Next-Generation Sequencing Analysis of Mutations in Circulating Tumor DNA from the Plasma of Patients with Head–Neck Cancer Undergoing Chemo-Radiotherapy Using a Pan-Cancer Cell-Free Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Treatment Technique
2.3. Assessment of Response
2.4. Plasma Collection
2.5. Extraction and Quantification of Plasma cfDNA
2.6. NGS Analysis
2.7. Statistical Analysis
3. Results
3.1. Gene Mutations
3.2. Associations with Histopathological Variables and Patient Age
3.3. Associations with Response to CRT
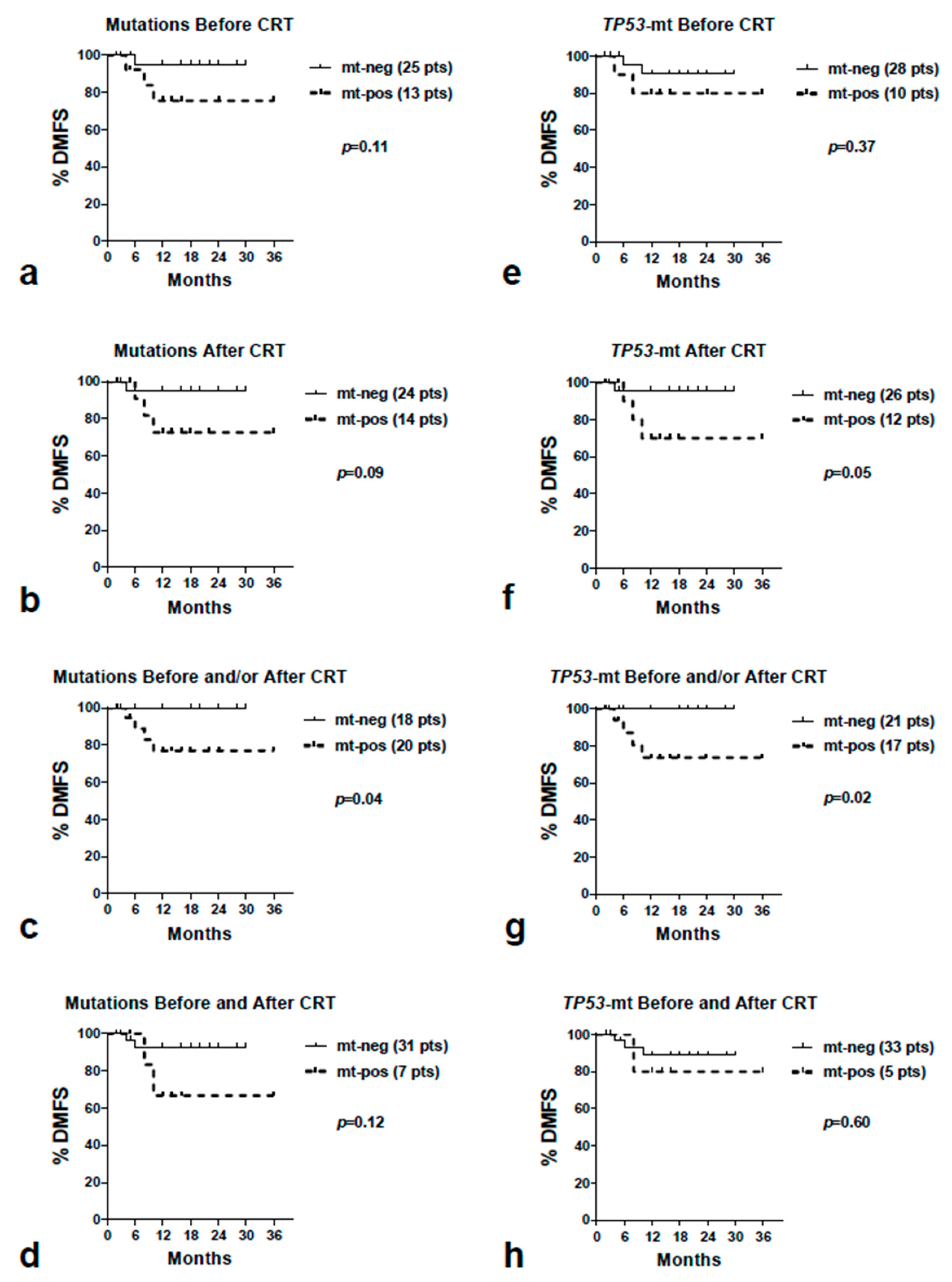
3.4. Survival Aanalysis
3.5. Specific Gene Mutations and Disease Progression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| GENE | FUNCTION |
|---|---|
| TP53 | A tumor suppressor gene. Encodes the tumor protein p53, a crucial regulator of apoptotic response, and guardian of the genome integrity. It also regulates DNA repair proteins and can induce cell cycle arrest at the G1/S cell cycle phase. Also involved in cellular senescence. |
| EGFR/ErbB1 | The Epidermal Growth Factor Receptor or ErbB-1 gene encodes a transmembrane receptor that is activated by specific ligands like EGF and TGF-α, initiating a cascade of signaling events involved in proliferation, metabolism and resistance to chemotherapy and radiotherapy. Amplification and mutations of the gene promote aberrant activation lading to carcinogenesis and tumor progression. |
| AR | Androgen receptor gene encodes ARs, transcription factors that, following their binding to testosterone, enter the nuclei to activate several genes involved in tumor progression. |
| FGFR3 | Encodes a member of the fibroblast growth factor receptor family, a membrane protein that binds to the fibroblast growth factors of the tumor stroma, promoting proliferation and differentiation. Mutations of the FGFR3 have been detected in bladder cancer and glioblastomas and are involved in cell proliferation and resistance to anti-cancer therapy. |
| FBXW7 | The F-box and WD repeat domain containing 7 gene encodes a member of the F-box protein family with critical tumor suppressor functions. It controls the degradation of several oncoproteins (c-myc, mcl-2, mTOR, jun, cycline E) through the proteasome pathway. Its mutations promote carcinogenesis and tumor growth. |
| mTOR | The mammalian target of rapamycin gene regulates cell proliferation, autophagy, apoptosis and metabolism pathways including glycolysis. Its mutations promote carcinogenesis. |
