A Pleiotropic Role of Long Non-Coding RNAs in the Modulation of Wnt/β-Catenin and PI3K/Akt/mTOR Signaling Pathways in Esophageal Squamous Cell Carcinoma: Implication in Chemotherapeutic Drug Response
Abstract
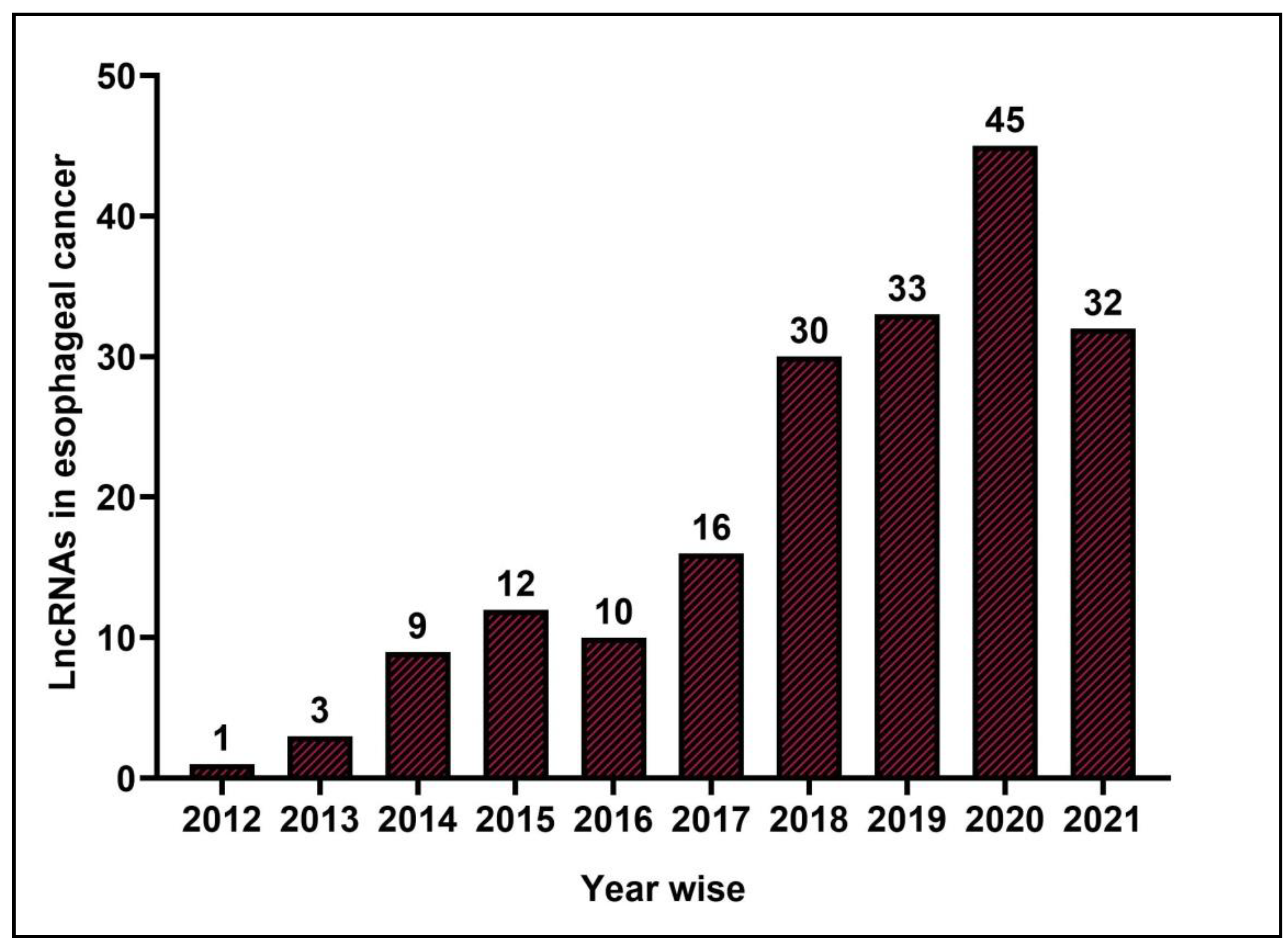
1. Introduction
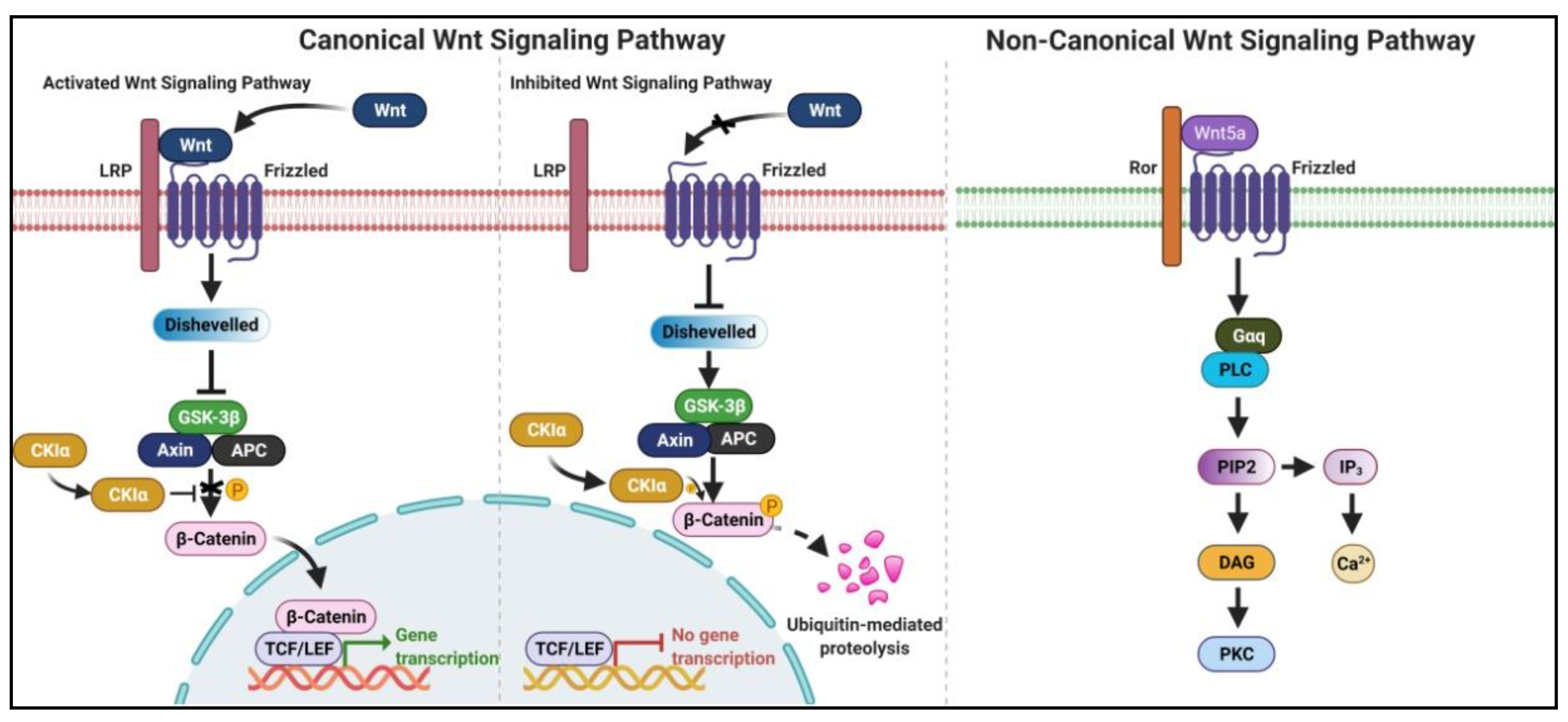
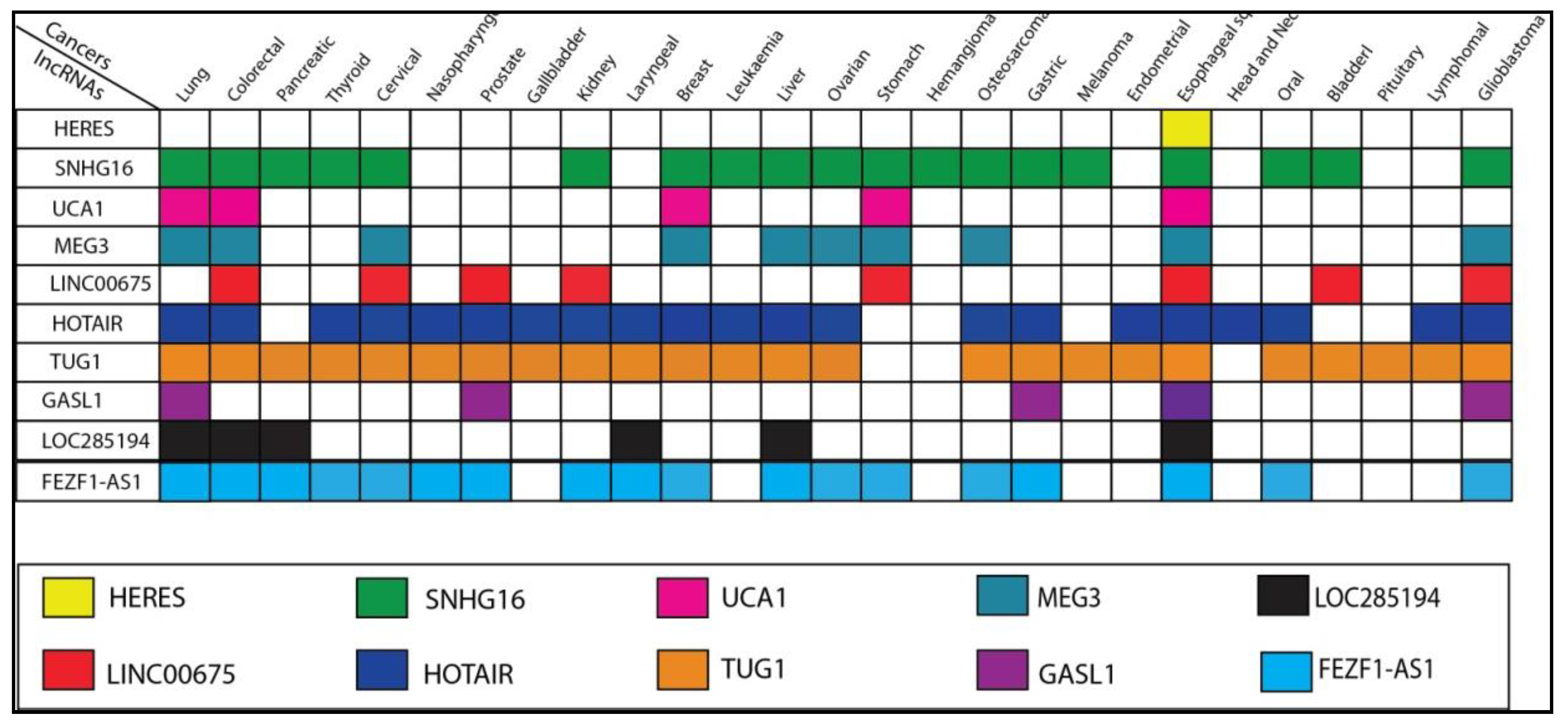
2. Wnt/β-Catenin Signaling Pathway-Related lncRNAs in ESCC
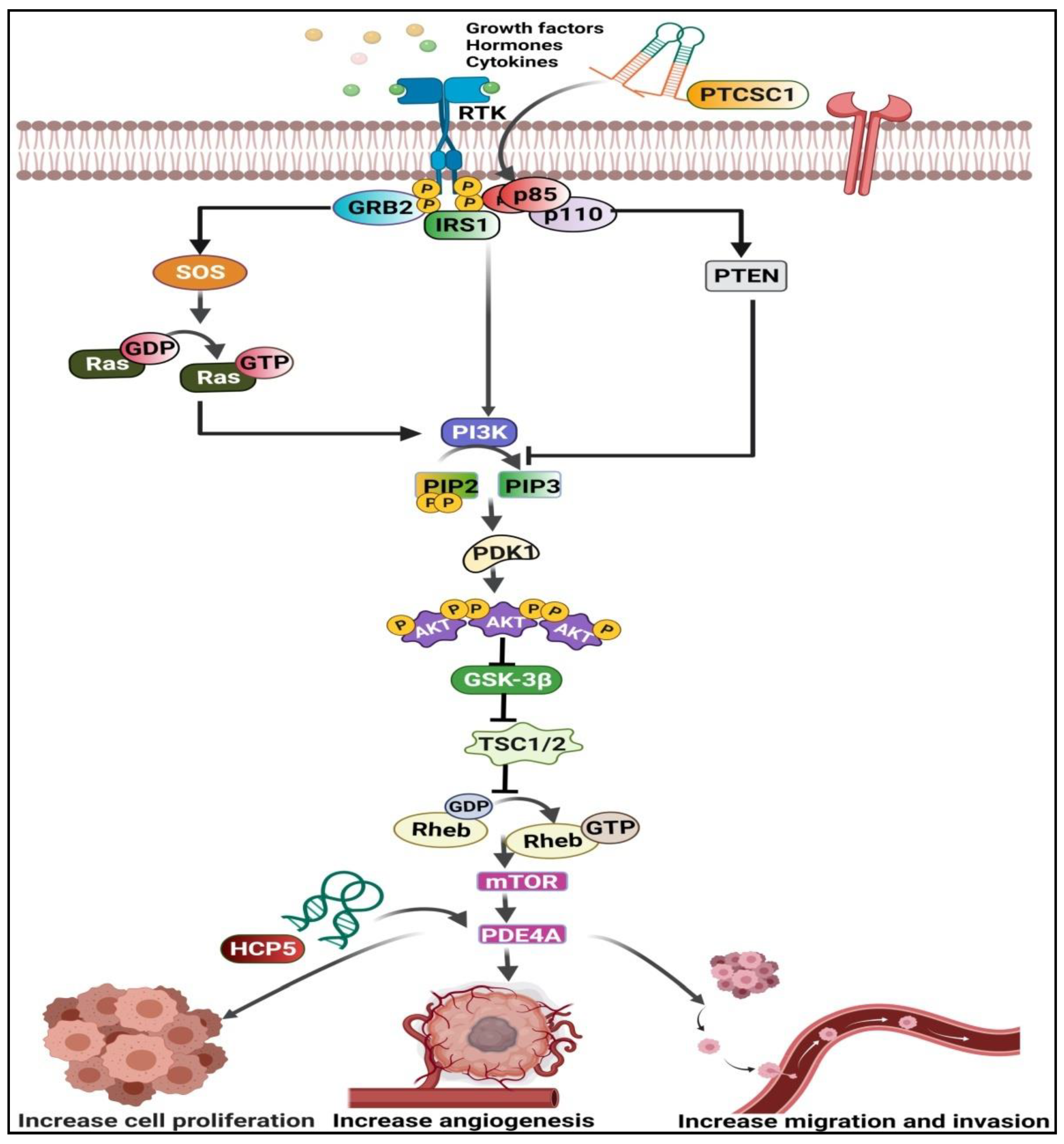
3. PI3K/Akt/mTOR Pathway-Related lncRNAs in ESCC
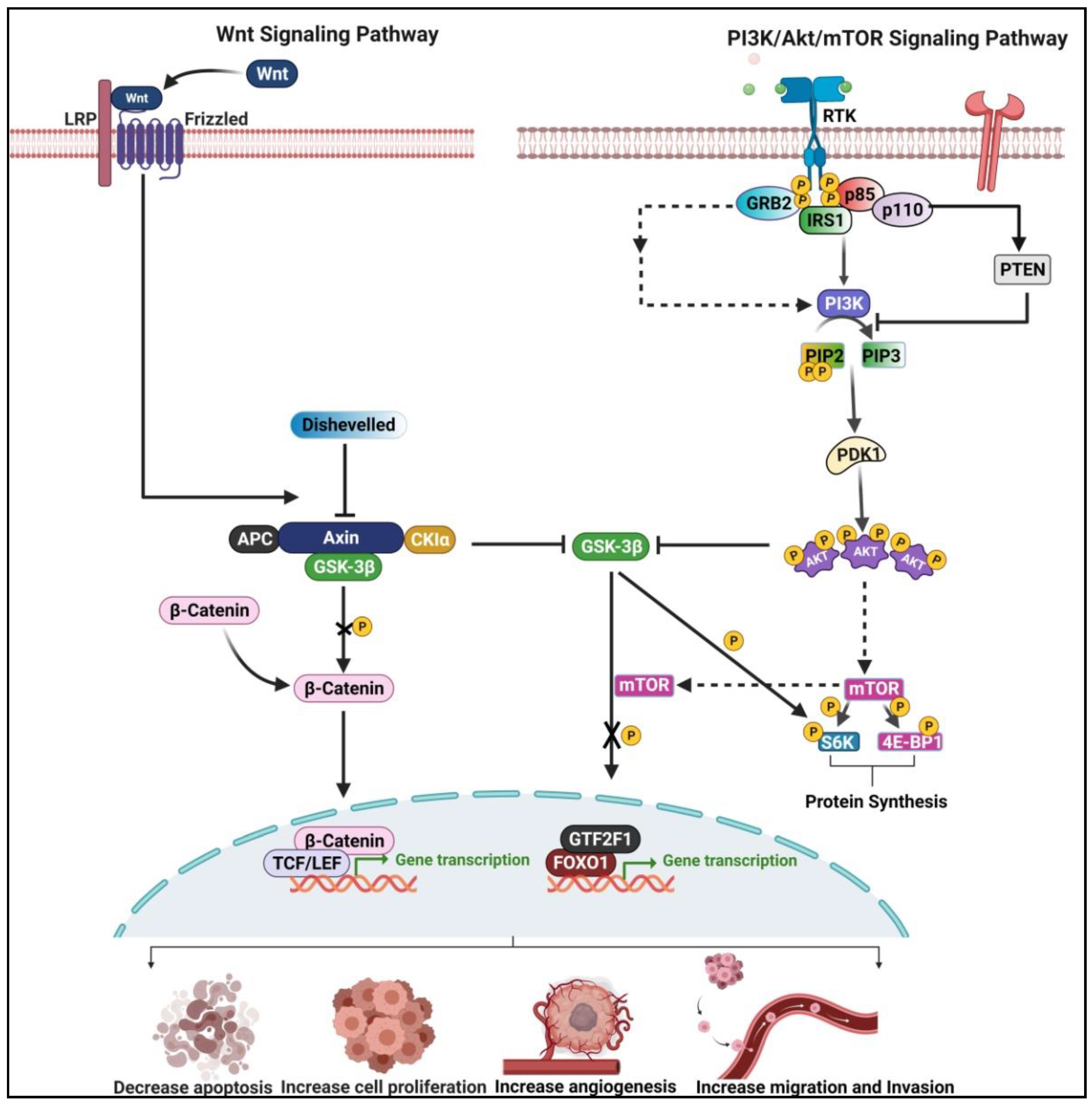
4. Crosstalk between Wnt/β-Catenin and PI3K/Akt/mTOR Pathway in ESCC
5. LncRNAs Regulate the Efficacy of Chemotherapeutic Drugs in ESCC
6. Conclusions and Future Aspects
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, P.; Liu, E.; Xing, C.; Zhu, D.; Zhang, J.; Wang, W.; Jiang, G. Prognostic value of a five-lncRNA signature in esophageal squamous cell carcinoma. Cancer Cell Int. 2020, 20, 386. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Barwal, T.; Acharya, V.; Singh, K.; Rana, M.; Singh, S.; Prakash, H.; Bishayee, A.; Jain, A. Long Non-Coding RNAs as Strategic Molecules to Augment the Radiation Therapy in Esophageal Squamous Cell Carcinoma. Int. J. Mol. Sci. 2020, 21, 6787. [Google Scholar] [CrossRef] [PubMed]
- Si, W.; Shen, J.; Zheng, H.; Fan, W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenet. 2019, 11, 25. [Google Scholar] [CrossRef]
- Wang, W.-T.; Han, C.; Sun, Y.-M.; Chen, T.-Q.; Chen, Y.-Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 1–15. [Google Scholar] [CrossRef]
- Tamang, S.; Acharya, V.; Roy, D.; Sharma, R.; Aryaa, A.; Sharma, U.; Khandelwal, A.; Prakash, H.; Vasquez, K.M.; Jain, A. SNHG12: An LncRNA as a Potential Therapeutic Target and Biomarker for Human Cancer. Front. Oncol. 2019, 9, 901. [Google Scholar] [CrossRef]
- Farooqi, A.A.; Nayyab, S.; Martinelli, C.; Berardi, R.; Katifelis, H.; Gazouli, M.; Cho, W.C. Regulation of Hippo, TGFβ/SMAD, Wnt/β-Catenin, JAK/STAT, and NOTCH by Long Non-Coding RNAs in Pancreatic Cancer. Front. Oncol. 2021, 11, 657965. [Google Scholar] [CrossRef]
- Sharma, U.; Barwal, T.S.; Acharya, V.; Tamang, S.; Vasquez, K.M.; Jain, A. Cancer Susceptibility Candidate 9 (CASC9): A Novel Targetable Long Noncoding RNA in Cancer Treatment. Transl. Oncol. 2020, 13, 100774. [Google Scholar] [CrossRef]
- You, B.-H.; Yoon, J.-H.; Kang, H.; Lee, E.K.; Lee, S.K.; Nam, J.-W. HERES, a lncRNA that regulates canonical and noncanonical Wnt signaling pathways via interaction with EZH2. Proc. Natl. Acad. Sci. USA 2019, 116, 24620–24629. [Google Scholar] [CrossRef]
- Han, G.H.; Lu, K.J.; Wang, P.; Ye, J.; Ye, Y.Y.; Huang, J.X. LncRNA SNHG16 predicts poor prognosis in ESCC and promotes cell proliferation and invasion by regulating Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3795–3803. [Google Scholar]
- Wang, X.; Gao, Z.; Liao, J.; Shang, M.; Li, X.; Yin, L.; Pu, Y.; Liu, R. lncRNA UCA1 inhibits esophageal squamous-cell carcinoma growth by regulating the Wnt signaling pathway. J. Toxicol. Environ. Health Part A 2016, 79, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, T.-F.; Han, X.-W.; Yuan, H.-F. Downregulated MEG3 contributes to tumour progression and poor prognosis in oesophagal squamous cell carcinoma by interacting with miR-4261, downregulating DKK2 and activating the Wnt/β-catenin signalling. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.B.; Shan, A.J.; Lv, W.; Wang, J.; Xu, J.Z. Long non-coding RNA LINC00675 inhibits tumorigenesis and EMT via repressing Wnt/beta-catenin signaling in esophageal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8288–8297. [Google Scholar] [PubMed]
- Ge, X.-S.; Ma, H.-J.; Zheng, X.-H.; Ruan, H.-L.; Liao, X.; Xue, W.-Q.; Chen, Y.-B.; Zhang, Y.; Jia, W.-H. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013, 104, 1675–1682. [Google Scholar] [CrossRef]
- Tang, Y.; Yang, P.; Zhu, Y.; Su, Y. LncRNA TUG1 contributes to ESCC progression via regulating miR-148a-3p/MCL-1/Wnt/beta-catenin axis in vitro. Thorac. Cancer 2020, 11, 82–94. [Google Scholar] [CrossRef]
- Ren, Y.; Guo, T.; Xu, J.; Liu, Y.; Huang, J. The novel target of esophageal squamous cell carcinoma: lncRNA GASL1 regulates cell migration, invasion and cell cycle stagnation by inactivating the Wnt3a/β-catenin signaling. Pathol.-Res. Pract. 2020, 217, 153289. [Google Scholar] [CrossRef]
- Zhong, B.; Wang, Q.; He, J.; Xiong, Y.; Cao, J. LncRNA LOC285194 modulates gastric carcinoma progression through activating Wnt/beta-catenin signaling pathway. Cancer Med. 2020, 9, 2181–2189. [Google Scholar] [CrossRef]
- Fu, X.; Cui, G.; Liu, S.; Zhao, S. Linc01014 regulates gefitinib resistance in oesophagus cancer via EGFR-PI3K-AKT-mTOR signalling pathway. J. Cell Mol. Med. 2020, 24, 1670–1675. [Google Scholar] [CrossRef]
- Xu, J.; Ma, J.; Guan, B.; Li, J.; Wang, Y.; Hu, S. LncRNA HCP5 promotes malignant cell behaviors in esophageal squamous cell carcinoma via the PI3K/AKT/mTOR signaling. Cell Cycle 2021, 20, 1374–1388. [Google Scholar] [CrossRef]
- Liu, T.; Liang, X.; Yang, S.; Sun, Y. Long noncoding RNA PTCSC1 drives esophageal squamous cell carcinoma progression through activating Akt signaling. Exp. Mol. Pathol. 2020, 117, 104543. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, S.; Yin, J.; Shen, Y.-X.; Wang, H.; Chen, X.-S.; Tang, H. LINC01535 promotes proliferation and inhibits apoptosis in esophageal squamous cell cancer by activating the JAK/STAT3 pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3694–3700. [Google Scholar] [PubMed]
- Cao, Y.; Wang, X.; Chen, L. [Knockdown of Fez family zinc finger protein 1 antisense ribonucleic acid 1 (FEZF1-AS1) inhibits invasion and migration of esophageal squamous cell carcinoma cells by blocking JAK2/STAT3 pathway]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2020, 36, 317–324. [Google Scholar] [PubMed]
- Tang, J.; Xu, H.; Liu, Q.; Zheng, J.; Pan, C.; Li, Z.; Wen, W.; Wang, J.; Zhu, Q.; Wang, Z.; et al. LncRNA LOC146880 promotes esophageal squamous cell carcinoma progression via miR-328-5p/FSCN1/MAPK axis. Aging 2021, 13, 14198–14218. [Google Scholar] [CrossRef]
- Sharma, U.; Barwal, T.S.; Khandelwal, A.; Rana, M.K.; Rana, A.P.S.; Singh, K.; Jain, A. Circulating Long Non-Coding RNAs LINC00324 and LOC100507053 as Potential Liquid Biopsy Markers for Esophageal Squamous Cell Carcinoma: A Pilot Study. Front. Oncol. 2022, 12. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Chen, Z.; Li, Y.; Wang, J.; Zhang, Z.; Che, Y.; Huang, J.; Sun, S.; Mao, S.; Lei, Y.; et al. TGF-beta-induced NKILA inhibits ESCC cell migration and invasion through NF-kappaB/MMP14 signaling. J. Mol. Med. 2018, 96, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Sun, K.; Chu, J.; Qu, Y.; Zhao, X.; Yin, H.; Ming, L.; Wan, J.; He, F. Long non-coding RNA FTH1P3 regulated metastasis and invasion of esophageal squamous cell carcinoma through SP1/NF-kB pathway. Biomed. Pharmacother. 2018, 106, 1570–1577. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, X.; Wang, Z.; Zhang, X.; Liu, S.; Liu, G. Downregulation of SNHG1 suppresses cell proliferation and invasion by regulating Notch signaling pathway in esophageal squamous cell cancer. Cancer Biomark. 2017, 21, 89–96. [Google Scholar] [CrossRef]
- Chen, M.; Xia, Z.; Chen, C.; Hu, W.; Yuan, Y. LncRNA MALAT1 promotes epithelial-to-mesenchymal transition of esophageal cancer through Ezh2-Notch1 signaling pathway. Anti-Cancer Drugs 2018, 29, 767–773. [Google Scholar] [CrossRef]
- Aggarwal, V.; Tuli, H.S.; Varol, M.; Tuorkey, M.; Sak, K.; Parashar, N.C.; Barwal, T.S.; Sharma, U.; Iqubal, A.; Parashar, G.; et al. NOTCH signaling: Journey of an evolutionarily conserved pathway in driving tumor progression and its modulation as a therapeutic target. Crit. Rev. Oncol. 2021, 164, 103403. [Google Scholar] [CrossRef]
- Zhang, X.; Xie, K.; Zhou, H.; Wu, Y.; Li, C.; Liu, Y.; Liu, Z.; Xu, Q.; Liu, S.; Xiao, D.; et al. Role of non-coding RNAs and RNA modifiers in cancer therapy resistance. Mol. Cancer 2020, 19, 1–26. [Google Scholar] [CrossRef]
