Application of Machine Learning in Predicting Hepatic Metastasis or Primary Site in Gastroenteropancreatic Neuroendocrine Tumors
Abstract
1. Introduction
2. Material and Methods
2.1. RNA-SEQ Datasets and Processing
2.2. Feature Selection, Model Building, and Performance Evaluation
2.3. Machine Learning Models and Performance Evaluation
2.4. Differential Expression Analyses
2.5. Statistical Software and Figures
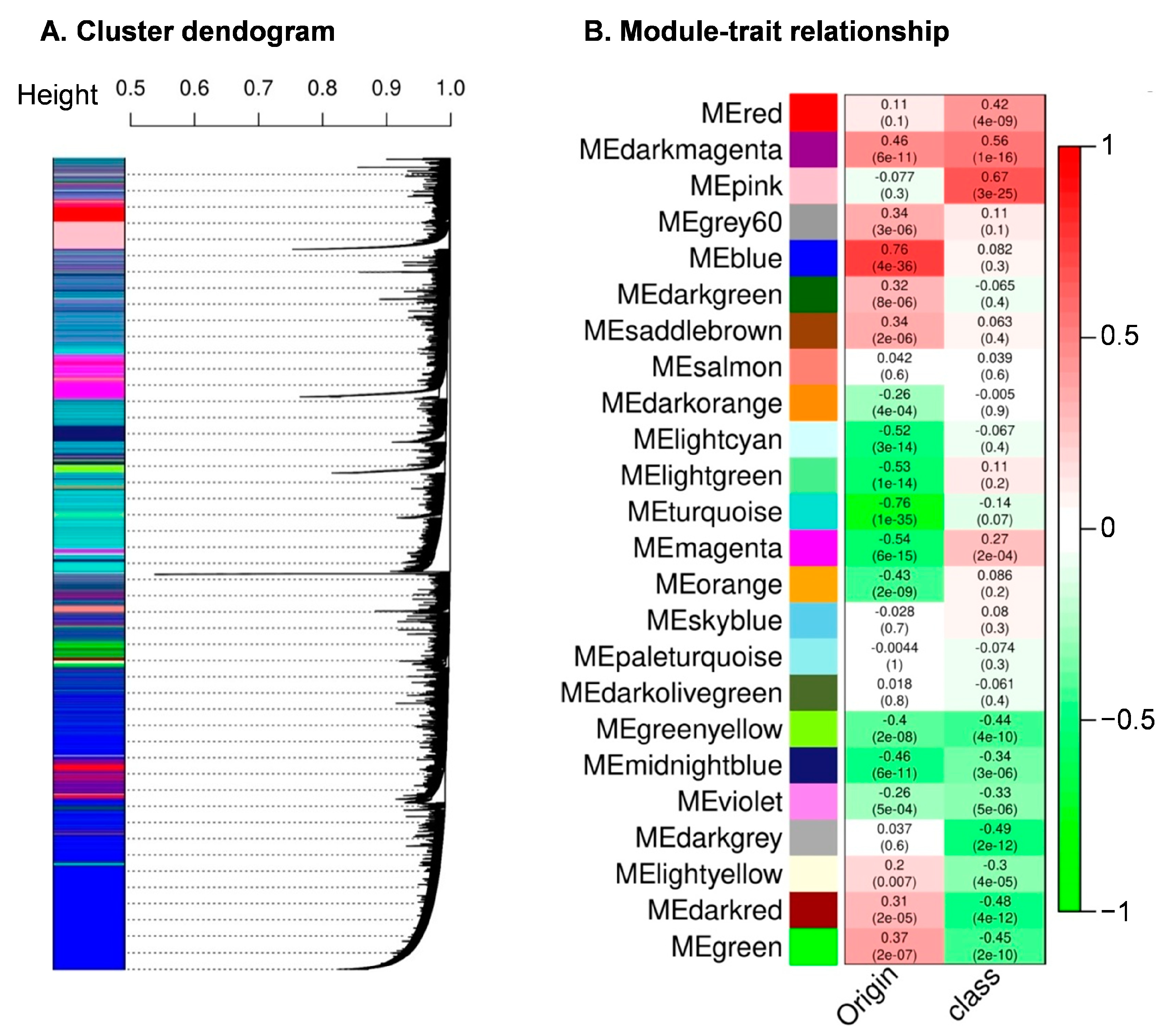
2.6. Weighted Gene Expression Network Analysis (WGCNA) Construction
3. Results
3.1. Alignment of RNA-SEQ Profiles with the Human Genome, Gene Quantification and Count Normalization
3.2. Hepatic Metastasis Model
3.2.1. Identification of Gene Features Relevant to Hepatic Metastasis
3.2.2. Development of Machine Learning Models and Importance of the Identified Features
3.2.3. Concise Gene Signatures Improve Classification Accuracy of the Hepatic Metastasis Model
3.3. Primary Site Model
3.3.1. Identification of Gene Features Relevant to the Primary Site
3.3.2. Development of Machine Learning Models and Importance of the Identified Features
3.3.3. Concise Gene Signatures Improve the Classification Accuracy of the Primary Site Model
3.4. A Multi-Label Model to Predict the Primary Site and Hepatic Metastasis of NETs
3.5. Weighted Gene Correlation Network Analysis
4. Discussion
5. Conclusions
6. Limitation of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rindi, G.; Wiedenmann, B. Neuroendocrine neoplasms of the gut and pancreas: New insights. Nat. Rev. Endocrinol. 2012, 8, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Man, D.; Wu, J.; Shen, Z.; Zhu, X. Prognosis of patients with neuroendocrine tumor: A SEER database analysis. Cancer Manag. Res. 2018, 10, 5629–5638. [Google Scholar] [CrossRef] [PubMed]
- Dasari, A.; Shen, C.; Halperin, D.; Zhao, B.; Zhou, S.; Xu, Y.; Shih, T.; Yao, J.C. Trends in the Incidence, Prevalence, and Survival Outcomes in Patients with Neuroendocrine Tumors in the United States. JAMA Oncol. 2017, 3, 1335–1342. [Google Scholar] [CrossRef] [PubMed]
- Díez, M.; Teulé, A.; Salazar, R. Gastroenteropancreatic neuroendocrine tumors: Diagnosis and treatment. Ann. Gastroenterol. 2013, 26, 29–36. [Google Scholar] [PubMed]
- Kyriakopoulos, G.; Mavroeidi, V.; Chatzellis, E.; Kaltsas, G.A.; Alexandraki, K.I. Histopathological, immunohistochemical, genetic and molecular markers of neuroendocrine neoplasms. Ann. Transl. Med. 2018, 6, 252. [Google Scholar] [CrossRef] [PubMed]
- Berner, A.M.; Pipinikas, C.; Ryan, A.; Dibra, H.; Moghul, I.; Webster, A.; Luong, T.V.; Thirlwell, C. Diagnostic Approaches to Neuroendocrine Neoplasms of Unknown Primary Site. Neuroendocrinology 2020, 110, 563–573. [Google Scholar] [CrossRef]
