The Clinically Actionable Molecular Profile of Early versus Late-Stage Non-Small Cell Lung Cancer, an Individual Age and Sex Propensity-Matched Pair Analysis
Abstract
:1. Introduction
1.1. Background
1.2. Objective
2. Methods
2.1. Study Design and Setting
2.2. Participants and Data Sources
2.3. Primary Outcomes
2.4. Targeted NGS Panels for NSCLC Genetic Alterations
2.5. Statistical Methods
3. Results
4. Discussion
4.1. Molecular Alteration Frequency Profiles—Early versus Late-Stage NSCLC
4.2. Study Limitations
4.3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AJCC | American Joint Commission on Cancer |
| ALK | Anaplastic lymphoma kinase |
| EGFR | Epidermal growth factor receptor |
| FDG-PET | Fluorodeoxyglucose positron emission tomography |
| FFPE | Formalin fixed paraffin embedded |
| MET | Met proto-oncogene |
| NSCLC | Non-small cell lung cancer |
| RET | Ret proto-oncogene |
| ROS1 | c-ROS proto- oncogene 1 |
| SCLC | Small cell lung cancer (SCLC) |
| TKI | Tyrosine kinase inhibitor (TKI) |
| VCC | Vancouver Cancer Centre |
| VGH | Vancouver General Hospital |
| USFDA | United States Food and Drug Administration |
| VAF | Variant allele frequency |
| WHO | World Health Organization |
References
- Canadian Cancer Statistics 2020. Available online: cancer.ca/Canadian-Cancer-Statistics-2020-EN (accessed on 1 September 2021).
- Travis, W.D.; Brambilla, E.; Noguchi, M.; Nicholson, A.G.; Geisinger, K.R.; Yatabe, Y.; Beer, D.G.; Powell, C.; Riely, G.J.; Van Schil, P.E.; et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011, 6, 244–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travis, W.D.; Brambilla, E.; Burke, A.P.; Marx, A.; Nicholson, A.G. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J. Thorac. Oncol. 2015, 10, 1240–1242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Yokoi, K. The IASLC Lung Cancer Staging Project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.-L.; Herbst, R.S.; Mann, H.; Rukazenkov, Y.; Marotti, M.; Tsuboi, M. ADAURA: Phase III, Double-blind, Randomized Study of Osimertinib Versus Placebo in EGFR Mutation-positive Early-stage NSCLC After Complete Surgical Resection. Clin. Lung Cancer 2018, 19, e533–e536. [Google Scholar] [CrossRef]
- Nishio, M.; Felip, E.; Orlov, S.; Park, K.; Yu, C.-J.; Tsai, C.-M.; Cobo, M.; McKeage, M.; Su, W.-C.; Mok, T.; et al. Final Overall Survival and Other Efficacy and Safety Results From ASCEND-3: Phase II Study of Ceritinib in ALKi-Naive Patients With ALK-Rearranged NSCLC. J. Thorac. Oncol. 2019, 15, 609–617. [Google Scholar] [CrossRef]
- Shaw, A.T.; Kim, D.-W.; Mehra, R.; Tan, D.S.; Felip, E.; Chow, L.Q.; Camidge, D.R.; Vansteenkiste, J.; Sharma, S.; De Pas, T.; et al. Ceritinib inALK-Rearranged Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2014, 370, 1189–1197. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Zou, Q.; Liu, H.; Qiu, B.; Li, Q.; Lin, Y.; Liang, Y. Management of Non-small Cell Lung Cancer Patients with MET Exon 14 Skipping Mutations. Curr. Treat. Options Oncol. 2020, 21, 33. [Google Scholar] [CrossRef]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in Non–Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef]
- Ackermann, C.J.; Stock, G.; Tay, R.; Dawod, M.; Gomes, F.; Califano, R. Targeted Therapy For RET-Rearranged Non-Small Cell Lung Cancer: Clinical Development And Future Directions. OncoTargets Ther. 2019, 12, 7857–7864. [Google Scholar] [CrossRef] [Green Version]
- Molina-Arcas, M.; Moore, C.; Rana, S.; van Maldegem, F.; Mugarza, E.; Romero-Clavijo, P.; Herbert, E.; Horswell, S.; Li, L.-S.; Janes, M.R.; et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci. Transl. Med. 2019, 11, eaaw7999. [Google Scholar] [CrossRef]
- Mitsudomi, T.; Morita, S.; Yatabe, Y.; Negoro, S.; Okamoto, I.; Tsurutani, J.; Seto, T.; Stouchi, M.; Tada, H.; Hirashima, T.; et al. Gefitinib versus cisplatin plus docetaxel in patients with non- small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010, 11, 121–128. [Google Scholar] [CrossRef]
- Rosell, R.; Carcereny, E.; Gervais, R.; Vergnenegre, A.; Massuti, B.; Felip, E.; Palmero, R.; Garcia-Gomez, R.; Pallares, C.; Sanchez, J.M.; et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012, 13, 239–246. [Google Scholar] [CrossRef]
- Suster, D.I.; Mino-Kenudson, M. Molecular Pathology of Primary Non-small Cell Lung Cancer. Arch. Med Res. 2020, 51, 784–798. [Google Scholar] [CrossRef]
- Non-Small Cell Lung Cancer NCCN Clinical Practice Guidelines in Oncology. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf (accessed on 30 August 2021).