- Yu, X.; Huang, M.; Yang, G. Long non-coding RNA BANCR promotes proliferation, invasion and migration in esophageal squamous cell carcinoma cells via the Raf/MEK/ERK signaling pathway. Mol. Med. Rep. 2021, 23, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, A.; Xu, R.; Zhou, F.; Hao, A.; Yang, H.; Yang, P. NR2F1-induced NR2F1-AS1 promotes esophageal squamous cell carcinoma progression via activating Hedgehog signaling pathway. Biochem. Biophys. Res. Commun. 2019, 519, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Miao, Y.; Shang, M.; Liu, M.; Liu, R.; Pan, E.; Pu, Y.; Yin, L. LincRNA-p21 leads to G1 arrest by p53 pathway in esophageal squamous cell carcinoma. Cancer Manag. Res. 2019, 11, 6201–6214. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zhang, Z.; Mao, C.; Zhou, Y.; Yu, L.; Yin, Y.; Wu, S.; Mou, X.; Zhu, Y. ANRIL inhibits p15(INK4b) through the TGFbeta1 signaling pathway in human esophageal squamous cell carcinoma. Cell Immunol. 2014, 289, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Yang, K.; Wan, L.; He, J.; Qi, L.; Wang, W.; Lu, Q.; Li, Z. The crosstalk between lncRNAs and the Hippo signalling pathway in cancer progression. Cell Prolif. 2020, 53. [Google Scholar] [CrossRef]
- Fu, P.-F.; Zheng, X.; Fan, X.; Lin, A.-F. Role of cytoplasmic lncRNAs in regulating cancer signaling pathways. J. Zhejiang Univ. Sci. B 2019, 20, 1–8. [Google Scholar] [CrossRef]
- Pai, S.G.; Carneiro, B.A.; Mota, J.M.; Costa, R.; Leite, C.A.; Barroso-Sousa, R.; Kaplan, J.B.; Chae, Y.K.; Giles, F.J. Wnt/beta-catenin pathway: Modulating anticancer immune response. J. Hematol. Oncol. 2017, 10, 1–12. [Google Scholar] [CrossRef]
- Yang, L.; Ye, Y.; Chu, J.; Jia, J.; Qu, Y.; Sun, T.; Yin, H.; Ming, L.; Wan, J.; He, F. Long noncoding RNA FEZF1-AS1 promotes the motility of esophageal squamous cell carcinoma through Wnt/beta-catenin pathway. Cancer Manag. Res. 2019, 11, 4425–4435. [Google Scholar] [CrossRef]
- Ruggero, D.; Sonenberg, N. The Akt of translational control. Oncogene 2005, 24, 7426–7434. [Google Scholar] [CrossRef]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 1–28. [Google Scholar] [CrossRef]
- Porta, C.; Paglino, C.; Mosca, A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Front. Oncol. 2014, 4, 64. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef] [PubMed]
- Myers, A.P.; Cantley, L.C. Targeting a Common Collaborator in Cancer Development. Sci. Transl. Med. 2010, 2, 48ps45. [Google Scholar] [CrossRef] [PubMed]
- Morgan, T.M.; Koreckij, T.D.; Corey, E. Targeted Therapy for Advanced Prostate Cancer: Inhibition of the PI3K/Akt/mTOR Pathway. Curr. Cancer Drug Targets 2009, 9, 237–249. [Google Scholar] [CrossRef]
- Sarris, E.G.; Saif, M.W.; Syrigos, K.N. The Biological Role of PI3K Pathway in Lung Cancer. Pharmaceuticals 2012, 5, 1236–1264. [Google Scholar] [CrossRef] [PubMed]
- Prossomariti, A.; Piazzi, G.; Alquati, C.; Ricciardiello, L. Are Wnt/beta-Catenin and PI3K/AKT/mTORC1 Distinct Pathways in Colorectal Cancer? Cell Mol. Gastroenterol. Hepatol. 2020, 10, 491–506. [Google Scholar] [CrossRef]
- Kaidanovich-Beilin, O.; Woodgett, J.R. GSK-3: Functional Insights from Cell Biology and Animal Models. Front. Mol. Neurosci. 2011, 4, 40. [Google Scholar] [CrossRef]
- Inoki, K.; Ouyang, H.; Zhu, T.; Lindvall, C.; Wang, Y.; Zhang, X.; Yang, Q.; Bennett, C.; Harada, Y.; Stankunas, K.; et al. TSC2 Integrates Wnt and Energy Signals via a Coordinated Phosphorylation by AMPK and GSK3 to Regulate Cell Growth. Cell 2006, 126, 955–968. [Google Scholar] [CrossRef]
- He, L.; Gomes, A.P.; Wang, X.; Yoon, S.O.; Lee, G.; Nagiec, M.J.; Cho, S.; Chavez, A.; Islam, T.; Yu, Y.; et al. mTORC1 Promotes Metabolic Reprogramming by the Suppression of GSK3-Dependent Foxk1 Phosphorylation. Mol. Cell 2018, 70, 949–960.e4. [Google Scholar] [CrossRef]
- He, L.; Fei, D.L.; Nagiec, M.J.; Mutvei, A.P.; Lamprakis, A.; Kim, B.Y.; Blenis, J. Regulation of GSK3 cellular location by FRAT modulates mTORC1-dependent cell growth and sensitivity to rapamycin. Proc. Natl. Acad. Sci. USA 2019, 116, 19523–19529. [Google Scholar] [CrossRef]
- Yang, C.; Shen, S.; Zheng, X.; Ye, K.; Ge, H.; Sun, Y.; Lu, Y. Long non-coding RNA LINC00337 induces autophagy and chemoresistance to cisplatin in esophageal squamous cell carcinoma cells via upregulation of TPX2 by recruiting E2F4. FASEB J. 2020, 34, 6055–6069. [Google Scholar] [CrossRef] [PubMed]
- Corrà, F.; Agnoletto, C.; Minotti, L.; Baldassari, F.; Volinia, S. The Network of Non-coding RNAs in Cancer Drug Resistance. Front. Oncol. 2018, 8, 327. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.-S.; Zhou, X.-L.; Wang, X.-W.; Wu, Q.-Q.; Yang, T.-X.; Lv, J.; Yang, J.-S.; Zhu, B.; Cao, X.-F. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J. Transl. Med. 2014, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.W.; Jia, Y.X.; Zhang, W.J.; Song, L.J.; Gao, M.; Li, M.J.; Zhao, R.H.; Li, J.; Zhong, Y.L.; Sun, Q.Z.; et al. LncRNA-TUSC7/miR-224 affected chemotherapy resistance of esophageal squamous cell carcinoma by competitively regulating DESC1. J. Exp. Clin. Cancer Res. 2018, 37, 56. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, W.; Li, G.; Sun, C.; Ren, Z.; Sheng, H.; Gao, H.; Wang, C.; Yu, H. High TUG1 expression is associated with chemotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. Cancer Chemother. Pharmacol. 2016, 78, 333–339. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiong, R.; Li, C.; Xu, M.; Guo, M. LncRNA TUG1 promotes cisplatin resistance in esophageal squamous cell carcinoma cells by regulating Nrf2. Acta Biochim. Biophys. Sin. 2019, 51, 826–833. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, W.; Zhu, W.; Yu, C.; Tao, G.; Wu, Q.; Song, Y.; Pan, P.; Tong, Y. High expression of long non-coding RNA AFAP1-AS1 predicts chemoradioresistance and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Mol. Carcinog. 2016, 55, 2095–2105. [Google Scholar] [CrossRef]
- Hu, M.; Zhang, Q.; Tian, X.H.; Wang, J.L.; Niu, Y.X.; Li, G. lncRNA CCAT1 is a biomarker for the proliferation and drug resistance of esophageal cancer via the miR-143/PLK1/BUBR1 axis. Mol. Carcinog. 2019, 58, 2207–2217. [Google Scholar] [CrossRef]
- Chen, J.-L.; Lin, Z.-X.; Qin, Y.-S.; She, Y.-Q.; Chen, Y.; Chen, C.; Qiu, G.-D.; Zheng, J.-T.; Chen, Z.-L.; Zhang, S.-Y. Overexpression of long noncoding RNA LINC01419 in esophageal squamous cell carcinoma and its relation to the sensitivity to 5-fluorouracil by mediating GSTP1 methylation. Ther. Adv. Med Oncol. 2019, 11, 1758835919838958. [Google Scholar] [CrossRef]
- Xue, W.; Shen, Z.; Li, L.; Zheng, Y.; Yan, D.; Kan, Q.; Zhao, J. Long non-coding RNAs MACC1-AS1 and FOXD2-AS1 mediate NSD2-induced cisplatin resistance in esophageal squamous cell carcinoma. Mol. Ther.-Nucleic Acids 2021, 23, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, J.; Luo, X.; Zeng, M.; Xu, L.; Zhang, Q.; Liu, H.; Guo, J.; Xu, L. Overexpression of the Long Noncoding RNA FOXD2-AS1 Promotes Cisplatin Resistance in Esophageal Squamous Cell Carcinoma Through the miR-195/Akt/mTOR Axis. Oncol. Res. 2020, 28, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Ren, M.; Li, Y.; Fu, Y.; Deng, M.; Li, C. Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J. Exp. Clin. Cancer Res. 2018, 37, 171. [Google Scholar] [CrossRef] [PubMed]
- Kalhori, M.; Khodayari, H.; Khodayari, S.; Vesovic, M.; Jackson, G.; Farzaei, M.; Bishayee, A. Regulation of Long Non-Coding RNAs by Plant Secondary Metabolites: A Novel Anticancer Therapeutic Approach. Cancers 2021, 13, 1274. [Google Scholar] [CrossRef]
- Tuli, H.S.; Mittal, S.; Aggarwal, D.; Parashar, G.; Parashar, N.C.; Upadhyay, S.K.; Barwal, T.S.; Jain, A.; Kaur, G.; Savla, R.; et al. Path of Silibinin from diet to medicine: A dietary polyphenolic flavonoid having potential anti-cancer therapeutic significance. Semin. Cancer Biol. 2020, 73, 196–218. [Google Scholar] [CrossRef]