- Alvarez, M.J.; Subramaniam, P.S.; Tang, L.H.; Grunn, A.; Aburi, M.; Rieckhof, G.; Komissarova, E.V.; Hagan, E.A.; Bodei, L.; Clemons, P.A.; et al. A precision oncology approach to the pharmacological targeting of mechanistic dependencies in neuroendocrine tumors. Nat. Genet. 2018, 50, 979–989. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.T.; Huang, X.; Huang, S.; Xie, S.J.; Xiao, Z.D.; Zhang, H. RNA sequencing: New technologies and applications in cancer research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef]
- Panarelli, N.; Tyryshkin, K.; Wong, J.J.M.; Majewski, A.; Yang, X.; Scognamiglio, T.; Kim, M.K.; Bogardus, K.; Tuschl, T.; Chen, Y.T.; et al. Evaluating gastroenteropancreatic neuroendocrine tumors through microRNA sequencing. Endocr. Relat. Cancer. 2019, 26, 47–57. [Google Scholar] [CrossRef]
- Siegel, M.B.; He, X.; Hoadley, K.A.; Hoyle, A.; Pearce, J.B.; Garrett, A.L.; Kumar, S.; Moylan, V.J.; Brady, C.M.; Van Swearingen, A.E.; et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J. Clin. Investig. 2018, 128, 1371–1383. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef]
- Mahendran, N.; Durai Raj Vincent, P.M.; Srinivasan, K.; Chang, C.Y. Machine Learning Based Computational Gene Selection Models: A Survey, Performance Evaluation, Open Issues, and Future Research Directions. Front. Genet. 2020, 11, 603808. [Google Scholar] [CrossRef] [PubMed]
- Malebary, S.J.; Khan, Y.D. Evaluating machine learning methodologies for identification of cancer driver genes. Sci. Rep. 2021, 11, 12281. [Google Scholar] [CrossRef] [PubMed]
- Wei, I.H.; Shi, Y.; Jiang, H.; Kumar-Sinha, C.; Chinnaiyan, A.M. RNA-Seq accurately identifies cancer biomarker signatures to distinguish tissue of origin. Neoplasia 2014, 16, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef]
- Tseng, Y.J.; Wang, H.Y.; Lin, T.W.; Lu, J.J.; Hsieh, C.H.; Liao, C.T. Development of a Machine Learning Model for Survival Risk Stratification of Patients with Advanced Oral Cancer. JAMA Netw. Open 2020, 3, e2011768. [Google Scholar] [CrossRef]
- Chan, C.S.; Laddha, S.V.; Lewis, P.W.; Koletsky, M.S.; Robzyk, K.; Da Silva, E.; Torres, P.J.; Torres, P.J.; Untch, B.R.; Li, J.; et al. ATRX, DAXX or MEN1 mutant pancreatic neuroendocrine tumors are a distinct alpha-cell signature subgroup. Nat. Commun. 2018, 9, 4158. [Google Scholar] [CrossRef]
- Andrews, S.a.K. Felix and {Segonds-Pichon}, Anne and Biggins, Laura and Krueger, Christel Wingett, Steven (0.11.9). “FastQC” Baraham Bioinfromatics). 2015. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 April 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Chiesa, M.; Colombo, G.I.; Piacentini, L. DaMiRseq-an R/Bioconductor package for data mining of RNA-Seq data: Normalization, feature selection and classification. Bioinformatics 2018, 34, 1416–1418. [Google Scholar] [CrossRef]