| ErbB3 | Encodes a member of the EGFR family protein. Activating mutations lead to resistance to anti-cancer therapy. |
| ALK | The anaplastic lymphoma kinase gene can be activated in a subgroup of solid tumors, driving cell growth and resistance to chemotherapy. Specific targeting drugs have been approved for the treatment of ALK-positive patients with lung cancer. |
| SF3B1 | It encodes subunit 1 of the splicing factor 3b protein complex. Mutations of the gene are linked with chronic lymphocytic leukemia, myelodysplastic syndromes, breast cancer, and orbital melanoma. |
References
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef]
- Skinner, H.D.; Sandulache, V.C.; Ow, T.J.; Meyn, R.E.; Yordy, J.S.; Beadle, B.M.; Fitzgerald, A.L.; Giri, U.; Ang, K.K.; Myers, J.N. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin. Cancer Res. 2012, 18, 290–300. [Google Scholar] [CrossRef]
- Kocakavuk, E.; Anderson, K.J.; Varn, F.S.; Johnson, K.C.; Amin, S.B.; Sulman, E.P.; Lolkema, M.P.; Barthel, F.P.; Verhaak, R.G.W. Radiotherapy is associated with a deletion signature that contributes to poor outcomes in patients with cancer. Nat. Genet. 2021, 53, 1088–1096. [Google Scholar] [CrossRef]
- Radiation Therapy-Induced Mutation Signatures Are Linked to Prognosis. Cancer Discov. 2021, 11, OF10. [CrossRef]
- Jing, C.; Mao, X.; Wang, Z.; Sun, K.; Ma, R.; Wu, J.; Cao, H. Next-generation sequencing-based detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her-2 and TP53 mutations in patients with non-small cell lung cancer. Mol. Med. Rep. 2018, 18, 2191–2197. [Google Scholar] [CrossRef]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef]
- Gormley, M.; Creaney, G.; Schache, A.; Ingarfield, K.; Conway, D.I. Reviewing the epidemiology of head and neck cancer: Definitions, trends and risk factors. Br. Dent. J. 2022, 233, 780–786. [Google Scholar] [CrossRef]
- Ganan, L.; Lopez, M.; Garcia, J.; Esteller, E.; Quer, M.; Leon, X. Management of recurrent head and neck cancer: Variables related to salvage surgery. Eur. Arch Otorhinolaryngol. 2016, 273, 4417–4424. [Google Scholar] [CrossRef]
- Petrelli, F.; Coinu, A.; Riboldi, V.; Borgonovo, K.; Ghilardi, M.; Cabiddu, M.; Lonati, V.; Sarti, E.; Barni, S. Concomitant platinum-based chemotherapy or cetuximab with radiotherapy for locally advanced head and neck cancer: A systematic review and meta-analysis of published studies. Oral Oncol. 2014, 50, 1041–1048. [Google Scholar] [CrossRef]