- Tuli, H.S.; Tuorkey, M.J.; Thakral, F.; Sak, K.; Kumar, M.; Sharma, A.K.; Sharma, U.; Jain, A.; Aggarwal, V.; Bishayee, A. Molecular Mechanisms of Action of Genistein in Cancer: Recent Advances. Front. Pharmacol. 2019, 10, 1336. [Google Scholar] [CrossRef]
- Chen, F.-J.; Sun, M.; Li, S.-Q.; Wu, Q.-Q.; Ji, L.; Liu, Z.-L.; Zhou, G.-Z.; Cao, G.; Jin, L.; Xie, H.-W.; et al. Upregulation of the long non-coding rna hotair promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol. Carcinog. 2012, 52, 908–915. [Google Scholar] [CrossRef]
- Wu, H.; Zheng, J.; Deng, J.; Hu, M.; You, Y.; Li, N.; Li, W.; Lu, J.; Zhou, Y. A genetic polymorphism in lincRNA-uc003opf.1 is associated with susceptibility to esophageal squamous cell carcinoma in Chinese populations. Carcinogenesis 2013, 34, 2908–2917. [Google Scholar] [CrossRef]
- Pan, F.; Yao, J.; Chen, Y.; Zhou, C.; Geng, P.; Mao, H.; Fang, X. A novel long non-coding RNA FOXCUT and mRNA FOXC1 pair promote progression and predict poor prognosis in esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 2838–2849. [Google Scholar]
- Gao, T.; He, B.; Pan, Y.; Gu, L.; Chen, L.; Nie, Z.; Xu, Y.; Li, R.; Wang, S. H19 DMR methylation correlates to the progression of esophageal squamous cell carcinoma through IGF2 imprinting pathway. Clin. Transl. Oncol. 2013, 16, 410–417. [Google Scholar] [CrossRef]
- Li, W.; Zheng, J.; Deng, J.; You, Y.; Wu, H.; Li, N.; Lu, J.; Zhou, Y. Increased Levels of the Long Intergenic Non–Protein Coding RNA POU3F3 Promote DNA Methylation in Esophageal Squamous Cell Carcinoma Cells. Gastroenterology 2014, 146, 1714–1726.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-M.; Wu, Q.-Q.; Li, S.-Q.; Chen, F.-J.; Tuo, L.; Xie, H.-W.; Tong, Y.-S.; Ji, L.; Zhou, G.-Z.; Cao, G.; et al. Upregulation of the Long Non-coding RNA PlncRNA-1 Promotes Esophageal Squamous Carcinoma Cell Proliferation and Correlates with Advanced Clinical Stage. Am. J. Dig. Dis. 2013, 59, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Shahryari, A.; Rafiee, M.R.; Fouani, Y.; Oliae, N.A.; Samaei, N.M.; Shafiee, M.; Semnani, S.; Vasei, M.; Mowla, S.J. Two Novel Splice Variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are Coupregulated with SOX2 and OCT4 in Esophageal Squamous Cell Carcinoma. Stem Cells 2013, 32, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.-W.; Wu, Q.-Q.; Zhu, B.; Chen, F.-J.; Ji, L.; Li, S.-Q.; Wang, C.-M.; Tong, Y.-S.; Tuo, L.; Wu, M.; et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumor Biol. 2014, 35, 7743–7754. [Google Scholar] [CrossRef]
- Li, J.-Y.; Ma, X.; Zhang, C.-B. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 7938–7944. [Google Scholar]
- Gao, T.; He, B.; Pan, Y.; Xu, Y.; Li, R.; Deng, Q.; Sun, H.; Wang, S. Long non-coding RNA 91H contributes to the occurrence and progression of esophageal squamous cell carcinoma by inhibiting IGF2 expression. Mol. Carcinog. 2014, 54, 359–367. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, H.; Hu, Z.; Peng, J.; Jiang, Z.; Song, T.; Wu, B.; Yue, J.; Zhou, R.; Xie, R.; et al. Targeting WISP1 to sensitize esophageal squamous cell carcinoma to irradiation. Oncotarget 2015, 6, 6218–6234. [Google Scholar] [CrossRef][Green Version]
- Zhang, X.; Xu, Y.; He, C.; Guo, X.; Zhang, J.; He, C.; Zhang, L.; Kong, M.; Chen, B.; Zhu, C. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J. Surg. Oncol. 2015, 111, 834–839. [Google Scholar] [CrossRef]
- Wu, H.; Zheng, J.; Deng, J.; Zhang, L.; Li, N.; Li, W.; Li, F.; Lu, J.; Zhou, Y. LincRNA-uc002yug.2 involves in alternative splicing of RUNX1 and serves as a predictor for esophageal cancer and prognosis. Oncogene 2014, 34, 4723–4734. [Google Scholar] [CrossRef]
- Kang, M.; Sang, Y.; Gu, H.; Zheng, L.; Wang, L.; Liu, C.; Shi, Y.; Shao, A.; Ding, G.; Chen, S.; et al. Long noncoding RNAs POLR2E rs3787016 C/T and HULC rs7763881 A/C polymorphisms are associated with decreased risk of esophageal cancer. Tumor Biol. 2015, 36, 6401–6408. [Google Scholar] [CrossRef]
- Hu, L.; Wu, Y.; Tan, D.; Meng, H.; Wang, K.; Bai, Y.; Yang, K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 7. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kong, J.; Ma, Z.; Gao, S.; Feng, X. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am. J. Cancer Res. 2015, 5, 2808–2815. [Google Scholar] [PubMed]
- Shi, W.-H.; Wu, Q.-Q.; Li, S.-Q.; Yang, T.-X.; Liu, Z.-H.; Tong, Y.-S.; Tuo, L.; Wang, S.; Cao, X.-F. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumor Biol. 2015, 36, 2501–2507. [Google Scholar] [CrossRef]
- Tong-Xin, Y.; Wang, X.-W.; Zhou, X.-L.; Liu, Z.-H.; Yang, T.-X.; Shi, W.-H.; Xie, H.-W.; Lv, J.; Wu, Q.-Q.; Cao, X.-F. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol. Cancer 2015, 14, 1–13. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, X.; Hu, L.; Tao, H.; Guan, X.; Zhang, K.; Bai, Y.; Yang, K. Copy number loss of variation_91720 in PIK3CA predicts risk of esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 14479–14485. [Google Scholar] [PubMed]
- Xu, Y.; Wang, J.; Qiu, M.; Xu, L.; Li, M.; Jiang, F.; Yin, R.; Xu, L. Upregulation of the long noncoding RNA TUG1 promotes proliferation and migration of esophageal squamous cell carcinoma. Tumor Biol. 2014, 36, 1643–1651. [Google Scholar] [CrossRef]
- Wang, Y.-L.; Bai, Y.; Yao, W.-J.; Guo, L.; Wang, Z.-M. Expression of long non-coding RNA ZEB1-AS1 in esophageal squamous cell carcinoma and its correlation with tumor progression and patient survival. Int. J. Clin. Exp. Pathol. 2015, 8, 11871–11876. [Google Scholar]
- Luo, H.; Huang, M.; Guo, J.; Fan, R.; Xia, X.; He, J.; Chen, X. AFAP1-AS1 is upregulated and promotes esophageal squamous cell carcinoma cell proliferation and inhibits cell apoptosis. Cancer Med. 2016, 5, 2879–2885. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, T.; Xu, Z.; Cao, X. Upregulation of the long non-coding RNA BANCR correlates with tumor progression and poor prognosis in esophageal squamous cell carcinoma. Biomed. Pharmacother. 2016, 82, 406–412. [Google Scholar] [CrossRef]
- Lu, C.; Yang, L.; Chen, H.; Shan, Z. Upregulated long non-coding RNA BC032469 enhances carcinogenesis and metastasis of esophageal squamous cell carcinoma through regulating hTERT expression. Tumor Biol. 2016, 37, 16065–16075. [Google Scholar] [CrossRef]
- Cao, X.-G.; Zhao, R.-H.; Zhu, C.-H.; Li, X.-K.; Cao, W.; Zong, H.; Hu, H.-Y. BC200 LncRNA a potential predictive marker of poor prognosis in esophageal squamous cell carcinoma patients. Onco Targets Ther. 2016, 9, 2221–2226. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Mao, W.; Bao, Y.; Zhang, M.; Su, X.; Xu, X. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016, 5, 2442–2447. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Han, H.; Li, Y.; Zhang, Q.; Mo, K.; Chen, S. Upregulation of long noncoding RNA HOTTIP promotes metastasis of esophageal squamous cell carcinoma via induction of EMT. Oncotarget 2016, 7, 84480–84485. [Google Scholar] [CrossRef] [PubMed]
- Sahebi, R.; Malakootian, M.; Balalaee, B.; Shahryari, A.; Khoshnia, M.; Abbaszadegan, M.R.; Moradi, A.; Mowla, S.J. Linc-ROR and its spliced variants 2 and 4 are significantly up-regulated in esophageal squamous cell carcinoma. Iran. J. Basic Med. Sci. 2016, 19, 1131–1135. [Google Scholar] [CrossRef]
- Guo, W.; Dong, Z.; Shi, Y.; Liu, S.; Liang, J.; Guo, Y.; Guo, X.; Shen, S.; Shan, B. Aberrant methylation-mediated downregulation of long noncoding RNA LOC100130476 correlates with malignant progression of esophageal squamous cell carcinoma. Dig. Liver Dis. 2016, 48, 961–969. [Google Scholar] [CrossRef]
- Lv, D.; Sun, R.; Yu, Q.; Zhang, X. The long non-coding RNA maternally expressed gene 3 activates p53 and is downregulated in esophageal squamous cell cancer. Tumor Biol. 2016, 37, 16259–16267. [Google Scholar] [CrossRef]
- Shafiee, M.; Aleyasin, S.A.; Vasei, M.; Semnani, S.S.; Mowla, S.J. Down-Regulatory Effects of miR-211 on Long Non-Coding RNA SOX2OT and SOX2 Genes in Esophageal Squamous Cell Carcinoma. Cell J. 2016, 17, 593–600. [Google Scholar] [CrossRef]