- Canon, J.; Rex, K.; Saiki, A.Y.; Mohr, C.; Cooke, K.; Bagal, D.; Gaida, K.; Holt, T.; Knutson, C.G.; Koppada, N.; et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019, 575, 217–223. [Google Scholar] [CrossRef]
- Nagasaka, M.; Li, Y.; Sukari, A.; Ou, S.-H.I.; Al-Hallak, M.N.; Azmi, A.S. KRAS G12C Game of Thrones, which direct KRAS inhibitor will claim the iron throne? Cancer Treat. Rev. 2020, 84, 101974. [Google Scholar] [CrossRef]
- Kim, I.A.; Hur, J.Y.; Kim, H.J.; Park, J.H.; Hwang, J.J.; Lee, S.A.; Lee, S.E.; Kim, W.S.; Lee, K.Y. Targeted Next-Generation Sequencing Analysis for Recurrence in Early-Stage Lung Adenocarcinoma. Ann. Surg. Oncol. 2020, 28, 3983–3993. [Google Scholar] [CrossRef]
- Lengel, H.; Connolly, J.; Jones, G.; Caso, R.; Zhou, J.; Sanchez-Vega, F.; Mastrogiacomo, B.; Isbell, J.; Li, B.; Liu, Y.; et al. The Emerging Importance of Tumor Genomics in Operable Non-Small Cell Lung Cancer. Cancers 2021, 13, 3656. [Google Scholar] [CrossRef]
- Saunders, C.T.; Wong, W.; Swamy, S.; Becq, J.; Murray, L.J.; Cheetham, R.K. Strelka: Accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 2012, 28, 1811–1817. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.C.-H.; Schuler, M.; Popat, S.; Miura, S.; Heeke, S.; Park, K.; Märten, A.; Kim, E.S. Afatinib for the Treatment of NSCLC Harboring Uncommon EGFR Mutations: A Database of 693 Cases. J. Thorac. Oncol. 2020, 15, 803–815. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.H.; Lim, S.H.; An, H.J.; Kim, K.H.; Park, K.U.; Kang, E.J.; Choi, Y.H.; Ahn, M.S.; Lee, M.H.; Sun, J.-M.; et al. Osimertinib for Patients With Non–Small-Cell Lung Cancer Harboring Uncommon EGFR Mutations: A Multicenter, Open-Label, Phase II Trial (KCSG-LU15-09). J. Clin. Oncol. 2020, 38, 488–495. [Google Scholar] [CrossRef]
- Chiu, C.-H.; Yang, C.-T.; Shih, J.-Y.; Huang, M.-S.; Su, W.-C.; Lai, R.-S.; Wang, C.-C.; Hsiao, S.-H.; Lin, Y.-C.; Ho, C.-L.; et al. Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Treatment Response in Advanced Lung Adenocarcinomas with G719X/L861Q/S768I Mutations. J. Thorac. Oncol. 2015, 10, 793–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.Y.; Choi, Y.J.; Shin, S.W. Osimertinib in EGFR Mutation–Positive Advanced NSCLC. N. Engl. J. Med. 2018, 378, 1261–1263. [Google Scholar] [CrossRef]