- Koukourakis, I.M.; Xanthopoulou, E.; Koukourakis, M.I. Using Liquid Biopsy to Predict Relapse After Radiotherapy in Squamous Cell Head-Neck and Esophageal Cancer. Cancer Diagn. Progn. 2023, 3, 403–410. [Google Scholar] [CrossRef]
- Koukourakis, M.I.; Xanthopoulou, E.; Koukourakis, I.M.; Fortis, S.P.; Kesesidis, N.; Karakasiliotis, I.; Baxevanis, C.N. Circulating Plasma Cell-free DNA (cfDNA) as a Predictive Biomarker for Radiotherapy: Results from a Prospective Trial in Head and Neck Cancer. Cancer Diagn. Progn. 2023, 3, 551–557. [Google Scholar] [CrossRef]
- Zidar, N.; Gale, N. Update from the 5th Edition of the World Health Organization Classification of Head and Neck Tumors: Hypopharynx, Larynx, Trachea and Parapharyngeal Space. Head Neck Pathol. 2022, 16, 31–39. [Google Scholar] [CrossRef]
- Shuryak, I.; Hall, E.J.; Brenner, D.J. Optimized Hypofractionation Can Markedly Improve Tumor Control and Decrease Late Effects for Head and Neck Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 272–278. [Google Scholar] [CrossRef]
- Koukourakis, M.I.; Tsoutsou, P.G.; Karpouzis, A.; Tsiarkatsi, M.; Karapantzos, I.; Daniilidis, V.; Kouskoukis, C. Radiochemotherapy with cetuximab, cisplatin, and amifostine for locally advanced head and neck cancer: A feasibility study. Int. J. Radiat. Oncol. Biol. Phys. 2010, 77, 9–15. [Google Scholar] [CrossRef]
- Therasse, P.; Arbuck, S.G.; Eisenhauer, E.A.; Wanders, J.; Kaplan, R.S.; Rubinstein, L.; Verweij, J.; Van Glabbeke, M.; van Oosterom, A.T.; Christian, M.C.; et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 2000, 92, 205–216. [Google Scholar] [CrossRef]
- Shah, M.; Takayasu, T.; Zorofchian Moghadamtousi, S.; Arevalo, O.; Chen, M.; Lan, C.; Duose, D.; Hu, P.; Zhu, J.J.; Roy-Chowdhuri, S.; et al. Evaluation of the Oncomine Pan-Cancer Cell-Free Assay for Analyzing Circulating Tumor DNA in the Cerebrospinal Fluid in Patients with Central Nervous System Malignancies. J. Mol. Diagn. 2021, 23, 171–180. [Google Scholar] [CrossRef]
- Mellert, H.; Reese, J.; Jackson, L.; Maxwell, V.; Tschida, C.; Pestano, G.A. Targeted Next-Generation Sequencing of Liquid Biopsy Samples from Patients with NSCLC. Diagnostics 2021, 11, 155. [Google Scholar] [CrossRef]