- Li, Z.; Wu, X.; Gu, L.; Shen, Q.; Luo, W.; Deng, C.; Zhou, Q.; Chen, X.; Li, Y.; Lim, Z.; et al. Long non-coding RNA ATB promotes malignancy of esophageal squamous cell carcinoma by regulating miR-200b/Kindlin-2 axis. Cell Death Dis. 2017, 8, e2888. [Google Scholar] [CrossRef]
- Zhang, E.; Han, L.; Yin, D.; He, X.; Hong, L.; Si, X.; Qiu, M.; Xu, T.; De, W.; Xu, L.; et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2016, 45, 3086–3101. [Google Scholar] [CrossRef]
- Cui, Y.; Wu, W.; Lv, P.; Zhang, J.; Bai, B.; Cao, W. Down-regulation of long non-coding RNA ESCCAL_1 inhibits tumor growth of esophageal squamous cell carcinoma in a xenograft mouse model. Oncotarget 2017, 9, 783–790. [Google Scholar] [CrossRef]
- Liu, H.; Zhen, Q.; Fan, Y. LncRNA GHET1 Promotes Esophageal Squamous Cell Carcinoma Cells Proliferation and Invasion via Induction of EMT. Int. J. Biol. Markers 2017, 32, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, W.; Gao, X.; Zhang, D.; Li, Y. HNF1A?AS1 promotes growth and metastasis of esophageal squamous cell carcinoma by sponging miR?214 to upregulate the expression of SOX-4. Int. J. Oncol. 2017, 51, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Da, C.; Zhan, Y.; Li, Y.; Tan, Y.; Li, R.; Wang, R. The expression and significance of HOX transcript antisense RNA and epithelial-mesenchymal transition-related factors in esophageal squamous cell carcinoma. Mol. Med. Rep. 2017, 15, 1853–1862. [Google Scholar] [CrossRef]
- Liang, Y.; Wu, Y.; Chen, X.; Zhang, S.; Wang, K.; Guan, X.; Yang, K.; Li, J.; Bai, Y. A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci. Rep. 2017, 37, BSR20171019. [Google Scholar] [CrossRef]
- Han, L.; Liu, S.; Liang, J.; Guo, Y.; Shen, S.; Guo, X.; Dong, Z.; Guo, W. A genetic polymorphism at miR-526b binding-site in the lincRNA-NR_024015 exon confers risk of esophageal squamous cell carcinoma in a population of North China. Mol. Carcinog. 2017, 56, 960–971. [Google Scholar] [CrossRef]
- Dong, Z.; Zhang, A.; Liu, S.; Lu, F.; Guo, Y.; Zhang, G.; Xu, F.; Shi, Y.; Shen, S.; Liang, J.; et al. Aberrant Methylation-Mediated Silencing of lncRNA MEG3 Functions as a ceRNA in Esophageal Cancer. Mol. Cancer Res. 2017, 15, 800–810. [Google Scholar] [CrossRef]
- Ren, Z.-P.; Chu, X.-Y.; Xue, Z.-Q.; Zhang, L.-B.; Wen, J.-X.; Deng, J.-Q.; Hou, X.-B. Down-regulation of lncRNA MIR31HG correlated with aggressive clinicopathological features and unfavorable prognosis in esophageal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3866–3870. [Google Scholar]
- Wu, X.; Lim, Z.-F.; Li, Z.; Gu, L.; Ma, W.; Zhou, Q.; Su, H.; Wang, X.; Yang, X.; Zhang, Z. NORAD Expression Is Associated with Adverse Prognosis in Esophageal Squamous Cell Carcinoma. Oncol. Res. Treat. 2017, 40, 370–374. [Google Scholar] [CrossRef]
- Feng, F.; Qiu, B.; Zang, R.; Song, P.; Gao, S. Pseudogene PHBP1 promotes esophageal squamous cell carcinoma proliferation by increasing its cognate gene PHB expression. Oncotarget 2017, 8, 29091–29100. [Google Scholar] [CrossRef]
- Li, P.-D.; Hu, J.-L.; Ma, C.; Ma, H.; Yao, J.; Chen, L.-L.; Chen, J.; Cheng, T.-T.; Yang, K.-Y.; Wu, G.; et al. Upregulation of the long non-coding RNA PVT1 promotes esophageal squamous cell carcinoma progression by acting as a molecular sponge of miR-203 and LASP1. Oncotarget 2017, 8, 34164–34176. [Google Scholar] [CrossRef]
- Yao, G.-L.; Pan, C.-F.; Xu, H.; Wei, K.; Liu, B.; Zhai, R.; Chen, Y.-J. Long noncoding RNA RP11-766N7.4 functions as a tumor suppressor by regulating epithelial-mesenchymal transition in esophageal squamous cell carcinoma. Biomed. Pharmacother. 2017, 88, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Chen, J.; Song, H.; Chen, L.-B. SNHG16/miR-140-5p axis promotes esophagus cancer cell proliferation, migration and EMT formation through regulating ZEB1. Oncotarget 2017, 9, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Dinglin, X.; Wang, X.; Luo, W.; Shen, Q.; Li, Y.; Gu, L.; Zhou, Q.; Zhu, H.; Li, Y.; et al. Long noncoding RNA XIST promotes malignancies of esophageal squamous cell carcinoma via regulation of miR-101/EZH2. Oncotarget 2017, 8, 76015–76028. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Liu, Z.; Pei, D.; Jiang, Y.; Zhu, H.; Chen, B. Development and validation of nomogram based on lncRNA ZFAS1 for predicting survival in lymph node-negative esophageal squamous cell carcinoma patients. Oncotarget 2017, 8, 59048–59057. [Google Scholar] [CrossRef]
- Liu, B.; Pan, C.-F.; Yao, G.-L.; Wei, K.; Xia, Y.; Chen, Y.-J. The long non-coding RNA AK001796 contributes to tumor growth via regulating expression of p53 in esophageal squamous cell carcinoma. Cancer Cell Int. 2018, 18, 38. [Google Scholar] [CrossRef]
- Shi, H.; Shi, J.; Zhang, Y.; Guan, C.; Zhu, J.; Wang, F.; Xu, M.; Ju, Q.; Fang, S.; Jiang, M. Long non-coding RNA DANCR promotes cell proliferation, migration, invasion and resistance to apoptosis in esophageal cancer. J. Thorac. Dis. 2018, 10, 2573–2582. [Google Scholar] [CrossRef]
- Wang, Z.; Ren, B.; Huang, J.; Yin, R.; Jiang, F.; Zhang, Q. LncRNA DUXAP10 modulates cell proliferation in esophageal squamous cell carcinoma through epigenetically silencing p21. Cancer Biol. Ther. 2018, 19, 998–1005. [Google Scholar] [CrossRef]
- Xu, L.-J.; Yu, X.-J.; Wei, B.; Hui, H.-X.; Sun, Y.; Dai, J.; Chen, X.-F. Long non-coding RNA DUXAP8 regulates proliferation and invasion of esophageal squamous cell cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2646–2652. [Google Scholar]
- Yao, J.; Shen, X.; Li, H.; Xu, J.; Shao, S.; Huang, J.X.; Lin, M. LncRNA-ECM is overexpressed in esophageal squamous cell carcinoma and promotes tumor metastasis. Oncol. Lett. 2018, 16, 3935–3942. [Google Scholar] [CrossRef]
- Zhang, X.-D.; Huang, G.-W.; Xie, Y.-H.; He, J.-Z.; Guo, J.-C.; Xu, X.-E.; Liao, L.-D.; Xie, Y.-M.; Song, Y.-M.; Lian-Di, L.; et al. The interaction of lncRNA EZR-AS1 with SMYD3 maintains overexpression of EZR in ESCC cells. Nucleic Acids Res. 2017, 46, 1793–1809. [Google Scholar] [CrossRef]
- Chen, M.; Liu, P.; Chen, Y.; Chen, Z.; Shen, M.; Liu, X.; Li, X.; Li, A.; Lin, Y.; Yang, R.; et al. Long Noncoding RNA FAM201A Mediates the Radiosensitivity of Esophageal Squamous Cell Cancer by Regulating ATM and mTOR Expression via miR-101. Front. Genet. 2018, 9, 611. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhang, C.-Q.; Li, H.-L.; Gu, J.; Miao, G.-Y.; Cai, H.-Y.; Wang, J.-K.; Zhang, L.-J.; Song, Y.-M.; Tian, Y.-H.; et al. LncRNA FER1L4 suppressed cancer cell growth and invasion in esophageal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2638–2645. [Google Scholar] [PubMed]
- Bao, J.; Zhou, C.; Zhang, J.; Mo, J.; Ye, Q.; He, J.; Diao, J. Upregulation of the long noncoding RNA FOXD2-AS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Biomark. 2018, 21, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Ke, K.; Sun, Z.; Wang, Z. Downregulation of long non-coding RNA GAS5 promotes cell proliferation, migration and invasion in esophageal squamous cell carcinoma. Oncol. Lett. 2018, 16, 1801–1808. [Google Scholar] [CrossRef]
- Sun, X.Y.; Wang, X.F.; Cui, Y.B.; Cao, X.G.; Zhao, R.H.; Wei, H.Y.; Cao, W.; Wu, W. [Expression level and clinical significance of LncRNA HOXA11-AS in esophageal squamous cell carcinoma patients]. Zhonghua Zhong Liu Za Zhi Chin. J. Oncol. 2018, 40, 186–190. [Google Scholar]
- Chen, Z.; Lin, J.; Wu, S.; Xu, C.; Chen, F.; Huang, Z. Up-regulated miR-548k promotes esophageal squamous cell carcinoma progression via targeting long noncoding RNA-LET. Exp. Cell Res. 2018, 362, 90–101. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, J.; Pan, S.; Yang, T.; Sun, X.; Wang, Y.; Shi, X.; Zhao, X.; Guo, J.; Zhang, X. LINC00657 played oncogenic roles in esophageal squamous cell carcinoma by targeting miR-615-3p and JunB. Biomed. Pharmacother. 2018, 108, 316–324. [Google Scholar] [CrossRef]