- Rebuzzi, S.E.; Alfieri, R.; La Monica, S.; Minari, R.; Petronini, P.G.; Tiseo, M. Combination of EGFR-TKIs and chemotherapy in advanced EGFR mutated NSCLC: Review of the literature and future perspectives. Crit. Rev. Oncol. 2019, 146, 102820. [Google Scholar] [CrossRef] [PubMed]
- Piotrowska, Z.; Fintelmann, F.J.; Sequist, L.V.; Jahagirdar, B. Response to Osimertinib in an EGFR Exon 20 Insertion-Positive Lung Adenocarcinoma. J. Thorac. Oncol. 2018, 13, e204–e206. [Google Scholar] [CrossRef] [Green Version]
- Brazel, D.; Nagasaka, M. Spotlight on Amivantamab (JNJ-61186372) for EGFR Exon 20 Insertions Positive Non-Small Cell Lung Cancer. Lung Cancer Targets Ther. 2021, 12, 133–138. [Google Scholar] [CrossRef]
- Nagasaka, M.; Balmanoukian, A.S.; Madison, R.; Zhang, S.S.; Klempner, S.J.; Ou, S.-H.I. Amivantamab (JNJ-61186372) induces clinical, biochemical, molecular, and radiographic response in a treatment-refractory NSCLC patient harboring amplified triple EGFR mutations (L858R/ T790M/G796S) in cis. Lung Cancer 2022, 164, 52–55. [Google Scholar] [CrossRef]
- Imran, M.; Alam Khan, S.; Alshammari, M.K.; Alreshidi, M.A.; Alreshidi, A.A.; Alghonaim, R.S.; Alanazi, F.A.; Alshehri, S.; Ghoneim, M.M.; Shakeel, F. Discovery, Development, Inventions, and Patent Trends on Mobocertinib Succinate: The First-in-Class Oral Treatment for NSCLC with EGFR Exon 20 Insertions. Biomedicines 2021, 9, 1938. [Google Scholar] [CrossRef]
- Scheffler, M.; Schultheis, A.; Teixido, C.; Michels, S.; Morales-Espinosa, D.; Viteri, S.; Hartmann, W.; Merkelbach-Bruse, S.; Fischer, R.; Schildhaus, H.-U.; et al. ROS1 rearrangements in lung adenocarcinoma: Prognostic impact, therapeutic options and genetic variability. Oncotarget 2015, 6, 10577–10585. [Google Scholar] [CrossRef] [Green Version]