- Errazquin, R.; Carrasco, E.; Del Marro, S.; Sunol, A.; Peral, J.; Ortiz, J.; Rubio, J.C.; Segrelles, C.; Duenas, M.; Garrido-Aranda, A.; et al. Early Diagnosis of Oral Cancer and Lesions in Fanconi Anemia Patients: A Prospective and Longitudinal Study Using Saliva and Plasma. Cancers 2023, 15, 1871. [Google Scholar] [CrossRef]
- Van Ginkel, J.H.; de Leng, W.W.; de Bree, R.; van Es, R.J.; Willems, S.M. Targeted sequencing reveals TP53 as a potential diagnostic biomarker in the post-treatment surveillance of head and neck cancer. Oncotarget 2016, 7, 61575–61586. [Google Scholar] [CrossRef]
- Huang, Q.; Li, F.; Ji, M.; Lin, L.; Hu, C. Evaluating the prognostic significance of p53 and TP53 mutations in HPV-negative hypopharyngeal carcinoma patients: A 5-year follow-up retrospective study. BMC Cancer 2023, 23, 324. [Google Scholar] [CrossRef]
- Flach, S.; Kumbrink, J.; Walz, C.; Hess, J.; Drexler, G.; Belka, C.; Canis, M.; Jung, A.; Baumeister, P. Analysis of genetic variants of frequently mutated genes in human papillomavirus-negative primary head and neck squamous cell carcinoma, resection margins, local recurrences and corresponding circulating cell-free DNA. J. Oral Pathol. Med. 2022, 51, 738–746. [Google Scholar] [CrossRef]
- Economopoulou, P.; Spathis, A.; Kotsantis, I.; Maratou, E.; Anastasiou, M.; Moutafi, M.K.; Kirkasiadou, M.; Pantazopoulos, A.; Giannakakou, M.; Edelstein, D.L.; et al. Next-generation sequencing (NGS) profiling of matched tumor and circulating tumor DNA (ctDNA) in head and neck squamous cell carcinoma (HNSCC). Oral Oncol. 2023, 139, 106358. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Mulvey, C.L.; Silliman, N.; Schaefer, J.; Sausen, M.; James, N.; Rettig, E.M.; Guo, T.; Pickering, C.R.; et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 2015, 7, 293ra104. [Google Scholar] [CrossRef]
- Porter, A.; Natsuhara, M.; Daniels, G.A.; Patel, S.P.; Sacco, A.G.; Bykowski, J.; Banks, K.C.; Cohen, E.E.W. Next generation sequencing of cell free circulating tumor DNA in blood samples of recurrent and metastatic head and neck cancer patients. Transl. Cancer Res. 2020, 9, 203–209. [Google Scholar] [CrossRef]
- Galot, R.; van Marcke, C.; Helaers, R.; Mendola, A.; Goebbels, R.M.; Caignet, X.; Ambroise, J.; Wittouck, K.; Vikkula, M.; Limaye, N.; et al. Liquid biopsy for mutational profiling of locoregional recurrent and/or metastatic head and neck squamous cell carcinoma. Oral Oncol. 2020, 104, 104631. [Google Scholar] [CrossRef]