- Zwiener, I.; Frisch, B.; Binder, H. Transforming RNA-Seq data to improve the performance of prognostic gene signatures. PLoS ONE 2014, 9, e85150. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- De Jay, N.; Papillon-Cavanagh, S.; Olsen, C.; El-Hachem, N.; Bontempi, G.; Haibe-Kains, B. mRMRe: An R package for parallelized mRMR ensemble feature selection. Bioinformatics 2013, 29, 2365–2368. [Google Scholar] [CrossRef]
- Yin, W.; Tang, G.; Zhou, Q.; Cao, Y.; Li, H.; Fu, X.; Wu, Z.; Jiang, X. Expression Profile Analysis Identifies a Novel Five-Gene Signature to Improve Prognosis Prediction of Glioblastoma. Front. Genet. 2019, 10, 419. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.W.; Dhahbi, J. Lung adenocarcinoma and lung squamous cell carcinoma cancer classification, biomarker identification, and gene expression analysis using overlapping feature selection methods. Sci. Rep. 2021, 11, 13323. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wickham, H. Getting Started with ggplot2. In ggplot2: Elegant Graphics for Data Analysis; Springer International Publishing: Cham, Switzerland, 2016; pp. 11–31. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Ren, Z.H.; Shang, G.P.; Wu, K.; Hu, C.Y.; Ji, T. WGCNA Co-Expression Network Analysis Reveals ILF3-AS1 Functions as a CeRNA to Regulate PTBP1 Expression by Sponging miR-29a in Gastric Cancer. Front. Genet. 2020, 11, 39. [Google Scholar] [CrossRef]
- Shi, G.; Shen, Z.; Liu, Y.; Yin, W. Identifying Biomarkers to Predict the Progression and Prognosis of Breast Cancer by Weighted Gene Co-expression Network Analysis. Front. Genet. 2020, 11, 597888. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, L.; Wen, Z.; Yao, J. Integrated analysis identifies oxidative stress genes associated with progression and prognosis in gastric cancer. Sci. Rep. 2021, 11, 3292. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct Target Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- Wang, Q.; Li, F.; Jiang, Q.; Sun, Y.; Liao, Q.; An, H.; Li, Y.; Li, Z.; Fan, L.; Guo, F.; et al. Gene Expression Profiling for Differential Diagnosis of Liver Metastases: A Multicenter, Retrospective Cohort Study. Front. Oncol. 2021, 11, 725988. [Google Scholar] [CrossRef] [PubMed]
- Brodt, P. Role of the Microenvironment in Liver Metastasis: From Pre- to Prometastatic Niches. Clin. Cancer Res. 2016, 22, 5971–5982. [Google Scholar] [CrossRef] [PubMed]
- Maitra, A. Molecular envoys pave the way for pancreatic cancer to invade the liver. Nature 2019, 567, 181–182. [Google Scholar] [CrossRef]
- van Loon, K.; Huijbers, E.J.M.; Griffioen, A.W. Secreted frizzled-related protein 2: A key player in noncanonical Wnt signaling and tumor angiogenesis. Cancer Metastasis Rev. 2021, 40, 191–203. [Google Scholar] [CrossRef]
- Veeck, J.; Noetzel, E.; Bektas, N.; Jost, E.; Hartmann, A.; Knuchel, R.; Dahl, E. Promoter hypermethylation of the SFRP2 gene is a high-frequent alteration and tumor-specific epigenetic marker in human breast cancer. Mol. Cancer 2008, 7, 83. [Google Scholar] [CrossRef]
