Khan, S. Differential expression of Vitamin D binding protein in thyroid cancer health disparities. Oncotarget 2021, 12, 596–607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.; Anderson, T. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Duchaine, T.F.; Fabian, M.R. Mechanistic Insights into MicroRNA-Mediated Gene Silencing. Cold Spring Harb. Perspect. Biol. 2019, 11, a032771. [Google Scholar] [CrossRef]
- Jung, H.J.; Suh, Y. Regulation of IGF -1 signaling by microRNAs. Front. Genet. 2015, 5, 472. [Google Scholar] [CrossRef]
- Müller, S.; Glaß, M.; Singh, A.K.; Haase, J.; Bley, N.; Fuchs, T.; Hüttelmaier, S. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic. Acids Res. 2019, 47, 375–390. [Google Scholar] [CrossRef]
- Li, M.; Glaß, M.; Singh, A.K.; Haase, J.; Bley, N.; Fuchs, T.; Hüttelmaier, S. Circulating miR-25-3p and miR-451a May Be Potential Biomarkers for the Diagnosis of Papillary Thyroid Carcinoma. PLoS ONE 2015, 10, e0132403. [Google Scholar]
- Rood, K.; Begum, K.; Wang, H.; Wangworawat, Y.C.; Davis, R.; Yamauchi, C.R.; Perez, M.C.; Simental, A.A.; Laxa, R.T.; Wang, C.; et al. Differential Expression of Non-Coding RNA Signatures in Thyroid Cancer between Two Ethnic Groups. Curr. Oncol. 2021, 28, 309. [Google Scholar] [CrossRef]
- Suresh, R.; Sethi, S.; Ali, S.; Giorgadze, T.; Sarkar, F.H. Differential Expression of MicroRNAs in Papillary Thyroid Carcinoma and Their Role in Racial Disparity. J. Cancer Sci. Ther. 2015, 7, 145–154. [Google Scholar] [CrossRef]
- Ma, Y.; Shen, N.; Wicha, M.S.; Luo, M. The Roles of the Let-7 Family of MicroRNAs in the Regulation of Cancer Stemness. Cells 2021, 10, 2415. [Google Scholar] [CrossRef] [PubMed]
- Perdas, E.; Stawski, R.; Nowak, D.; Zubrzycka, M. The Role of miRNA in Papillary Thyroid Cancer in the Context of miRNA Let-7 Family. Int. J. Mol. Sci. 2016, 17, 909. [Google Scholar] [CrossRef] [PubMed]
- Loh, K.-C.; Greenspan, F.S.; Gee, L.; Miller, T.R.; Yeo, P.P.B. Pathological Tumor-Node-Metastasis (pTNM) Staging for Papillary and Follicular Thyroid Carcinomas: A Retrospective Analysis of 700 Patients. J. Clin. Endocrinol. Metab. 1997, 82, 3553–3562. [Google Scholar] [CrossRef] [PubMed]


| Pearson Correlation Coefficients, N = 87 Prob > |r| under HO: Rho = 0 | ||
|---|---|---|
| DDCT_PDLIM7 | Staging | |
| DDCT_PDLIMZ | 1.00000 | 0.17866 |
| DDCT_PDLIM7 | 0.0978 | |
| Staging | 0.17866 | 1.00000 |
| Staging | 0.0978 | |
| Spearman Correlation Coefficients, N = 87 Prob > |r| under HO: Rho = 0 | ||
| DDCT_PDLIM7 | Staging | |
| DDCT_PDLIM7 | 1.00000 | 0.14464 |
| DDCT_PDLIM7 | 0.1813 | |
| Staging | 0.14464 | 1.00000 |
| Staging | 0.1813 | |
| N | t-Value | Mean & SD | 95% CI |
|---|---|---|---|
| 87 | 1.96 | 1.10 ± 5.26 | (4.58, 6.18) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rood, K.; Yamauchi, C.R.; Sharma, U.; Laxa, R.T.; Robins, C.; Lanza, G.; Sánchez-Ruiz, K.; Khan, A.; Kim, H.S.; Shields, A.; et al. Regulatory and Interacting Partners of PDLIM7 in Thyroid Cancer. Curr. Oncol. 2023, 30, 10450-10462. https://doi.org/10.3390/curroncol30120761
Rood K, Yamauchi CR, Sharma U, Laxa RT, Robins C, Lanza G, Sánchez-Ruiz K, Khan A, Kim HS, Shields A, et al. Regulatory and Interacting Partners of PDLIM7 in Thyroid Cancer. Current Oncology. 2023; 30(12):10450-10462. https://doi.org/10.3390/curroncol30120761
Chicago/Turabian StyleRood, Kristiana, Celina Romi Yamauchi, Umang Sharma, Ria T. Laxa, Collin Robins, Gerardo Lanza, Kidianys Sánchez-Ruiz, Aminah Khan, Hae Soo Kim, Andrea Shields, and et al. 2023. "Regulatory and Interacting Partners of PDLIM7 in Thyroid Cancer" Current Oncology 30, no. 12: 10450-10462. https://doi.org/10.3390/curroncol30120761
APA StyleRood, K., Yamauchi, C. R., Sharma, U., Laxa, R. T., Robins, C., Lanza, G., Sánchez-Ruiz, K., Khan, A., Kim, H. S., Shields, A., Kennedy, K., Mirshahidi, S., Perez, M. C., Firek, A., Munir, I., Simental, A. A., & Khan, S. (2023). Regulatory and Interacting Partners of PDLIM7 in Thyroid Cancer. Current Oncology, 30(12), 10450-10462. https://doi.org/10.3390/curroncol30120761