- Clavé, S.; Gimeno, J.; Muñoz-Mármol, A.M.; Vidal, J.; Reguart, N.; Carcereny, E.; Pijuan, L.; Menéndez, S.; Taus, Á.; Mate, J.L.; et al. ROS1 copy number alterations are frequent in non-small cell lung cancer. Oncotarget 2016, 7, 8019–8028. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.K.; Kim, K.A.; Lee, C.Y.; Kim, S.; Chang, S.; Cho, B.C.; Shim, H.S. Molecular Diagnostic Assays and Clinicopathologic Implications of MET Exon 14 Skipping Mutation in Non–small-cell Lung Cancer. Clin. Lung Cancer 2018, 20, e123–e132. [Google Scholar] [CrossRef]
- Takeuchi, K.; Soda, M.; Togashi, Y.; Suzuki, R.; Sakata, S.; Hatano, S.; Asaka, R.; Hamanaka, W.; Ninomiya, H.; Uehara, H.; et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012, 18, 378–381. [Google Scholar] [CrossRef]
- Gautschi, O.; Milia, J.; Filleron, T.; Wolf, J.; Carbone, D.P.; Owen, D.H.; Camidge, R.; Narayanan, V.; Doebele, R.C.; Besse, B.; et al. Targeting RET in Patients With RET-Rearranged Lung Cancers: Results From the Global, Multicenter RET Registry. J. Clin. Oncol. 2017, 35, 1403–1410. [Google Scholar] [CrossRef] [Green Version]
- Ferrara, R.; Auger, N.; Auclin, E.; Besse, B. Clinical and Translational Implications of RET Rearrangements in Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2018, 13, 27–45. [Google Scholar] [CrossRef] [Green Version]
- Loh, Z.; Mitchell, P.; John, T.; Arulananda, S. RET-rearranged non-small-cell lung cancer and therapeutic implications. Intern. Med. J. 2019, 49, 1541–1545. [Google Scholar] [CrossRef] [Green Version]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
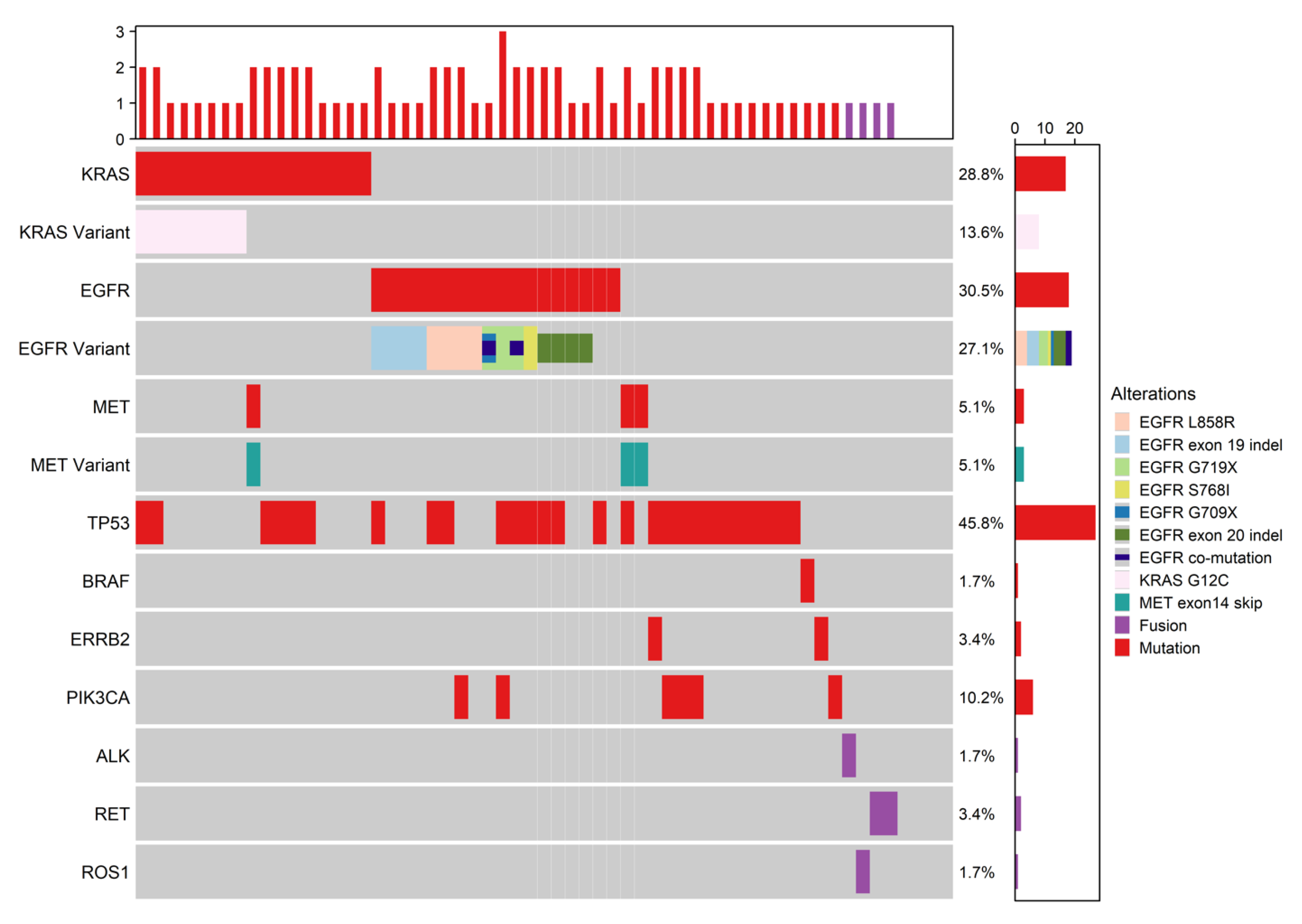
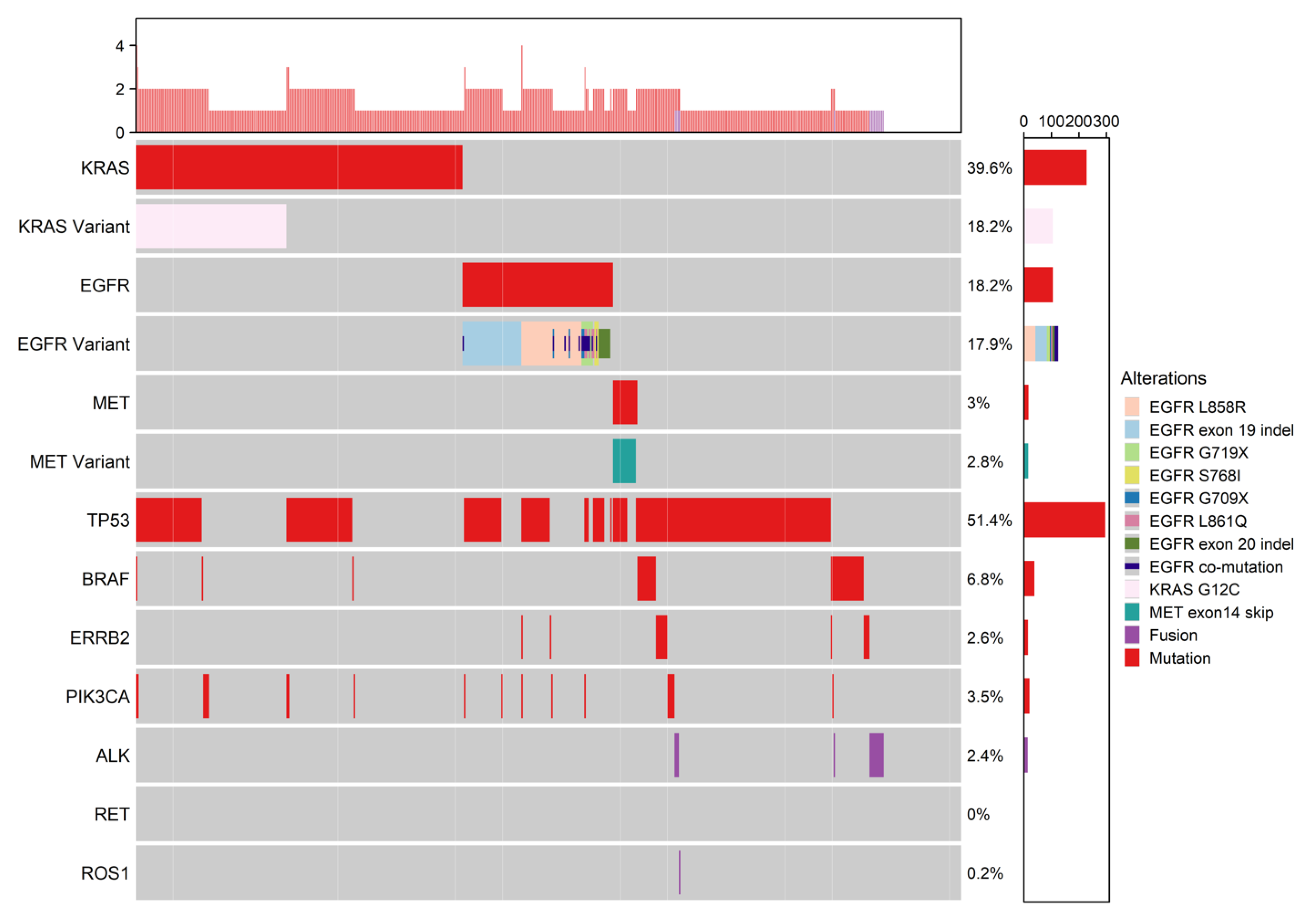
| Demographics/Diagnosis | All n = 635 | Early Stage n = 59 | Late Stage n = 576 | p Value | Standardized Mean Difference (SMD) |
|---|---|---|---|---|---|
| Age (years): mean ± sd | 70.9 ± 10.2 | 68.0 ± 10.3 | 71.2 ± 10.2 | 0.020 | −31.2 |
| Sex (female): n (%) | 276 (43.5) | 22 (37.3) | 254 (44.1) | 0.32 | −11.4 |
| Smoking status: n (%) | 0.027 | 20.4 | |||
| Never smoker | 140 (22.0) | 21 (35.6) | 119 (20.7) | ||
| Former smoker | 385 (60.6) | 31 (52.5) | 354 (61.5) | ||
| Current smoker | 110 (17.3) | 7 (11.9) | 103 (17.9) | ||
| Histology: n (%) | <0.001 | 72.4 | |||
| Adenocarcinoma | 527 (83.0) | 49 (83.1) | 478 (83.0) | ||