- O’Hurley, G.; Perry, A.S.; O’Grady, A.; Loftus, B.; Smyth, P.; O’Leary, J.J.; Sheils, O.; Fitzpatrick, J.M.; Hewitt, S.M.; Lawler, M.; et al. The role of secreted frizzled-related protein 2 expression in prostate cancer. Histopathology 2011, 59, 1240–1248. [Google Scholar] [CrossRef]
- Fotouhi, O.; Adel Fahmideh, M.; Kjellman, M.; Sulaiman, L.; Hoog, A.; Zedenius, J.; Hashemi, J.; Larsson, C. Global hypomethylation and promoter methylation in small intestinal neuroendocrine tumors: An in vivo and in vitro study. Epigenetics 2014, 9, 987–997. [Google Scholar] [CrossRef]
- Muller, W.A. Getting leukocytes to the site of inflammation. Vet. Pathol. 2013, 50, 7–22. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Zeng, Q.; Zhou, G.; Mu, X. Identification of novel transcription factor-microRNA-mRNA co-regulatory networks in pulmonary large-cell neuroendocrine carcinoma. Ann. Transl. Med. 2021, 9, 133. [Google Scholar] [CrossRef]
- Yang, K.C.; Kalloger, S.E.; Aird, J.J.; Lee, M.K.; Rushton, C.; Mungall, K.L.; Mungall, A.J.; Gao, D.; Chow, C.; Xu, J.; et al. Proteotranscriptomic classification and characterization of pancreatic neuroendocrine neoplasms. Cell Rep. 2021, 37, 109817. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.-D.; Wang, H.-B.; Liu, X.-P.; Jin, L.-Y.; Shen, R.-W.; Wang, X.-G.; Kong, B.; Qu, H.-L.; Li, F.-N.; Yang, Q.-F. Silencing of Prrx2 Inhibits the Invasion and Metastasis of Breast Cancer both In Vitro and In Vivo by Reversing Epithelial-Mesenchymal Transition. Cell Physiol. Biochem. 2017, 42, 1847–1856. [Google Scholar] [CrossRef] [PubMed]
- Juang, Y.-L.; Jeng, Y.-M.; Chen, C.-L.; Lien, H.C. PRRX2 as a novel TGF—Induced factor enhances invasion and migration in mammary epithelial cell and correlates with poor prognosis in breast cancer. Mol. Carcinog. 2016, 55, 2247–2259. [Google Scholar] [CrossRef]
- Chai, W.X.; Sun, L.G.; Dai, F.H.; Shao, H.S.; Zheng, N.G.; Cai, H.Y. Inhibition of PRRX2 suppressed colon cancer liver metastasis via inactivation of Wnt/β-catenin signaling pathway. Pathol. Res. Pract. 2019, 215, 152593. [Google Scholar] [CrossRef]
- Larsen, S.; Yokochi, T.; Isogai, E.; Nakamura, Y.; Ozaki, T.; Nakagawara, A. LMO3 interacts with p53 and inhibits its transcriptional activity. Biochem. Biophys. Res. Commun. 2010, 392, 252–257. [Google Scholar] [CrossRef]
- Isogai, E.; Ohira, M.; Ozaki, T.; Oba, S.; Nakamura, Y.; Nakagawara, A. Oncogenic LMO3 collaborates with HEN2 to enhance neuroblastoma cell growth through transactivation of Mash1. PLoS ONE 2011, 6, e19297. [Google Scholar] [CrossRef]
- Colao, A.; de Nigris, F.; Modica, R.; Napoli, C. Clinical Epigenetics of Neuroendocrine Tumors: The Road Ahead. Front. Endocrinol. 2020, 11, 604341. [Google Scholar] [CrossRef]
- Yang, Q.; Huang, T.; Ye, G.; Wang, B.; Zhang, X. Methylation of SFRP2 gene as a promising noninvasive biomarker using feces in colorectal cancer diagnosis: A systematic meta-analysis. Sci. Rep. 2016, 6, 33339. [Google Scholar] [CrossRef]