- Yang, X.-Z.; He, Q.-J.; Cheng, T.-T.; Chi, J.; Lei, Z.-Y.; Tang, Z.; Liao, Q.-X.; Zhang, H.; Zeng, L.-S.; Cui, S.-Z. Predictive Value of LINC01133 for Unfavorable Prognosis was Impacted by Alcohol in Esophageal Squamous Cell Carcinoma. Cell. Physiol. Biochem. 2018, 48, 251–262. [Google Scholar] [CrossRef]
- Wang, B.; Liang, T.; Li, J. Long noncoding RNA LINC01296 is associated with poor prognosis in ESCC and promotes ESCC cell proliferation, migration and invasion. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4524–4531. [Google Scholar]
- Xie, J.-J.; Jiang, Y.-Y.; Jiang, Y.; Li, C.; Lim, M.-C.; An, O.; Mayakonda, A.; Ding, L.-W.; Long, L.; Sun, C.; et al. Super-Enhancer-Driven Long Non-Coding RNA LINC01503, Regulated by TP63, Is Over-Expressed and Oncogenic in Squamous Cell Carcinoma. Gastroenterology 2018, 154, 2137–2151. [Google Scholar] [CrossRef]
- Niu, G.; Zhuang, H.; Li, B.; Cao, G. Long noncoding RNA linc-UBC1 promotes tumor invasion and metastasis by regulating EZH2 and repressing E-cadherin in esophageal squamous cell carcinoma. JBUON Off. J. Balk. Union Oncol. 2018, 23, 157–162. [Google Scholar]
- Guo, W.; Liu, S.; Dong, Z.; Guo, Y.; Ding, C.; Shen, S.; Liang, J.; Shan, B. Aberrant methylation-mediated silencing of lncRNA CTC-276P9.1 is associated with malignant progression of esophageal squamous cell carcinoma. Clin. Exp. Metastasis 2018, 35, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; You, B.-H.; Park, C.H.; Kim, Y.J.; Nam, J.-W.; Kil Lee, S. The long noncoding RNA LUCAT1 promotes tumorigenesis by controlling ubiquitination and stability of DNA methyltransferase 1 in esophageal squamous cell carcinoma. Cancer Lett. 2018, 417, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Luo, M.; Zhou, C.; Shi, X.; Yang, W.; Lu, Z.; Chen, Z.; Sun, N.; He, J. Novel long noncoding RNA NMR promotes tumor progression via NSUN2 and BPTF in esophageal squamous cell carcinoma. Cancer Lett. 2018, 430, 57–66. [Google Scholar] [CrossRef]
- Chen, R.; Xia, W.; Wang, X.; Qiu, M.; Yin, R.; Wang, S.; Xi, X.; Wang, J.; Xu, Y.; Dong, G.; et al. Upregulated long non-coding RNA SBF2-AS1 promotes proliferation in esophageal squamous cell carcinoma. Oncol. Lett. 2018, 15, 5071–5080. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Guo, J.; Yan, W.; Huang, M.; Zhu, C.; Yin, Y.; Chen, X. Small nucleolar host gene 6 promotes esophageal squamous cell carcinoma cell proliferation and inhibits cell apoptosis. Oncol. Lett. 2018, 15, 6497–6502. [Google Scholar] [CrossRef]
- Lin, C.; Zhang, S.; Wang, Y.; Wang, Y.; Nice, E.; Guo, C.; Zhang, E.; Chenyu, L.; Li, M.; Liu, C.; et al. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin. Cancer Res. 2017, 24, 486–498. [Google Scholar] [CrossRef]
- Kang, K.; Huang, Y.-H.; Li, H.-P.; Guo, S.-M. Expression of UCA1 and MALAT1 long-chain non-coding RNAs in esophageal squamous cell carcinoma tissues is predictive of patient prognosis. Arch. Med. Sci. 2018, 14, 752–759. [Google Scholar] [CrossRef]
- Sun, K.; Zhang, G. Long noncoding RNA CASC2 suppresses esophageal squamous cell carcinoma progression by increasing SOCS1 expression. Cell Biosci. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, L.; Yang, J.; Fu, Y.; Li, H.; Xie, L.; Cui, Y. MicroRNA-33a-5p suppresses esophageal squamous cell carcinoma progression via regulation of lncRNA DANCR and ZEB1. Eur. J. Pharmacol. 2019, 861, 172590. [Google Scholar] [CrossRef]
- Wang, M.; Li, Y.; Yang, Y.; Liu, X.; Zang, M.; Li, Y.; Yang, K.; Yang, W.; Zhang, S. Long non-coding RNA DLX6-AS1 is associated with malignant progression and promotes proliferation and invasion in esophageal squamous cell carcinoma. Mol. Med. Rep. 2018, 19, 1942–1950. [Google Scholar] [CrossRef]
- Zhang, H.; Hua, Y.; Jiang, Z.; Yue, J.; Shi, M.; Zheng, X.; Zhang, X.; Yang, L.; Zhou, R.; Wu, S. Cancer-associated Fibroblast–promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res. 2019, 25, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Wang, R.; Wang, B.; Hua, P.; Li, J. LncRNA Erbb4-IR promotes esophageal squamous cell carcinoma (ESCC) by downregulating miR-145. J. Cell. Biochem. 2019, 120, 17566–17572. [Google Scholar] [CrossRef]
- Li, W.; Zhang, L.; Guo, B.; Deng, J.; Wu, S.; Li, F.; Wang, Y.; Lu, J.; Zhou, Y. Exosomal FMR1-AS1 facilitates maintaining cancer stem-like cell dynamic equilibrium via TLR7/NFkappaB/c-Myc signaling in female esophageal carcinoma. Mol. Cancer 2019, 18, 22. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Li, S.; Wang, S.; Rubegni, P.; Tognetti, L.; Zhang, J.; Yan, L. Long noncoding RNA HAND2-AS1 inhibits cancer cell proliferation, migration, and invasion in esophagus squamous cell carcinoma by regulating microRNA-21. J. Cell Biochem. 2019, 120, 9564–9571. [Google Scholar] [CrossRef]
- Huang, J.; Li, J.; Li, Y.; Lu, Z.; Che, Y.; Mao, S.; Lei, Y.; Zang, R.; Zheng, S.; Liu, C.; et al. Interferon-inducible lncRNA IRF1-AS represses esophageal squamous cell carcinoma by promoting interferon response. Cancer Lett. 2019, 459, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, W.; Pan, T.; Wang, H.; Zhang, Y.; Li, C. LBX2-AS1 is activated by ZEB1 and promotes the development of esophageal squamous cell carcinoma by interacting with HNRNPC to enhance the stability of ZEB1 and ZEB2 mRNAs. Biochem. Biophys. Res. Commun. 2019, 511, 566–572. [Google Scholar] [CrossRef]
- Zong, M.-Z.; Feng, W.-T.; Du, N.; Yu, X.-J.; Yu, W.-Y. Upregulation of long noncoding RNA LEF1-AS1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7929–7934. [Google Scholar]
- Yang, Y.; Sun, X.; Chi, C.; Liu, Y.; Lin, C.; Xie, D.; Shen, X.; Lin, X. Upregulation of long noncoding RNA LINC00152 promotes proliferation and metastasis of esophageal squamous cell carcinoma. Cancer Manag. Res 2019, 11, 4643–4654. [Google Scholar] [CrossRef]
- He, Z. LINC00473/miR-497-5p Regulates Esophageal Squamous Cell Carcinoma Progression Through Targeting PRKAA1. Cancer Biother. Radiopharm. 2019, 34, 650–659. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, H.; Wang, X.; Hu, B.; Zhang, F.; Wei, H.; Li, L. LINC01518 knockdown inhibits tumorigenicity by suppression of PIK3CA/Akt pathway in oesophageal squamous cell carcinoma. Artif. Cells Nanomed. Biotechnol. 2019, 47, 4284–4292. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liang, Y.; Wu, Y.; Chen, X.; Wang, K.; Li, J.; Guan, X.; Xiong, G.; Yang, K.; Bai, Y. Upregulation of a novel lncRNA LINC01980 promotes tumor growth of esophageal squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2019, 513, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-M.; Li, S.-Y.; Hao-Bin, Y.; Lin-Yan, X.; Sheng, X. IL-11 activated by lnc-ATB promotes cell proliferation and invasion in esophageal squamous cell cancer. Biomed. Pharmacother. 2019, 114, 108835. [Google Scholar] [CrossRef] [PubMed]
- Li, F.-Z.; Zang, W.-Q. Knockdown of lncRNAXLOC_001659 inhibits proliferation and invasion of esophageal squamous cell carcinoma cells. World J. Gastroenterol. 2019, 25, 6299–6310. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Lin, J.; Zhao, Y.; Liu, G.-J.; Liu, Y.-B.; Feng, L.; Yang, H.-Y.; Cui, W.-X.; Zhang, X.-H. Long noncoding RNA LSINCT5 is upregulated and promotes the progression of esophageal squamous cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5195–5205. [Google Scholar]
- Chu, J.; Li, H.; Xing, Y.; Jia, J.; Sheng, J.; Yang, L.; Sun, K.; Qu, Y.; Zhang, Y.; Yin, H.; et al. LncRNA MNX1-AS1 promotes progression of esophageal squamous cell carcinoma by regulating miR-34a/SIRT1 axis. Biomed. Pharmacother. 2019, 116, 109029. [Google Scholar] [CrossRef]
- Shi, W.; Wang, Q.; Bian, Y.; Fan, Y.; Zhou, Y.; Feng, T.; Li, Z.; Cao, X. Long noncoding RNA PANDA promotes esophageal squamous carcinoma cell progress by dissociating from NF-YA but interact with SAFA. Pathol.-Res. Pr. 2019, 215, 152604. [Google Scholar] [CrossRef]
- Zang, B.; Zhao, J.; Chen, C. LncRNA PCAT-1 Promoted ESCC Progression via Regulating ANXA10 Expression by Sponging miR-508-3p. Cancer Manag. Res. 2019, 11, 10841–10849. [Google Scholar] [CrossRef]