| Squamous cell carcinoma | 19 (3.0) | 9 (15.3) | 10 (1.7) | ||
| Other lung carcinoma | 89 (14.0) | 1 (1.7) | 88 (15.3) |
| Demographics/Diagnosis | All n = 212 | Early Stage n = 53 | Late Stage n = 159 | p Value | Standardized Mean Difference (SMD) |
|---|---|---|---|---|---|
| Age (years): mean ± sd | 68.5 ± 11.2 | 68.5 ± 10.2 | 68.5 ± 11.5 | 0.99 | 0.0 |
| Sex (female): n (%) | 143 (67.5) | 34 (64.2) | 109 (68.6) | 0.43 | −7.6 |
| Smoking status: n (%) | 0.83 | 9.8 | |||
| Never smoker | 83 (39.2) | 21 (39.6) | 62 (39.0) | ||
| Former smoker | 108 (50.9) | 26 (49.1) | 82 (51.6) | ||
| Current smoker | 21 (9.9) | 6 (11.3) | 15 (9.4) | ||
| Histology: n (%) | 0.99 | 3.1 | |||
| Adenocarcinoma | 197 (92.9) | 49 (92.5) | 148 (93.1) | ||
| Squamous cell carcinoma | 11 (5.2) | 3 (5.7) | 8 (5.0) | ||
| Other lung carcinoma | 4 (1.9) | 1 (1.9) | 3 (1.9) |
| Outcome Variable | All n = 635 | Early Stage n = 59 | Late Stage n = 576 | p Value |
|---|---|---|---|---|
| Any alteration mutation or fusion | 594 (93.5) | 55 (93.2) | 539 (93.6) | 0.92 |
| Potential therapeutic target | 274 (43.1) | 29 (49.2) | 245 (42.5) | 0.33 |
| EGFR mutation present | 123 (19.4) | 18 (30.5) | 105 (18.2) | 0.023 |
| EGFRm common sensitizing present | 91 (14.3) | 8 (13.6) | 83 (14.4) | 0.86 |
| EGFR exon 19 deletion | 45 (7.1) | 4 (6.8) | 41 (7.1) | 0.92 |
| EGFR L858R | 46 (7.2) | 4 (6.8) | 42 (7.3) | 0.89 |
| EGFRm uncommon sensitizing present ** | 23 (3.6) | 5 (8.5) | 18 (3.1) | 0.05 |
| EGFR G709X * | 5 (0.8) | 1 (1.7) | 4 (0.7) | 0.39 |
| EGFR G719X * | 11 (1.7) | 3 (5.1) | 8 (1.4) | 0.07 |
| EGFR S768I * | 4 (0.6) | 1 (1.7) | 3 (0.5) | 0.99 |
| EGFR L861Q/R * | 6 (0.9) | 0 (0.0) | 6 (1.0) | 0.32 |
| EGFR co-mutation | 15 (2.4) | 2 (3.4) | 13 (2.3) | 0.64 |
| EGFRm uncommon non-sensitizing present * | 13 (2.0) | 4 (6.8) | 9 (1.6) | 0.025 |
| EGFR exon 20 insertion * | 12 (1.9) | 4 (6.8) | 8 (1.4) | 0.019 |
| KRAS any mutation | 245 (38.6) | 17 (28.8) | 228 (39.6) | 0.11 |
| KRAS G12C | 113 (17.8) | 8 (13.6) | 105 (18.2) | 0.37 |
| Met present | 20 (3.1) | 3 (5.1) | 17 (3.0) | 0.42 |
| MET exon14 skip | 19 (3.0) | 3 (5.1) | 16 (2.8) | 0.41 |