- Watts, G.S.; Futscher, B.W.; Holtan, N.; DeGeest, K.; E Domann, F.; Rose, S.L. DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC Med. Genomics 2008, 1, 47. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Lei, Q.; Yu, Z.; Xu, G.; Tang, H.; Wang, W.; Wang, Z.; Li, G.; Wu, M. MiR-101 reverses the hypomethylation of the LMO3 promoter in glioma cells. Oncotarget 2015, 6, 7930–7943. [Google Scholar] [CrossRef] [PubMed]
- Marcinkiewicz, K.M.; Gudas, L.J. Altered epigenetic regulation of homeobox genes in human oral squamous cell carcinoma cells. Exp. Cell Res. 2014, 320, 128–143. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, S.; Zhang, Y.; Zhang, S.; Qiu, L.; Zhang, B.; Han, J. Identification of Hub Genes Related to Liver Metastasis of Colorectal Cancer by Integrative Analysis. Front. Oncol. 2021, 11, 714866. [Google Scholar] [CrossRef]
- Tai, C.-S.; Lin, Y.-R.; Teng, T.-H.; Lin, P.-Y.; Tu, S.-J.; Chou, C.-H.; Huang, Y.-R.; Huang, W.-C.; Weng, S.-L.; Huang, H.-D.; et al. Haptoglobin expression correlates with tumor differentiation and five-year overall survival rate in hepatocellular carcinoma. PLoS ONE 2017, 12, e0171269. [Google Scholar] [CrossRef]
- Papiernik, D.; Urbaniak, A.; Kłopotowska, D.; Nasulewicz-Goldeman, A.; Ekiert, M.; Nowak, M.; Jarosz, J.; Cuprych, M.; Strzykalska, A.; Ugorski, M.; et al. Retinol-Binding Protein 4 Accelerates Metastatic Spread and Increases Impairment of Blood Flow in Mouse Mammary Gland Tumors. Cancers 2020, 12, 623. [Google Scholar] [CrossRef]
- Yuan, Y.-M.; Ma, N.; Zhang, E.-B.; Chen, T.-W.; Jiang, H.; Yin, F.-F.; Wang, J.-J.; Zhang, F.-K.; Ni, Q.-Z.; Wang, X.; et al. BMP10 suppresses hepatocellular carcinoma progression via PTPRS–STAT3 axis. Oncogene 2019, 38, 7281–7293. [Google Scholar] [CrossRef]
- Chen, Y.; Xiao, D.; Zhang, L.; Cai, C.-L.; Li, B.-Y.; Liu, Y. The Role of Tbx20 in Cardiovascular Development and Function. Front. Cell Dev. Biol. 2021, 9, 638542. [Google Scholar] [CrossRef]
- Lichtenauer, M.; Jung, C. TBX20 and the PROK2-PROKR1 pathway-new kid on the block in angiogenesis research. Ann. Transl. Med. 2018, 6, S8. [Google Scholar] [CrossRef]
- Scott, A.T.; Weitz, M.; Breheny, P.J. Gene Expression Signatures Identify Novel Therapeutics for Metastatic Pancreatic Neuroendocrine Tumors. Clin. Cancer Res. 2020, 26, 2011–2021. [Google Scholar] [CrossRef]
- Shuwen, H.; Xi, Y.; Qing, Z.; Jing, Z.; Wei, W. Predicting biomarkers from classifier for liver metastasis of colorectal adenocarcinomas using machine learning models. Cancer Med. 2020, 9, 6667–6678. [Google Scholar] [CrossRef]
- Rickman, C.; Davletov, B. Mechanism of calcium-independent synaptotagmin binding to target SNAREs. J. Biol. Chem. 2003, 278, 5501–5504. [Google Scholar] [CrossRef] [PubMed]