- Beckta, J.M.; Ahmad, S.F.; Yang, H.; Valerie, K. Revisiting p53 for cancer-specific chemo- and radiotherapy: Ten years after. Cell Cycle 2014, 13, 710–713. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Y.; Lu, X.; Ding, H.; Han, B.; Song, X.; Miao, H.; Cui, X.; Wei, S.; Liu, W.; et al. CDC20 regulates the cell proliferation and radiosensitivity of P53 mutant HCC cells through the Bcl-2/Bax pathway. Int. J. Biol. Sci. 2021, 17, 3608–3621. [Google Scholar] [CrossRef]
- Cui, D.; Xiong, X.; Shu, J.; Dai, X.; Sun, Y.; Zhao, Y. FBXW7 Confers Radiation Survival by Targeting p53 for Degradation. Cell Rep. 2020, 30, 497–509.e494. [Google Scholar] [CrossRef]
- Azad, T.D.; Chaudhuri, A.A.; Fang, P.; Qiao, Y.; Esfahani, M.S.; Chabon, J.J.; Hamilton, E.G.; Yang, Y.D.; Lovejoy, A.; Newman, A.M.; et al. Circulating Tumor DNA Analysis for Detection of Minimal Residual Disease After Chemoradiotherapy for Localized Esophageal Cancer. Gastroenterology 2020, 158, 494–505.e496. [Google Scholar] [CrossRef]
- Morrissy, A.S.; Garzia, L.; Shih, D.J.; Zuyderduyn, S.; Huang, X.; Skowron, P.; Remke, M.; Cavalli, F.M.; Ramaswamy, V.; Lindsay, P.E.; et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 2016, 529, 351–357. [Google Scholar] [CrossRef]
- Wilson, H.L.; D’Agostino, R.B., Jr.; Meegalla, N.; Petro, R.; Commander, S.; Topaloglu, U.; Zhang, W.; Porosnicu, M. The Prognostic and Therapeutic Value of the Mutational Profile of Blood and Tumor Tissue in Head and Neck Squamous Cell Carcinoma. Oncologist 2021, 26, e279–e289. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.; Zou, J.; Magalhaes, M.; Oliva, M.; Spreafico, A.; Hansen, A.R.; McDade, S.S.; Coyle, V.M.; Lawler, M.; Elimova, E.; et al. Circulating tumour DNA kinetics in recurrent/metastatic head and neck squamous cell cancer patients. Eur. J. Cancer 2023, 188, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, N.; Cheng, G.; Zhang, T.; Wang, J.; Deng, L.; Li, J.; Zhao, X.; Xu, Y.; Yang, P.; et al. Prognostic value of circulating tumour DNA during post-radiotherapy surveillance in locally advanced esophageal squamous cell carcinoma. Clin. Transl. Med. 2022, 12, e1116. [Google Scholar] [CrossRef] [PubMed]
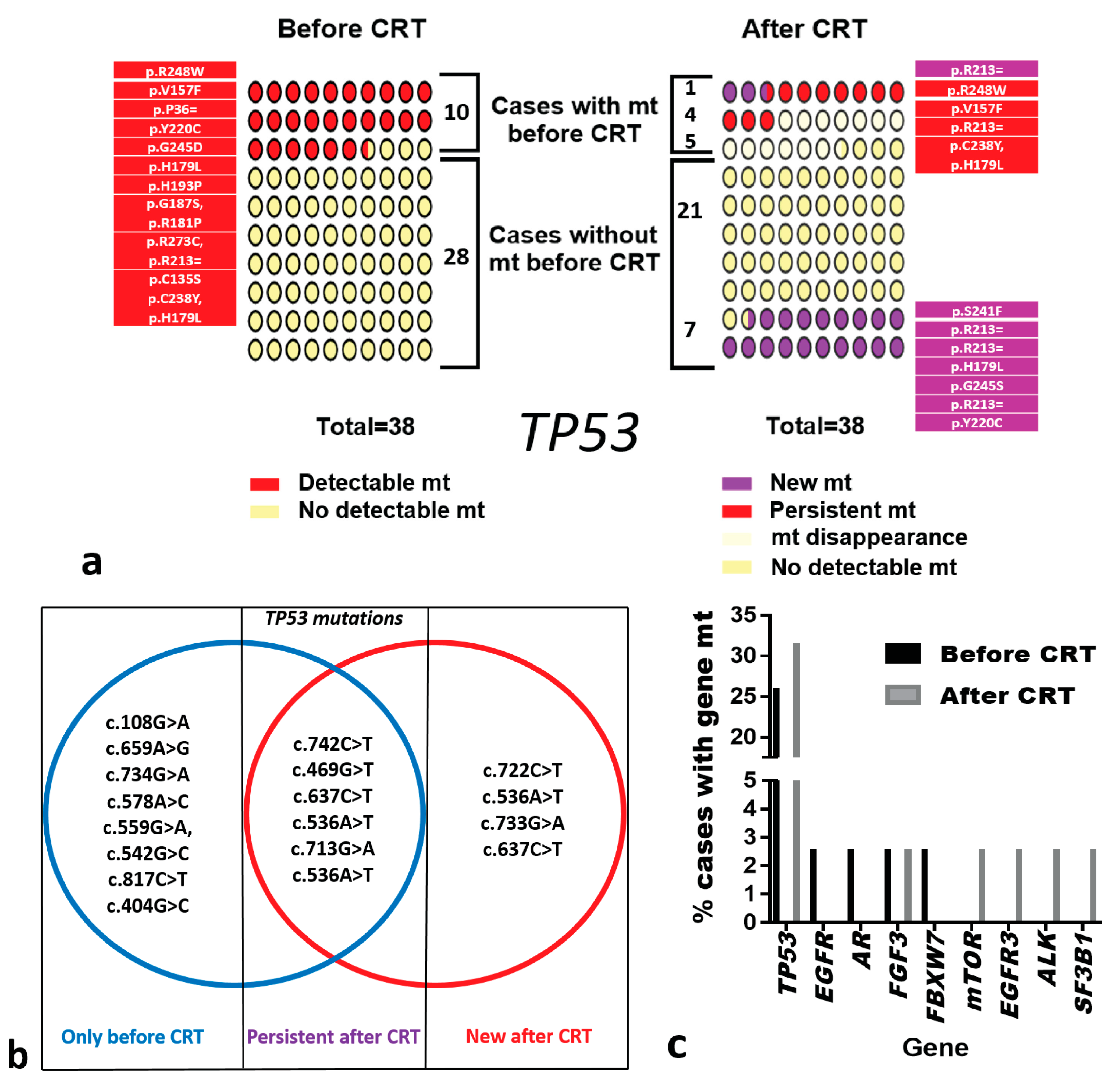

| Before CRT | After CRT | |||||||
|---|---|---|---|---|---|---|---|---|