| TP53 mutation | 323 (50.9) | 27 (45.8) | 296 (51.4) | 0.41 |
| BRAF mutation * | 40 (6.3) | 1 (1.7) | 39 (6.8) | 0.16 |
| ERRB2 mutation | 17 (2.7) | 2 (3.4) | 15 (2.6) | 0.67 |
| PIK3CA mutation | 26 (4.1) | 6 (10.2) | 20 (3.5) | 0.026 |
| FUSION present | 19 (3.0) | 4 (6.8) | 15 (2.6) | 0.09 |
| ALK fusion * | 15 (2.4) | 1 (1.7) | 14 (2.4) | 0.99 |
| RET fusion * | 2 (0.3) | 2 (3.4) | 0 (0.0) | 0.009 |
| ROS1 fusion * | 2 (0.3) | 1 (1.7) | 1 (0.2) | 0.18 |
| Alteration | All Matched n = 212 | Early Stage n = 53 | Late Stage n = 159 | p Value |
|---|---|---|---|---|
| Any alteration mutation or fusion | 199 (93.9) | 50 (94.3) | 149 (93.7) | 0.87 |
| Potential therapeutic target | 106 (50.0) | 29 (54.7) | 77 (48.4) | 0.44 |
| EGFR mutation present | 63 (29.7) | 17 (32.1) | 46 (28.9) | 0.65 |
| EGFRm common sensitizing present | 44 (20.8) | 8 (15.1) | 36 (22.6) | 0.23 |
| EGFR exon 19 deletion | 26 (12.3) | 4 (7.5) | 22 (13.8) | 0.24 |
| EGFR L858R | 18 (8.5) | 4 (7.5) | 14 (8.8) | 0.78 |
| EGFRm uncommon sensitizing present ** | 12 (5.7) | 5 (9.4) | 7 (4.4) | 0.17 |
| EGFR G709X * | 1 (0.5) | 1 (1.9) | 0 (0.0) | - |
| EGFR G719X * | 6 (2.8) | 3 (5.7) | 3 (1.9) | - |
| EGFR S768I * | 1 (0.5) | 1 (1.9) | 0 (0.0) | - |
| EGFR L861Q * | 3 (1.4) | 0 (0.0) | 3 (1.9) | - |
| EGFR co-mutation | 6 (2.8) | 2 (3.8) | 4 (2.5) | 0.64 |
| EGFRm uncommon non-sensitizing present * | 8 (3.8) | 4 (7.5) | 4 (2.5) | - |
| EGFR exon 20 insertion * | 8 (3.8) | 4 (7.5) | 4 (2.5) | - |
| KRAS any mutation | 60 (28.3) | 17 (32.1) | 43 (27) | 0.46 |
| KRAS G12C | 25 (11.8) | 8 (15.1) | 17 (10.7) | 0.40 |
| Met present | 8 (3.8) | 3 (5.7) | 5 (3.1) | 0.40 |
| MET exon14 skip | 8 (3.8) | 3 (5.7) | 5 (3.1) | 0.40 |
| TP53 mutation | 96 (45.3) | 23 (43.4) | 73 (45.9) | 0.75 |
| BRAF mutation * | 16 (7.5) | 1 (1.9) | 15 (9.4) | - |
| ERRB2 mutation | 7 (3.3) | 2 (3.8) | 5 (3.1) | 0.81 |
| PIK3CA mutation | 9 (4.2) | 3 (5.7) | 6 (3.8) | 0.55 |
| FUSION present | 13 (6.1) | 4 (7.5) | 9 (5.7) | 0.62 |
| ALK fusion * | 9 (4.2) | 1 (1.9) | 8 (5) | - |
| RET fusion * | 2 (0.9) | 2 (3.8) | 0 (0.0) | - |
| ROS1 fusion * | 2 (0.9) | 1 (1.9) | 1 (0.6) | - |