- Rachdi, L.; Maugein, A.; Pechberty, S.; Armanet, M.; Hamroune, J.; Ravassard, P.; Marullo, S.; Albagli, O.; Scharfmann, R. Regulated expression and function of the GABAB receptor in human pancreatic beta cell line and islets. Sci. Rep. 2020, 10, 13469. [Google Scholar] [CrossRef] [PubMed]
- Balhuizen, A.; Massa, S.; Mathijs, I.; Turatsinze, J.-V.; De Vos, J.; Demine, S.; Xavier, C.; Villate, O.; Millard, I.; Egrise, D.; et al. A nanobody-based tracer targeting DPP6 for non-invasive imaging of human pancreatic endocrine cells. Sci. Rep. 2017, 7, 15130. [Google Scholar] [CrossRef]
- Taguchi, Y.; Allende, M.L.; Mizukami, H.; Cook, E.K.; Gavrilova, O.; Tuymetova, G.; Clarke, B.A.; Chen, W.; Olivera, A.; Proia, R.L. Sphingosine-1-phosphate Phosphatase 2 Regulates Pancreatic Islet beta-Cell Endoplasmic Reticulum Stress and Proliferation. J. Biol. Chem. 2016, 291, 12029–12038. [Google Scholar] [CrossRef]
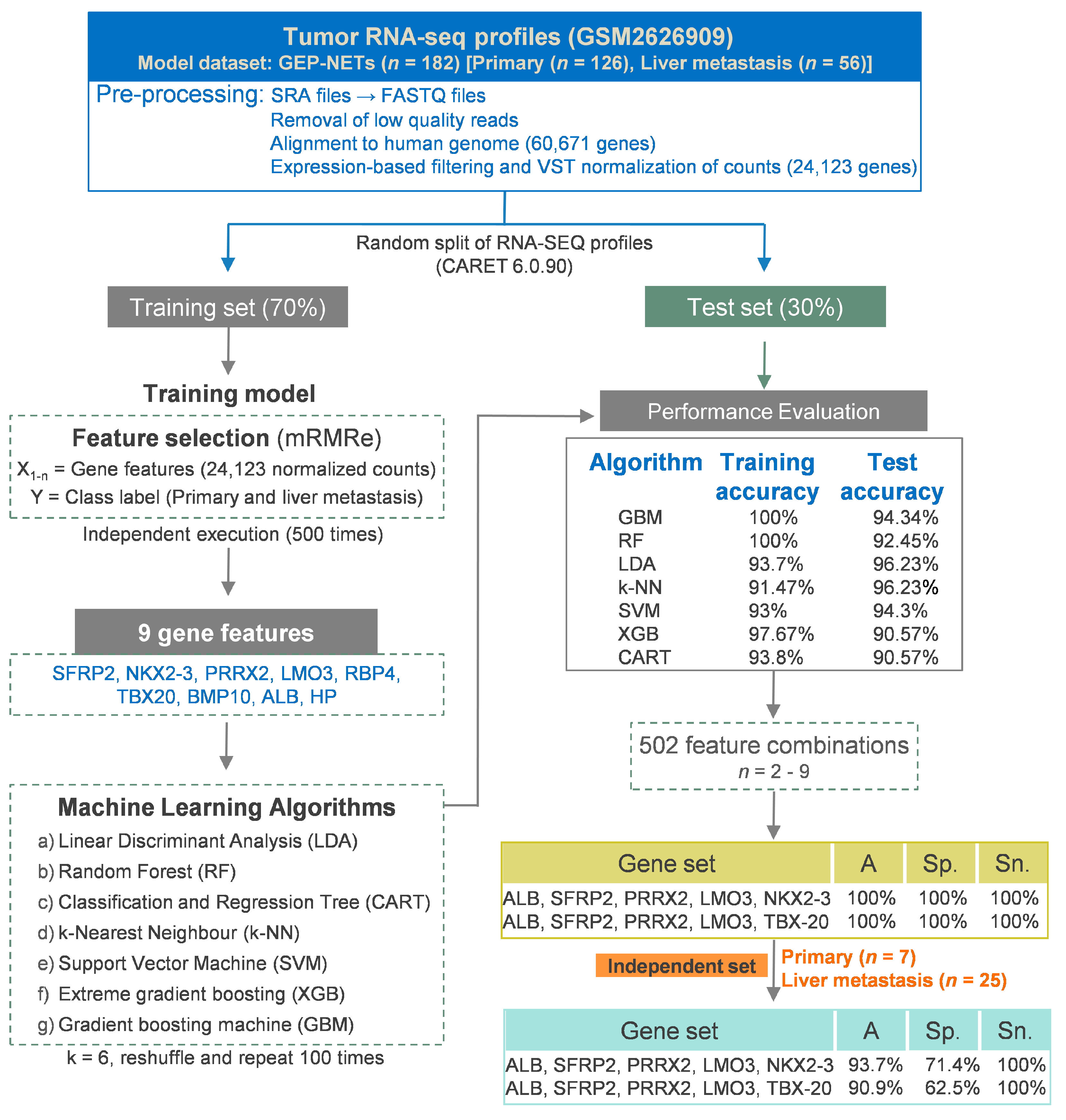
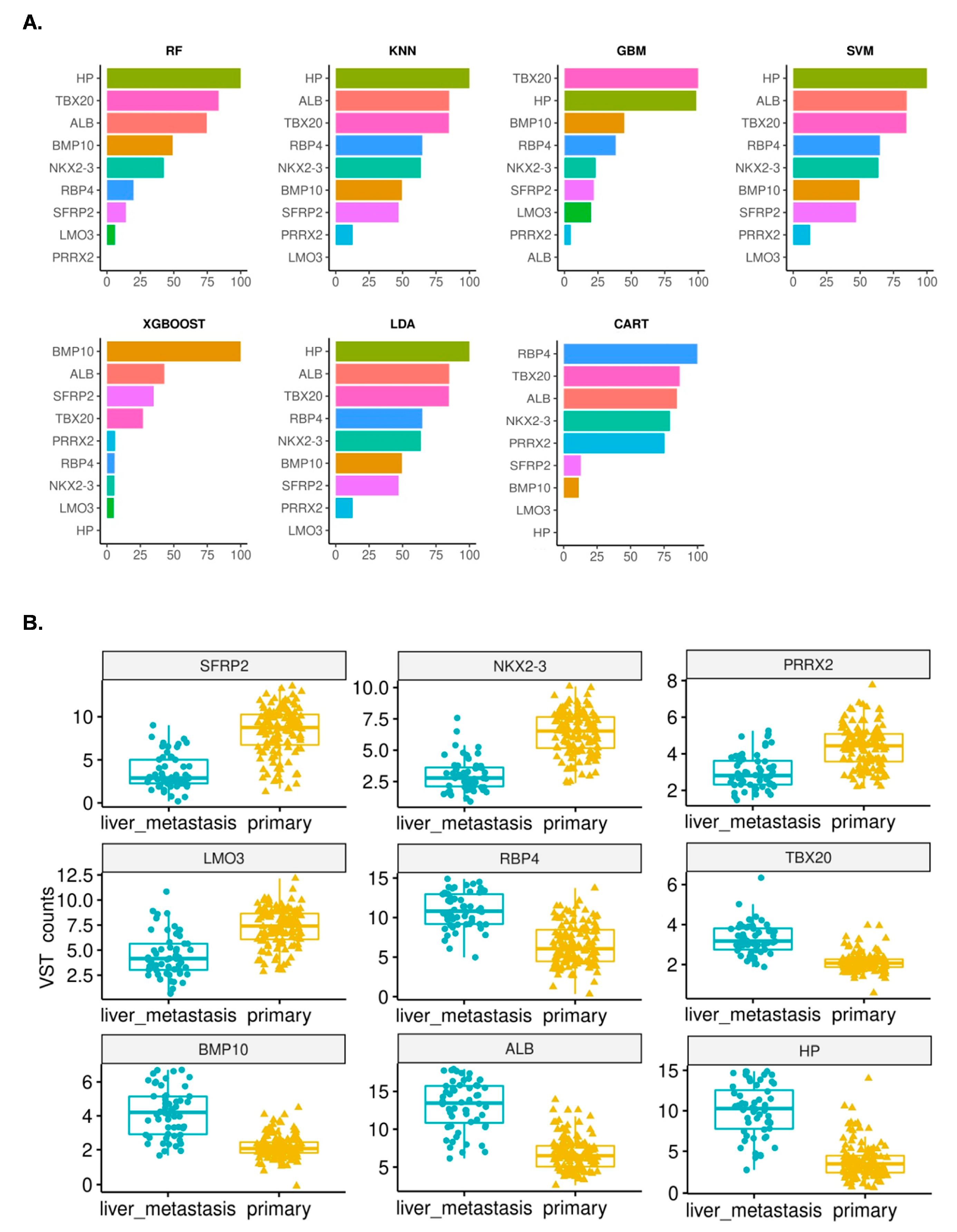


| GEO Accession | pNETs | siNETs | Purpose | Reference |
|---|---|---|---|---|
| GSE98894 | n = 113 | n = 69 | Training and Test sets | [7] |
| GSE118014 | n = 32 | n = 0 | Independent validation set | [17] |
| Sr. No. | Symbol | Description | LOG2FC (Primary/Liver mets) | padj |
|---|---|---|---|---|
| 1. | SFRP2 | Secreted frizzled related protein 2 | 5.51 | 8.35 × 10−34 |
| 2. | NKX2-3 | NK2 homeobox 3 | 4.33 | 1.36 × 10−33 |
| 3. | PRRX2 | Paired related homeobox 2 | 1.94 | 1.74 × 10−7 |
| 4. | LMO3 | LIM domain only 3 | 1.86 | 2.95 × 10−5 |
| 5. | RBP4 | Retinol binding protein 4 | −2.83 | 4.17 × 10−6 |
| 6. | TBX20 | T-box transcription factor 20 | −3.8 | 2.53 × 10−15 |
| 7. | BMP10 | Bone morphogenetic protein 10 | −8.09 | 2.36 × 10−43 |
| 8. | ALB | Albumin | −10.06 | 7.71 × 10−106 |
| 9. | HP | Haptoglobin | −10.28 | 4.69 × 10−87 |
| Gene Sets | Accuracy | Specificity | Sensitivity |
|---|---|---|---|
| HM-RF1: ALB, SFRP2, PRRX2, LMO3, NKX2-3 | 100% | 100% | 100% |
| HM-RF2: ALB, SFRP2, PRRX2, LMO3, TBX20 | 100% | 100% | 100% |
| Models | Accuracy | Sensitivity | Specificity | 95% Confidence Interval |
|---|---|---|---|---|
| HM-RF1 | 93.75% | 71.43% | 100% | 0.7567–0.9923 |
| HM-RF2 | 90.91% | 62.5% | 100% | 0.7567–0.9808 |
| Sr. No. | Symbol | Description | Log2FC (siNET/pNET) | padj |
|---|---|---|---|---|
| 1. | SYT16 | Synaptotagmin 16 | 3.01 | 9.88 × 10−6 |
| 2. | FAR2 | Fatty acyl-CoA reductase 2 | 1.78 | 1.62 × 10−8 |
| 3. | SIDT1 | SID1 transmembrane family member 1 | 1.48 | 1.34 × 10−5 |
| 4. | GABBR2 | Gamma-amino butyric acid type B receptor subunit 2 | 1.32 | 0.02 |
| 5. | OGG1 | 8-oxoguanine DNA glycosylase | 1.02 | 2.85 × 10−9 |
| 6. | TAF1A-AS1 | TAF1A antisense RNA 1 | 0.79 | 0 |
| 7. | ENSG00000259081 | lncRNA | −0.59 | 0.02 |
| 8. | SGPP1 | Sphingosine-1-phosphate phosphatase 1 | −0.67 | 3.09 × 105 |