| No. Patient | Gene (No) | AA Mutation | CDS Mutation | Molecular Frequency % | Gene (No) | AA Mutation | CDS Mutation | Molecular Frequency % |
| TP53 | TP53 | |||||||
| 1 | 1 | p.R248W | c.742C>T | 1.2 | 1 | p.R248W | c.742C>T | 0.49 |
| 11 | 1 | p.V157F | c.469G>T | 54 | 1 | p.V157F | c.469G>T | 126 |
| 13 | 1 | p.P36= | c.108G>A | 52 | 0 | ----- | ---- | ----- |
| 17 | 1 | p.Y220C | c.659A>G | 1.18 | 0 | ----- | ---- | ----- |
| 18 | 1 | p.G245D | c.734G>A | 1.68 | 1 | p.R213= | c.637C>T | 52 |
| 29 | 1 | p.H179L | c.536A>T | 0.42 | 0 | ----- | ---- | ----- |
| 38 | 1 | p.H193P | c.578A>C | 2.14 | 0 | ----- | ---- | ----- |
| 30 | 2 | p.G187S p.R181P | c.559G>A, c.542G>C | 1.60 2.40 | 0 | ----- ----- | ----- ----- | ----- ----- |
| 35 | 2 | p.R273C p.R213= | c.817C>T c.637C>T | 0.17 0.32 | 1 | ----- p.R213= | ----- c.637C>T | 0.48 |
| 22 | 3 | p.C135S p.C238Y p.H179L | c.404G>C c.713G>A c.536A>T | 1.34 0.39 41 | 2 | ----- p.C238Y p.H179L | ----- c.713G>A c.536A>T | ----- 0.34 3.70 |
| 10 | 0 | ----- | ---- | ---- | 1 | p.S241F | c.722C>T | 15 |
| 20 | 0 | ----- | ---- | ---- | 1 | p.R213= | c.637C>T | 1.08 |
| 23 | 0 | ----- | ---- | ---- | 1 | p.R213= | c.637C>T | 51 |
| 25 | 0 | ----- | ---- | ---- | p.H179L | c.536A>T | 0.39 | |
| 26 | 0 | ----- | ---- | ---- | 1 | p.G245S | c.733G>A | 0.27 |
| 27 | 0 | ----- | ---- | ---- | 1 | p.R213= | c.637C>T | 0.27 |
| 33 | 0 | ----- | ---- | ---- | 1 | p.Y220C | c.659A>G | 1.17 |
| EGFR | EGFR | |||||||
| 28 | 1 | p.P848L | c.2543C>T | 0.10 | 0 | ----- | ---- | ---- |
| AR | AR | |||||||
| 10 | 1 | p.E894K | c.2680G>A | 0.49 | 0 | ----- | ---- | ---- |
| FGFR3 | FGFR3 | |||||||
| 16 | 1 | p.F384L | c.1150T>C | 48 | 1 | p.F384L | c.1150T>C | 46 |
| FBXW7 | FBXW7 | |||||||
| 10 | 1 | p.R505C | c.1513C>T | 269 | 0 | ---- | ---- | ---- |
| mTOR | mTOR | |||||||
| 23 | 0 | ---- | ---- | ---- | 1 | p.R2217W | unknown | 0.24 |
| EGFR3 | EGFR3 | |||||||
| 27 | 0 | ---- | ---- | ---- | 1 | p.V104M | c.310G>A | 0.16 |
| ALK | ALK | |||||||
| 26 | 0 | ---- | ---- | ---- | 1 | p.R1275Q | c.382G>A | 0.17 |
| SF3B1 | SF3B1 | |||||||
| 9 | 0 | ----- | ---- | ---- | 1 | p.K700E | c.2098A>G | 0.62 |
| All Mutations | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| LRFS | OS | DMFS | |||||||
| Yes vs. No | p-value | HR | 95%CI | No | Yes | p-value | No | Yes | p-value (*) |
| B | 0.17 | 2.20 | 0.7–6.8 | 0.93 | 1.05 | 0.3–3.6 | 0.11 | 5.12 | 0.6–39 |