| All Patients | Matched Patients | |||||
|---|---|---|---|---|---|---|
| Univariate Regression Analysis | Multiple Regression Analysis | Univariate Regression Analysis | ||||
| Alteration | Odds Ratio (95% CI) Late vs. Early | p Value | Odds Ratio (95% CI) Late vs. Early | p Value | Odds Ratio (95% CI) Late vs. Early | p Value |
| Any alteration mutation or fusion | 1.059 (0.364, 3.083) | 0.92 | 0.993 (0.354, 2.784) | 0.99 | 0.895 (0.238, 3.359) | 0.87 |
| Potential target | 0.766 (0.448, 1.309) | 0.33 | 0.790 (0.439, 1.423) | 0.43 | 0.788 (0.430, 1.445) | 0.44 |
| EGFR mutation present | 0.508 (0.281, 0.919) | 0.025 | 0.656 (0.34, 1.263) | 0.21 | 0.849 (0.420, 1.719) | 0.65 |
| EGFRm common sensitizing present | 1.073 (0.492, 2.343) | 0.86 | 1.571 (0.687, 3.590) | 0.28 | 1.715 (0.711, 4.137) | 0.23 |
| EGFR exon 19 deletion | 1.054 (0.364, 3.052) | 0.92 | 1.489 (0.512, 4.329) | 0.46 | 1.940 (0.643, 5.853) | 0.24 |
| EGFR L858R | 1.081 (0.374, 3.129) | 0.89 | 1.282 (0.454, 3.621) | 0.64 | 1.180 (0.373, 3.731) | 0.78 |
| EGFRm uncommon sensitizing present | 0.348 (0.124, 0.975) | 0.045 | 0.328 (0.121, 0.891) | 0.029 | 0.411 (0.116, 1.460) | 0.17 |
| EGFR G709X * | - | - | - | - | - | - |
| EGFR G719X * | - | - | - | - | - | - |
| EGFR S768I * | - | - | - | - | - | - |
| EGFR L861Q * | - | - | - | - | - | - |
| EGFR co-mutation | 0.658 (0.145, 2.989) | 0.59 | 0.551 (0.138, 2.205) | 0.40 | 0.667 (0.122, 3.640) | 0.64 |
| EGFRm uncommon non-sensitizing present ** | - | - | - | - | - | - |
| EGFR exon 20 insertion * | - | - | - | - | - | - |
| KRAS any mutation | 1.618 (0.899, 2.913) | 0.11 | 1.071 (0.554, 2.070) | 0.84 | 0.766 (0.376, 1.560) | 0.46 |
| KRAS G12C | 1.421 (0.655, 3.083) | 0.37 | 1.099 (0.501, 2.412) | 0.81 | 0.685 (0.283, 1.659) | 0.40 |
| MET present | 0.568 (0.161, 1.997) | 0.38 | 0.505 (0.154, 1.657) | 0.26 | 0.521 (0.113, 2.390) | 0.40 |
| MET exon14 skip | 0.533 (0.151, 1.886) | 0.33 | 0.475 (0.144, 1.569) | 0.22 | 0.521 (0.113, 2.390) | 0.40 |
| TP53 mutation | 1.252 (0.732, 2.144) | 0.41 | 1.241 (0.697, 2.207) | 0.46 | 1.110 (0.589, 2.091) | 0.75 |
| BRAF mutation * | - | - | - | - | - | - |