| 9. | C19orf12 | Chromosome 19 open reading frame 12 | −0.69 | 0 |
| 10. | DRAM1 | DNA damage-regulated autophagy modulator 1 | −0.78 | 2.94 × 10−6 |
| 11. | LOC100129434 | Uncharacterized LOC100129434 | −2.01 | 4.66 × 10−9 |
| 12. | DPP6 | Dipeptidyl peptidase like 6 | −3.63 | 1.29 × 10−7 |
| Sr. No. | Model *: Gene Features |
|---|---|
| 1 | PS-RF1: DPP6, GABBR2, SYT16, SGPP1 |
| 2 | PS-RF2: DPP6, GABBR2, SYT16, SGPP1, TAF1A-AS1 |
| 3 | PS-RF3: DPP6, GABBR2, SYT16, SGPP1, LOC100129434 |
| 4 | PS-GBM1: GABBR2, FAR2 |
| 5 | PS-GBM2: SYT16, SGPP1, C19orf12 |
| 6 | PS-GBM3: TAF1A-AS1, GABBR2, FAR2, SYT16 |
| 7 | PS-GBM4: TAF1A-AS1, GABBR2, FAR2, SGPP1 |
| 8 | PS-GBM5: LOC100129434, GABBR2, SGPP1, C19orf12 |
| 9 | PS-GBM6: LOC100129434, SYT16, SGPP1, C19orf12 |
| 10 | PS-GBM7: SIDT1, DPP6, DRAM1, SYT16 |
| 11 | PS-GBM8: GABBR2, FAR2, SYT16, SGPP1 |
| 12 | PS-GBM9: GABBR2, SYT16, SGPP1, C19orf12 |
| 13 | PS-GBM10: TAF1A-AS1, GABBR2, SYT16, SGPP1, C19orf12 |
| 14 | PS-GBM11: LOC100129434, OCG1, GABBR2, SYT16, SGPP1, C19orf12 |
| 15 | PS-GBM12: LOC100129434, DPP6, GABBR2, SYT16, SGPP1, C19orf12 |
| 16 | PS-GBM13: LOC100129434, GABBR2, SYT16, SGPP1, ENSG00000259081, C19orf12 |
| 17 | PS- XGB1: SIDT1, DPP6, SYT16, SGPP1 |
| 18 | PS-XGB2: LOC100129434, DPP6, GABBR2, SYT16, ENSG00000259081 |
| 19 | PS- XGB3: DPP6, DRAM1, SYT16, SGPP1, C19orf12 |
| 20 | PS- XGB4: DPP6, GABBR2, SYT16, SGPP1, ENSG00000259081, C19orf12 |
| 21 | PS- XGB5: TAF1A-AS1, SIDT1, DPP6, GABBR2, FAR2, DRAM1, SYT16, SGPP1, ENSG00000259081 |
| Sr. No. | Models | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| 1 | PS-RF1: DPP6, GABBR2, SYT16, SGPP1 | 100% | 100% | 100% |
| 2 | PS-RF3: DPP6, GABBR2, SYT16, SGPP1, LOC100129434 | 100% | 100% | 100% |
| 3 | PS-GBM2: SYT16, SGPP1, C19orf12 | 100% | 100% | 100% |
| 4 | PS-GBM5: LOC100129434, GABBR2, SGPP1, C19orf12 | 100% | 100% | 100% |
| 5 | PS-GBM6: LOC100129434, SYT16, SGPP1, C19orf12 | 100% | 100% | 100% |
| 6 | PS-GBM9: GABBR2, SYT16, SGPP1, C19orf12 | 100% | 100% | 100% |
| 7 | PS-GBM10: TAF1A-AS1, GABBR2, SYT16, SGPP1, C19orf12 | 100% | 100% | 100% |
| 8 | PS-GBM11: LOC100129434, OCG1, GABBR2, SYT16, SGPP1, C19orf12 | 100% | 100% | 100% |
| 9 | PS-GBM12: LOC100129434, DPP6, GABBR2, SYT16, SGPP1, C19orf12 | 100% | 100% | 100% |
| 10 | PS-GBM13: LOC100129434, GABBR2, SYT16, SGPP1, ENSG00000259081, C19orf12 | 100% | 100% | 100% |
| 11 | PS- XGB2: LOC100129434, DPP6, GABBR2, SYT16, ENSG00000259081 | 100% | 100% | 100% |
| 12 | PS- XGB3: DPP6, DRAM1, SYT16, SGPP1, C19orf12 | 100% | 100% | 100% |
| 13 | PS- XGB4: DPP6, GABBR2, SYT16, SGPP1, ENSG00000259081, C19orf12 | 100% | 100% | 100% |
| Model_Name Accuracy (%) | Training Metastasis | Test Metastasis | Training Origin | Test Origin | Independent Metastasis | Independent Origin |
|---|---|---|---|---|---|---|
| Multi-label-16 | 100 | 94.55 | 100 | 89.09 | 93.75 | 96.88 |