| A | 0.08 | 3.10 | 0.9–9.6 | 0.51 | 1.52 | 0.4–5.2 | 0.09 | 5.75 | 0.7–44 |
| B and/or A | 0.19 | 2.28 | 0.8–6.0 | 0.43 | 1.60 | 0.4–5.2 | 0.04 | 6.75 | 0.9–48 |
| B and A | 0.02 | 5.38 | 1.2–24 | 0.93 | 0.93 | 0.2–4.2 | 0.12 | 6.82 | 0.5–80 |
| Yes vs. No | TP53 mutations | ||||||||
| B | 0.90 | 1.42 | 0.4–1.6 | 0.98 | 0.98 | 024–3.7 | 0.37 | 2.66 | 0.3–22 |
| A | 0.15 | 3.01 | 0.9–9.9 | 0.71 | 1.27 | 0.3–4.0 | 0.05 | 7.75 | 0.9–64 |
| B and/or A | 0.22 | 2.42 | 0.8–7.0 | 0.49 | 1.51 | 0.4–5.0 | 0.02 | 9.19 | 1.2–65 |
| B and A | 0.41 | 1.90 | 0.4–8.9 | 0.61 | 0.64 | 0.1–3.6 | 0.60 | 2.03 | 0.1–30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koukourakis, M.I.; Xanthopoulou, E.; Koukourakis, I.M.; Fortis, S.P.; Kesesidis, N.; Kakouratos, C.; Karakasiliotis, I.; Baxevanis, C.N. Next-Generation Sequencing Analysis of Mutations in Circulating Tumor DNA from the Plasma of Patients with Head–Neck Cancer Undergoing Chemo-Radiotherapy Using a Pan-Cancer Cell-Free Assay. Curr. Oncol. 2023, 30, 8902-8915. https://doi.org/10.3390/curroncol30100643
Koukourakis MI, Xanthopoulou E, Koukourakis IM, Fortis SP, Kesesidis N, Kakouratos C, Karakasiliotis I, Baxevanis CN. Next-Generation Sequencing Analysis of Mutations in Circulating Tumor DNA from the Plasma of Patients with Head–Neck Cancer Undergoing Chemo-Radiotherapy Using a Pan-Cancer Cell-Free Assay. Current Oncology. 2023; 30(10):8902-8915. https://doi.org/10.3390/curroncol30100643
Chicago/Turabian StyleKoukourakis, Michael I., Erasmia Xanthopoulou, Ioannis M. Koukourakis, Sotirios P. Fortis, Nikolaos Kesesidis, Christos Kakouratos, Ioannis Karakasiliotis, and Constantin N. Baxevanis. 2023. "Next-Generation Sequencing Analysis of Mutations in Circulating Tumor DNA from the Plasma of Patients with Head–Neck Cancer Undergoing Chemo-Radiotherapy Using a Pan-Cancer Cell-Free Assay" Current Oncology 30, no. 10: 8902-8915. https://doi.org/10.3390/curroncol30100643
APA StyleKoukourakis, M. I., Xanthopoulou, E., Koukourakis, I. M., Fortis, S. P., Kesesidis, N., Kakouratos, C., Karakasiliotis, I., & Baxevanis, C. N. (2023). Next-Generation Sequencing Analysis of Mutations in Circulating Tumor DNA from the Plasma of Patients with Head–Neck Cancer Undergoing Chemo-Radiotherapy Using a Pan-Cancer Cell-Free Assay. Current Oncology, 30(10), 8902-8915. https://doi.org/10.3390/curroncol30100643