| ERRB2 mutation | 0.762 (0.170, 3.414) | 0.72 | 0.635 (0.161, 2.507) | 0.52 | 0.795 (0.125, 5.077) | 0.81 |
| PIK3CA mutation | 0.318 (0.122, 0.826) | 0.019 | 0.430 (0.154, 1.201) | 0.11 | 0.642 (0.150, 2.755) | 0.55 |
| FUSION present | 0.368 (0.118, 1.146) | 0.08 | 0.420 (0.138, 1.283) | 0.13 | 0.737 (0.219, 2.485) | 0.62 |
| ALK fusion * | - | - | - | - | - | - |
| RET fusion * | - | - | - | - | - | - |
| ROS1 fusion * | - | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McGuire, A.L.; McConechy, M.K.; Melosky, B.L.; English, J.C.; Choi, J.J.; Peng, D.; Yee, J.; Furman, B.L.S.; Aguirre Hernandez, R.; Feijao, P.; et al. The Clinically Actionable Molecular Profile of Early versus Late-Stage Non-Small Cell Lung Cancer, an Individual Age and Sex Propensity-Matched Pair Analysis. Curr. Oncol. 2022, 29, 2630-2643. https://doi.org/10.3390/curroncol29040215
McGuire AL, McConechy MK, Melosky BL, English JC, Choi JJ, Peng D, Yee J, Furman BLS, Aguirre Hernandez R, Feijao P, et al. The Clinically Actionable Molecular Profile of Early versus Late-Stage Non-Small Cell Lung Cancer, an Individual Age and Sex Propensity-Matched Pair Analysis. Current Oncology. 2022; 29(4):2630-2643. https://doi.org/10.3390/curroncol29040215
Chicago/Turabian StyleMcGuire, Anna L., Melissa K. McConechy, Barb L. Melosky, John C. English, James J. Choi, Defen Peng, John Yee, Benjamin L. S. Furman, Rosalia Aguirre Hernandez, Pedro Feijao, and et al. 2022. "The Clinically Actionable Molecular Profile of Early versus Late-Stage Non-Small Cell Lung Cancer, an Individual Age and Sex Propensity-Matched Pair Analysis" Current Oncology 29, no. 4: 2630-2643. https://doi.org/10.3390/curroncol29040215
APA StyleMcGuire, A. L., McConechy, M. K., Melosky, B. L., English, J. C., Choi, J. J., Peng, D., Yee, J., Furman, B. L. S., Aguirre Hernandez, R., Feijao, P., Mulder, D., Hughesman, C., & Yip, S. (2022). The Clinically Actionable Molecular Profile of Early versus Late-Stage Non-Small Cell Lung Cancer, an Individual Age and Sex Propensity-Matched Pair Analysis. Current Oncology, 29(4), 2630-2643. https://doi.org/10.3390/curroncol29040215