| Multi-label-36 | 100 | 94.55 | 100 | 89.09 | 96.88 | 93.75 |
| Multi-label-27 | 100 | 92.73 | 100 | 87.27 | 84.38 | 34.38 |
| Multi-label-38 | 100 | 92.73 | 100 | 87.27 | 90.63 | 53.13 |
| Multi-label-40 | 100 | 92.73 | 100 | 87.27 | 90.63 | 78.13 |
| Multi-label-29 | 100 | 92.73 | 100 | 85.45 | 90.63 | 62.50 |
| Multi-label-21 | 100 | 92.73 | 100 | 83.64 | 93.75 | 81.25 |
| Multi-label-18 | 100 | 90.91 | 100 | 90.91 | 93.75 | 62.50 |
| Multi-label-31 | 100 | 90.91 | 100 | 90.91 | 90.63 | 75.00 |
| Multi-label-30 | 100 | 90.91 | 100 | 89.09 | 90.63 | 28.13 |
| Multi-label-32 | 100 | 90.91 | 100 | 89.09 | 87.50 | 100 |
| Multi-label-41 | 100 | 90.91 | 100 | 89.09 | 87.50 | 87.50 |
| Multi-label-17 | 100 | 90.91 | 100 | 87.27 | 87.50 | 59.38 |
| Multi-label-28 | 100 | 90.91 | 100 | 87.27 | 90.63 | 53.13 |
| Ensemble ID | Gene Name | Module | GSE | GSP | GMM |
|---|---|---|---|---|---|
| ENSG00000167157 | PRRX2 | Midnight blue | −0.50 | 3.67 × 10−13 | 0.66 |
| ENSG00000163631 | ALB | Pink | 0.75 | 1.96 × 10−34 | 0.85 |
| ENSG00000164532 | TBX20 | Pink | 0.68 | 2.43 × 10−26 | 0.70 |
| ENSG00000119919 | NKX2-3 | Dark red | −0.70 | 1.24 × 10−28 | 0.71 |
| ENSG00000145423 | SFRP2 | Midnight blue | −0.66 | 6.91 × 10−24 | 0.47 |
| ENSG00000048540 | LMO3 | Green-yellow | −0.54 | 2.25 × 10−15 | 0.46 |
| Ensemble ID | Gene Name | Module | GSE | GSP | GMM |
|---|---|---|---|---|---|
| ENSG00000136928 | GABBR2 | Blue | 0.43 | 2.13 × 10−9 | 0.55 |
| ENSG00000139973 | SYT16 | Blue | 0.64 | 3.40 × 10−22 | 0.74 |
| ENSG00000126821 | SGPP1 | Light green | −0.45 | 2.04 × 10−10 | 0.53 |
| ENSG00000225265 | TAF1A-AS1 | Blue | 0.28 | 1.38 × 10−4 | 0.33 |
| ENSG00000233251 | LOC100129434 | Midnight blue | −0.29 | 5.79 × 10−5 | 0.41 |
| ENSG00000130226 | DPP6 | Turquoise | −0.63 | 2.18 × 10−21 | 0.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padwal, M.K.; Basu, S.; Basu, B. Application of Machine Learning in Predicting Hepatic Metastasis or Primary Site in Gastroenteropancreatic Neuroendocrine Tumors. Curr. Oncol. 2023, 30, 9244-9261. https://doi.org/10.3390/curroncol30100668
Padwal MK, Basu S, Basu B. Application of Machine Learning in Predicting Hepatic Metastasis or Primary Site in Gastroenteropancreatic Neuroendocrine Tumors. Current Oncology. 2023; 30(10):9244-9261. https://doi.org/10.3390/curroncol30100668
Chicago/Turabian StylePadwal, Mahesh Kumar, Sandip Basu, and Bhakti Basu. 2023. "Application of Machine Learning in Predicting Hepatic Metastasis or Primary Site in Gastroenteropancreatic Neuroendocrine Tumors" Current Oncology 30, no. 10: 9244-9261. https://doi.org/10.3390/curroncol30100668
APA StylePadwal, M. K., Basu, S., & Basu, B. (2023). Application of Machine Learning in Predicting Hepatic Metastasis or Primary Site in Gastroenteropancreatic Neuroendocrine Tumors. Current Oncology, 30(10), 9244-9261. https://doi.org/10.3390/curroncol30100668