- Zhihua, Z.; Weiwei, W.; Lihua, N.; Jianying, Z.; Jiang, G. p53-induced long non-coding RNA PGM5-AS1 inhibits the progression of esophageal squamous cell carcinoma through regulating miR-466/PTEN axis. IUBMB Life 2019, 71, 1492–1502. [Google Scholar] [CrossRef]
- Dong, Z.; Liang, X.; Wu, X.; Kang, X.; Guo, Y.; Shen, S.; Liang, J.; Guo, W. Promoter hypermethylation-mediated downregulation of tumor suppressor gene SEMA3B and lncRNA SEMA3B-AS1 correlates with progression and prognosis of esophageal squamous cell carcinoma. Clin. Exp. Metastasis 2019, 36, 225–241. [Google Scholar] [CrossRef]
- Zhang, C.; Jiang, F.; Su, C.; Xie, P.; Xu, L. Upregulation of long noncoding RNA SNHG20 promotes cell growth and metastasis in esophageal squamous cell carcinoma via modulating ATM-JAK-PD-L1 pathway. J. Cell. Biochem. 2019, 120, 11642–11650. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, R.; Ding, X.; Zhang, K.; Qin, W. Upregulation of long non-coding RNA SNHG6 promote esophageal squamous cell carcinoma cell malignancy and its diagnostic value. Am. J. Transl. Res. 2019, 11, 1084–1091. [Google Scholar] [PubMed]
- Song, H.; Song, J.; Lu, L.; Li, S. SNHG8 is upregulated in esophageal squamous cell carcinoma and directly sponges microRNA-411 to increase oncogenicity by upregulating KPNA2. Onco Targets Ther. 2019, 12, 6991–7004. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.-F.; Pan, Y.; Yang, H.-J.; Li, J.-K.; Zhao, F.; Su, J.-F.; Li, Y.-Y.; Tian, L.-Q.; Yu, P.-T.; Cao, Y.-T.; et al. Decreased expression of SPINT1-AS1 and SPINT1 mRNA might be independent unfavorable prognostic indicators in esophageal squamous cell carcinoma. Onco Targets Ther. 2019, 12, 4755–4763. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Shu, L.; Zou, W. Role of long non-coding RNA TP73-AS1 in cancer. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Dong, Z.; Li, S.; Wu, X.; Niu, Y.; Liang, X.; Yang, L.; Guo, Y.; Shen, S.; Liang, J.; Guo, W. Aberrant hypermethylation-mediated downregulation of antisense lncRNA ZNF667-AS1 and its sense gene ZNF667 correlate with progression and prognosis of esophageal squamous cell carcinoma. Cell Death Dis. 2019, 10, 1–18. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Z.-X.; Wu, Q.-N.; Lu, Y.-X.; Wong, C.-W.; Miao, L.; Wang, Y.; Wang, Z.; Jin, Y.; He, M.-M.; et al. Long noncoding RNA AGPG regulates PFKFB3-mediated tumor glycolytic reprogramming. Nat. Commun. 2020, 11, 1507–1516. [Google Scholar] [CrossRef]
- Sang, Y.; Gu, H.; Chen, Y.; Shi, Y.; Liu, C.; Lv, L.; Sun, Y.; Zhang, Y. Long non-coding RNA CASC8 polymorphisms are associated with the risk of esophageal cancer in a Chinese population. Thorac. Cancer 2020, 11, 2852–2857. [Google Scholar] [CrossRef]
- Feng, Z.; Li, X.; Qiu, M.; Luo, R.; Lin, J.; Liu, B. LncRNA EGFR-AS1 Upregulates ROCK1 by Sponging miR-145 to Promote Esophageal Squamous Cell Carcinoma Cell Invasion and Migration. Cancer Biother. Radiopharm. 2020, 35, 66–71. [Google Scholar] [CrossRef]
- Zhang, C.; Luo, Y.; Cao, J.; Wang, X.; Miao, Z.; Shao, G. Exosomal lncRNA FAM225A accelerates esophageal squamous cell carcinoma progression and angiogenesis via sponging miR-206 to upregulate NETO2 and FOXP1 expression. Cancer Med. 2020, 9, 8600–8611. [Google Scholar] [CrossRef]
- Feng, B.; Wang, G.; Liang, X.; Wu, Z.; Wang, X.; Dong, Z.; Guo, Y.; Shen, S.; Liang, J.; Guo, W. LncRNA FAM83H-AS1 promotes oesophageal squamous cell carcinoma progression via miR-10a-5p/Girdin axis. J Cell Mol Med. 2020, 24, 8962–8976. [Google Scholar]
- Gao, J.; Zhang, Z.; Su, H.; Zong, L.; Li, Y. Long Noncoding RNA FGD5-AS1 Acts as a Competing Endogenous RNA on microRNA-383 to Enhance the Malignant Characteristics of Esophageal Squamous Cell Carcinoma by Increasing SP1 Expression. Cancer Manag. Res. 2020, 12, 2265–2278. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, T.; Yang, Y.; Kang, W.; Dong, S.; Cheng, S. YY1-induced upregulation of FOXP4-AS1 and FOXP4 promote the proliferation of esophageal squamous cell carcinoma cells. Cell Biol. Int. 2020, 44, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiao, X.; Chang, R.; Zhang, C. Comprehensive bioinformatics analysis identifies lncRNA HCG22 as a migration inhibitor in esophageal squamous cell carcinoma. J. Cell. Biochem. 2019, 121, 468–481. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; You, D.; Pan, Y.; Liu, P. Downregulation of lncRNA-HEIH curbs esophageal squamous cell carcinoma progression by modulating miR-4458/PBX3. Thorac. Cancer 2020, 11, 1963–1971. [Google Scholar] [CrossRef]
- Wang, B.; Hua, P.; Zhang, L.; Li, J.; Zhang, Y. LncRNA-IUR up-regulates PTEN by sponging miR-21 to regulate cancer cell proliferation and apoptosis in esophageal squamous cell carcinoma. Esophagus 2020, 17, 298–304. [Google Scholar] [CrossRef]
- Liu, J.-Q.; Deng, M.; Xue, N.-N.; Li, T.-X.; Guo, Y.-X.; Gao, L.; Zhao, D.; Fan, R.-T. lncRNA KLF3-AS1 Suppresses Cell Migration and Invasion in ESCC by Impairing miR-185-5p-Targeted KLF3 Inhibition. Mol. Ther.-Nucleic Acids 2020, 20, 231–241. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, L.; Deng, J.; Guo, B.; Li, F.; Wang, Y.; Wu, R.; Zhang, S.; Lu, J.; Zhou, Y. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res. 2020, 80, 2790–2803. [Google Scholar] [CrossRef]
- Zhang, Z.; Liang, X.; Ren, L.; Zhang, S.; Li, S.; Wan, T.; Xu, D.; Lv, S. LINC00662 promotes cell viability and metastasis in esophageal squamous cell carcinoma by sponging miR -340-5p and upregulating HOXB2. Thorac. Cancer 2020, 11, 2306–2315. [Google Scholar] [CrossRef]
- Zhou, M.; Mao, Y.; Yu, S.; Li, Y.; Yin, R.; Zhang, Q.; Lu, T.; Sun, R.; Lin, S.; Qian, Y.; et al. LINC00673 Represses CDKN2C and Promotes the Proliferation of Esophageal Squamous Cell Carcinoma Cells by EZH2-Mediated H3K27 Trimethylation. Front. Oncol. 2020, 10, 1546. [Google Scholar] [CrossRef]
- Liu, H.-F.; Zhen, Q.; Fan, Y.-K. LINC00963 predicts poor prognosis and promotes esophageal cancer cells invasion via targeting miR-214-5p/RAB14 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 164–173. [Google Scholar] [PubMed]
- Zhao, M.; Cui, H.; Zhao, B.; Li, M.; Man, H. Long intergenic non-coding RNA LINC01232 contributes to esophageal squamous cell carcinoma progression by sequestering microRNA-654-3p and consequently promoting hepatoma-derived growth factor expression. Int. J. Mol. Med. 2020, 46, 2007–2018. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Liu, Y.-T.; Wu, C.-P.; Jiang, J.-T.; Zhang, L.; Wang, Z.-L.; Wang, Q.-Y. Long non-coding RNA linc01433 promotes tumorigenesis and progression in esophageal squamous cell carcinoma by sponging miR-1301. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4785–4792. [Google Scholar] [PubMed]
- Du, J.; Zhang, G.; Qiu, H.; Yu, H.; Yuan, W. A novel positive feedback loop of linc02042 and c-Myc mediated by YBX1 promotes tumorigenesis and metastasis in esophageal squamous cell carcinoma. Cancer Cell Int. 2020, 20, 1–10. [Google Scholar] [CrossRef]
- Lu, T.; Ma, K.; Zhan, C.; Yang, X.; Shi, Y.; Jiang, W.; Wang, H.; Wang, S.; Wang, Q.; Tan, L. Downregulation of long non-coding RNA LINP1 inhibits the malignant progression of esophageal squamous cell carcinoma. Ann. Transl. Med. 2020, 8, 675. [Google Scholar] [CrossRef]
- Ma, J.; Xiao, Y.; Tian, B.; Chen, S.; Zhang, B.; Wu, J.; Wu, Z.; Li, X.; Tang, J.; Yang, D.; et al. Long noncoding RNA lnc-ABCA12-3 promotes cell migration, invasion, and proliferation by regulating fibronectin 1 in esophageal squamous cell carcinoma. J. Cell. Biochem. 2019, 121, 1374–1387. [Google Scholar] [CrossRef]
- Liu, G.; Guo, W.; Chen, G.; Li, W.; Cui, Y.; Qin, J.; Peng, J. Lnc-MCEI mediated the chemosensitivity of esophageal squamous cell carcinoma via miR-6759-5p to competitively regulate IGF2. Int. J. Biol. Sci. 2020, 16, 2938–2950. [Google Scholar] [CrossRef]
- Wang, P.; Yang, Z.; Ye, T.; Shao, F.; Li, J.; Sun, N.; He, J. lncTUG1/miR-144-3p affect the radiosensitivity of esophageal squamous cell carcinoma by competitively regulating c-MET. J. Exp. Clin. Cancer Res. 2020, 39, 7. [Google Scholar] [CrossRef]
- Guan, Z.; Wang, Y.; Wang, Y.; Liu, X.; Wang, Y.; Zhang, W.; Chi, X.; Dong, Y.; Liu, X.; Shao, S.; et al. Long non-coding RNA LOC100133669 promotes cell proliferation in oesophageal squamous cell carcinoma. Cell Prolif. 2020, 53, e12750. [Google Scholar] [CrossRef]
- Wang, G.; Feng, B.; Niu, Y.; Wu, J.; Yang, Y.; Shen, S.; Guo, Y.; Liang, J.; Guo, W.; Dong, Z. A novel long noncoding RNA, LOC440173, promotes the progression of esophageal squamous cell carcinoma by modulating the miR-30d-5p/HDAC9 axis and the epithelial-mesenchymal transition. Mol. Carcinog. 2020, 59, 1392–1408. [Google Scholar] [CrossRef]
- Hu, W.; Chen, Z.; Chen, J.; Cai, D.; Chen, C.; Fang, T. LOC441178 Overexpression Inhibits the Proliferation and Migration of Esophageal Carcinoma Cells via Methylation of miR-182. Onco Targets Ther. 2020, 13, 11253–11263. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.J.; Xu, Z.R.; Chen, L.Y.; Wang, Y.C.; Yao, J. LncRNA MAFG-AS1 Accelerates Cell Migration, Invasion and Aerobic Glycolysis of Esophageal Squamous Cell Carcinoma Cells via miR-765/PDX1 Axis. Cancer Manag. Res. 2020, 12, 6895–6908. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xie, L.; Fu, Y.; Yang, J.; Cui, Y. lncRNA MIAT promotes esophageal squamous cell carcinoma progression by regulating miR-1301-3p/INCENP axis and interacting with SOX2. J. Cell. Physiol. 2020, 235, 7933–7944. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jia, J.; Yang, L.; Chu, J.; Sheng, J.; Wang, C.; Meng, W.; Jia, Z.; Yin, H.; Wan, J.; et al. LncRNA MIR205HG Drives Esophageal Squamous Cell Carcinoma Progression by Regulating miR-214/SOX4 Axis. Onco Targets Ther. 2020, 13, 13097–13109. [Google Scholar] [CrossRef]
- Li, D.; Li, D.; Meng, L.; Liu, J.; Huang, C.; Sun, H. LncRNA NLIPMT Inhibits Tumorigenesis in Esophageal Squamous-Cell Carcinomas by Regulating miR-320/Survivin Axis. Cancer Manag. Res. 2020, 12, 12603–12612. [Google Scholar] [CrossRef]
- Qiu, B.Q.; Lin, X.H.; Ye, X.D.; Huang, W.; Pei, X.; Xiong, D.; Long, X.; Zhu, S.Q.; Lu, F.; Lin, K.; et al. Long non-coding RNA PSMA3-AS1 promotes malignant phenotypes of esophageal cancer by modulating the miR-101/EZH2 axis as a ceRNA. Aging (Albany NY) 2020, 12, 1843–1856. [Google Scholar] [CrossRef]
- Li, Z.W.; Zhang, T.Y.; Yue, G.J.; Tian, X.; Wu, J.Z.; Feng, G.Y.; Wang, Y.S. Small nucleolar RNA host gene 22 (SNHG22) promotes the progression of esophageal squamous cell carcinoma by miR-429/SESN3 axis. Ann. Transl. Med. 2020, 8, 1007. [Google Scholar] [CrossRef]
- Liang, M.; Pan, Z.; Yu, F.; Chen, C. Long noncoding RNA SNHG12 suppresses esophageal squamous cell carcinoma progression through competing endogenous RNA networks. Clin. Transl. Oncol. 2020, 22, 1786–1795. [Google Scholar] [CrossRef]
- Wang, W.; Yang, J. Long noncoding RNA TTTY15 promotes growth and metastasis of esophageal squamous cell carcinoma by sponging microRNA-337-3p to upregulate the expression of JAK2. Anti-Cancer Drugs 2020, 31, 1038–1045. [Google Scholar] [CrossRef]
- Zhao, K.; Guo, Y.; Huo, Z.; Ma, G.; Zhang, G.; Xing, Y.; Xu, Q. [Serum level of lncRNA TUSC7 in patients with esophageal squamous cell carcinoma and its role in promoting tumor cell migration and invasion]. Nan Fang Yi Ke Da Xue Xue Bao 2020, 40, 661–669. [Google Scholar]
- Yao, J.; Zhang, H.; Li, H.; Qian, R.; Liu, P.; Huang, J. P53-regulated lncRNA uc061hsf.1 inhibits cell proliferation and metastasis in human esophageal squamous cell cancer. IUBMB Life 2019, 72, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Du, F.; Guo, T.; Cao, C. Restoration of UPK1A-AS1 Expression Suppresses Cell Proliferation, Migration, and Invasion in Esophageal Squamous Cell Carcinoma Cells Partially by Sponging microRNA-1248. Cancer Manag. Res. 2020, ume 12, 2653–2662. [Google Scholar] [CrossRef]
- Zhang, Q.; Guan, F.; Fan, T.; Li, S.; Ma, S.; Zhang, Y.; Guo, W.; Liu, H. LncRNA WDFY3-AS2 suppresses proliferation and invasion in oesophageal squamous cell carcinoma by regulating miR-2355-5p/SOCS2 axis. J. Cell. Mol. Med. 2020, 24, 8206–8220. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-H.; Chen, R.-Z.; Liu, L.-Y.; Li, X.-M.; Wu, C.-P.; Zhou, Y.-T.; Yan, J.-D.; Zhang, Z.-Y. LncRNA ZEB2-AS1 promotes the proliferation, migration and invasion of esophageal squamous cell carcinoma cell through miR-574-3p/HMGA2 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 5391–5403. [Google Scholar]
- Sun, G.; Wu, C. ZFPM2-AS1 facilitates cell growth in esophageal squamous cell carcinoma via up-regulating TRAF4. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.; Zhang, W.; Wu, Q.; Fan, J.; Zhan, Q. BAALC-AS1/G3BP2/c-Myc feedback loop promotes cell proliferation in esophageal squamous cell carcinoma. Cancer Commun. 2021, 41, 240–257. [Google Scholar] [CrossRef]
- Yan, S.; Xu, J.; Liu, B.; Ma, L.; Feng, H.; Tan, H.; Fang, C. Long non-coding RNA BCAR4 aggravated proliferation and migration in esophageal squamous cell carcinoma by negatively regulating p53/p21 signaling pathway. Bioengineered 2021, 12, 682–696. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Dong, M.; Wang, Z.; Wan, J.; Xie, Y.; Jiao, Y.; Yan, D. Long non-coding RNA CASC15 facilitates esophageal squamous cell carcinoma tumorigenesis via decreasing SIM2 stability via FTO-mediated demethylation. Oncol. Rep. 2020, 45, 1059–1071. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Jia, Y.; Wang, J.; Liu, T.; Cheng, Z.; Sang, M.; Lv, W.; Qin, J.; Liu, L. Long noncoding RNA DGCR5 involves in tumorigenesis of esophageal squamous cell carcinoma via SRSF1-mediated alternative splicing of Mcl-1. Cell Death Dis. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, R.; Deng, M.; Xue, N.; Li, T.; Guo, Y.; Gao, L.; Fan, R.; Zhao, D. Long non-coding RNA DIO3OS binds to microRNA-130b to restore radiosensitivity in esophageal squamous cell carcinoma by upregulating PAX9. Cancer Gene Ther. 2021, 1–12. [Google Scholar] [CrossRef]
- Jia, J.; Li, H.; Chu, J.; Sheng, J.; Wang, C.; Jia, Z.; Meng, W.; Yin, H.; Wan, J.; He, F. LncRNA FAM83A-AS1 promotes ESCC progression by regulating miR-214/CDC25B axis. J. Cancer 2021, 12, 1200–1211. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yang, S.; Dai, S.; Ni, Q.; Liu, H.; Yu, L.; Lu, K.; Han, G.; Huang, J. Expression and Clinical Value of LncRNA GAPLINC in Esophageal Squamous Cell Carcinoma. Onco Targets Ther. 2021, 14, 4039–4045. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Miao, J.; Liu, S.; Liu, H.; Zhang, L.; Zhang, Q. Long non-coding RNA KCNQ1 overlapping transcript 1 promotes the progression of esophageal squamous cell carcinoma by adsorbing microRNA-133b. Clinics 2021, 76, e2175. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Lu, J.; Wu, Z.; Guo, Y.; Shen, S.; Liang, J.; Dong, Z.; Guo, W. LINC00239 Interacts with C-Myc Promoter-Binding Protein-1 (MBP-1) to Promote Expression of C-Myc in Esophageal Squamous Cell Carcinoma. Mol. Cancer Res. 2021, 19, 1465–1475. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Yan, P.; Liu, Y.; Jiang, X. LINC00261 Suppresses Cisplatin Resistance of Esophageal Squamous Cell Carcinoma Through miR-545-3p/MT1M Axis. Front. Cell Dev. Biol. 2021, 9, 1915. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, S.; Chen, X.; Dong, S.; Zhou, S.; Xu, S. LncRNA LINC00467 acted as an oncogene in esophageal squamous cell carcinoma by accelerating cell proliferation and preventing cell apoptosis via the miR-485-5p / DPAGT1 axis. J. Gastroenterol. Hepatol. 2020, 36, 721–730. [Google Scholar] [CrossRef]
- Ye, H.; Shrestha, S.M.; Zhu, J.; Ding, Y.; Shi, R. Long non-coding RNA LINC00491 promotes proliferation and inhibits apoptosis in esophageal squamous cell carcinoma. Int. J. Mol. Med. 2021, 47, 1-1. [Google Scholar] [CrossRef]
- Peng, X.; Zhou, Y.; Chen, Y.; Tang, L.; Wang, G.; Jiang, H.; Wang, X.; Tao, Y.; Zhuang, W. Reduced LINC00551 expression promotes proliferation and invasion of esophageal squamous cancer by increase in HSP27 phosphorylation. J. Cell. Physiol. 2020, 236, 1418–1431. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, H.; Sun, N.; Zhang, X.; Liang, G.; Zhu, J.; Xia, L.; Kou, Y.; Lu, J. Linc00941 regulates esophageal squamous cell carcinoma via functioning as a competing endogenous RNA for miR-877-3p to modulate PMEPA1 expression. Aging 2021, 13, 17830–17846. [Google Scholar] [CrossRef]
- Wang, B.; Tang, D.; Liu, Z.; Wang, Q.; Xue, S.; Zhao, Z.; Feng, D.; Sheng, C.; Li, J.; Zhou, Z. LINC00958 promotes proliferation, migration, invasion, and epithelial-mesenchymal transition of oesophageal squamous cell carcinoma cells. PLoS ONE 2021, 16, e0251797. [Google Scholar] [CrossRef]
- Huang, G.-W.; Chen, Q.-Q.; Ma, C.-C.; Xie, L.-H.; Gu, J. linc01305 promotes metastasis and proliferation of esophageal squamous cell carcinoma through interacting with IGF2BP2 and IGF2BP3 to stabilize HTR3A mRNA. Int. J. Biochem. Cell Biol. 2021, 136, 106015. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Zhou, P.; Zhu, Z.; Wang, Y.; Guo, Z.; Shen, M.; Xiao, Y.; Shen, W.; Wu, D. Upregulated long noncoding RNA LincIN promotes tumor progression via the regulation of nuclear factor 90/microRNA7/HOXB13 in esophageal squamous cell carcinoma. Int. J. Mol. Med. 2021, 47, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rong, H.; Chen, B.; Ma, K.; Wei, X.; Peng, J.; Zhu, J. Downregulation of lncRNA LINC-PINT Participates in the Recurrence of Esophageal Squamous Cell Carcinoma Possibly by Interacting miRNA-21. Cancer Biother. Radiopharm. 2021, 36, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Long, D.; Liu, B.; Pei, D.; Cao, N.; Zhang, G.; Xia, Z.; Luo, M. Downregulation of long non-coding RNA LOC101928477 correlates with tumor progression by regulating the epithelial-mesenchymal transition in esophageal squamous cell carcinoma. Thorac. Cancer 2021, 12, 1303–1311. [Google Scholar] [CrossRef]
- Luo, J.; Xie, K.; Gao, X.; Yao, Y.; Wang, G.; Shao, C.; Li, X.; Xu, Y.; Ren, B.; Hu, L.; et al. Long Noncoding RNA Nuclear Paraspeckle Assembly Transcript 1 Promotes Progression and Angiogenesis of Esophageal Squamous Cell Carcinoma through miR-590-3p/MDM2 Axis. Front. Oncol. 2021, 10. [Google Scholar] [CrossRef]
- Wang, Y.; Bao, D.; Wan, L.; Zhang, C.; Hui, S.; Guo, H. Long non-coding RNA small nucleolar RNA host gene 7 facilitates the proliferation, migration, and invasion of esophageal cancer cells by regulating microRNA-625. J. Gastrointest. Oncol. 2021, 12, 423–432. [Google Scholar] [CrossRef]
- Li, H.; Chu, J.; Jia, J.; Sheng, J.; Zhao, X.; Xing, Y.; He, F. LncRNA LOXL1-AS1 promotes esophageal squamous cell carcinoma progression by targeting DESC1. J. Cancer 2021, 12, 530–538. [Google Scholar] [CrossRef]
- Li, J.; Han, X.; Gu, Y.; Wu, J.; Song, J.; Shi, Z.; Chang, H.; Liu, M.; Zhang, Y. LncRNA MTX2-6 Suppresses Cell Proliferation by Acting as ceRNA of miR-574-5p to Accumulate SMAD4 in Esophageal Squamous Cell Carcinoma. Front. Cell Dev. Biol. 2021, 9, 496. [Google Scholar] [CrossRef]
- Gu, S.; Qian, L.; Liu, Y.; Miao, J.; Shen, H.; Zhang, S.; Mao, G. Upregulation of long non-coding RNA MYU promotes proliferation, migration and invasion of esophageal squamous cell carcinoma cells. Exp. Ther. Med. 2021, 21, 1–9. [Google Scholar] [CrossRef]
- Wei, S.; Sun, S.; Zhou, X.; Zhang, C.; Li, X.; Dai, S.; Wang, Y.; Zhao, L.; Shan, B. SNHG5 inhibits the progression of EMT through the ubiquitin-degradation of MTA2 in oesophageal cancer. Carcinogenesis 2020, 42, 315–326. [Google Scholar] [CrossRef]
- Cheng, J.; Ma, H.; Yan, M.; Xing, W. THAP9-AS1/miR-133b/SOX4 positive feedback loop facilitates the progression of esophageal squamous cell carcinoma. Cell Death Dis. 2021, 12, 401. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Li, B.; Zhang, J.; Du, X.; Gu, D. LncRNA THAP9-AS1 accelerates cell growth of esophageal squamous cell carcinoma through sponging miR-335–5p to regulate SGMS2. Pathol.-Res. Pr. 2021, 224, 153526. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Li, G.; Li, Z.; Liu, J.; Tang, Y. TMEM161B-AS1 suppresses proliferation, invasion and glycolysis by targeting miR-23a-3p/HIF1AN signal axis in oesophageal squamous cell carcinoma. J. Cell Mol. Med. 2021, 25, 6535–6549. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, W.; Liu, W.; Huang, L.; Wang, Y.; Li, D.; Wang, G.; Zhao, Z.; Chi, X.; Xue, Y.; et al. Long Noncoding RNA VESTAR Regulates Lymphangiogenesis and Lymph Node Metastasis of Esophageal Squamous Cell Carcinoma by Enhancing VEGFC mRNA Stability. Cancer Res. 2021, 81, 3187–3199. [Google Scholar] [CrossRef]


| LncRNA | Expression Pattern (Up/Down Regulation) | Drug | Conc. of Drugs Used | Time Points of Treatment | Patient Tissue/Cell Line/In Vivo Model | Clinical Endpoint | Pathological Response | Cohort Size | References |
|---|---|---|---|---|---|---|---|---|---|
| LOC285194 | Down | Cisplatin | NA | NA | Tissue and cell lines | DFS and OS | CR = 15% | Female = 48 | [54] |
| Male = 94 | |||||||||
| TUG1 | Up | Cisplatin | 1 μg/mL | 48 h | Tissue and cells | OS | NA | Male = 171 Female = 47 | [56] |
| [57] | |||||||||
| AFAP1-AS1 | Up | 5-Fluorouracial Cisplatin Paclitaxel | 2, 4, 8, 16, 32, 64, 128, 256 μM 0.3125, 0.625, 1.25, 2.5 5, 10, 25, 50 μM 0.03125, 0.0652, 0.125, 0.25, 0.5, 1, 2, 4, 8 μM | 24 h on days 1–4 | Tissue and cells | OS and PFS | CR = 19.8% PR = 40.7% NC = 37.7% PD = 1.8% | Male = 123 Female = 39 | [58] |
| PART1 | Up | Gefitinib | 0.01–10 μM | 48 h | Serum | NA | NA | 79 | [63] |
| TUSC7 | Down | Cisplatin 5-Fluorouracial | 1, 2, 4, 8, 16 μM 1, 4, 16, 32, 64 μM | 48 h | Tissue and cell lines | OS | NA | Male = 43 Female= 19 | [55] |
| CCAT1 | Up | Cisplatin | 0.1, 0.2, 0.5, 1, 2, 5 μM | 48 h | Cell lines | NA | NA | NA | [59] |
| LINC01419 | Up | 5-fluorouracil | 10 μg/mL | 48 h | Tissue and cell lines | NA | NA | 76 | [60] |
| LINC00337 | Up | Cisplatin | 0.5, 1, 2, 3 μg/mL | 48 h | Tissue and cell lines | NA | NA | Male = 48 Female = 26 | [51] |
| Linc01014 | Up | Gefitinib | 10 μM | 48 h | Cell lines | NA | NA | NA | [18] |
| MACC1-AS1 | Up | Cisplatin | 20, 40, 60, 80, 100 µM | NA | Tissue and cell lines | NA | NA | Male = 62 Female = 8 | [61] |
| FOXD2-AS1 | Up | Cisplatin | 20, 40, 60, 80, 100 µM 6.25, 12.5, 25, 50, 100 µg/mL | NA | Tissue and cell lines | NA | NA | Male = 62 Female = 8 | [61] |
| [62] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, U.; Murmu, M.; Barwal, T.S.; Tuli, H.S.; Jain, M.; Prakash, H.; Kaceli, T.; Jain, A.; Bishayee, A. A Pleiotropic Role of Long Non-Coding RNAs in the Modulation of Wnt/β-Catenin and PI3K/Akt/mTOR Signaling Pathways in Esophageal Squamous Cell Carcinoma: Implication in Chemotherapeutic Drug Response. Curr. Oncol. 2022, 29, 2326-2349. https://doi.org/10.3390/curroncol29040189
Sharma U, Murmu M, Barwal TS, Tuli HS, Jain M, Prakash H, Kaceli T, Jain A, Bishayee A. A Pleiotropic Role of Long Non-Coding RNAs in the Modulation of Wnt/β-Catenin and PI3K/Akt/mTOR Signaling Pathways in Esophageal Squamous Cell Carcinoma: Implication in Chemotherapeutic Drug Response. Current Oncology. 2022; 29(4):2326-2349. https://doi.org/10.3390/curroncol29040189
Chicago/Turabian StyleSharma, Uttam, Masang Murmu, Tushar Singh Barwal, Hardeep Singh Tuli, Manju Jain, Hridayesh Prakash, Tea Kaceli, Aklank Jain, and Anupam Bishayee. 2022. "A Pleiotropic Role of Long Non-Coding RNAs in the Modulation of Wnt/β-Catenin and PI3K/Akt/mTOR Signaling Pathways in Esophageal Squamous Cell Carcinoma: Implication in Chemotherapeutic Drug Response" Current Oncology 29, no. 4: 2326-2349. https://doi.org/10.3390/curroncol29040189
APA StyleSharma, U., Murmu, M., Barwal, T. S., Tuli, H. S., Jain, M., Prakash, H., Kaceli, T., Jain, A., & Bishayee, A. (2022). A Pleiotropic Role of Long Non-Coding RNAs in the Modulation of Wnt/β-Catenin and PI3K/Akt/mTOR Signaling Pathways in Esophageal Squamous Cell Carcinoma: Implication in Chemotherapeutic Drug Response. Current Oncology, 29(4), 2326-2349. https://doi.org/10.3390/curroncol29040189







