CD36 and Its Role in Regulating the Tumor Microenvironment
Abstract
1. Introduction
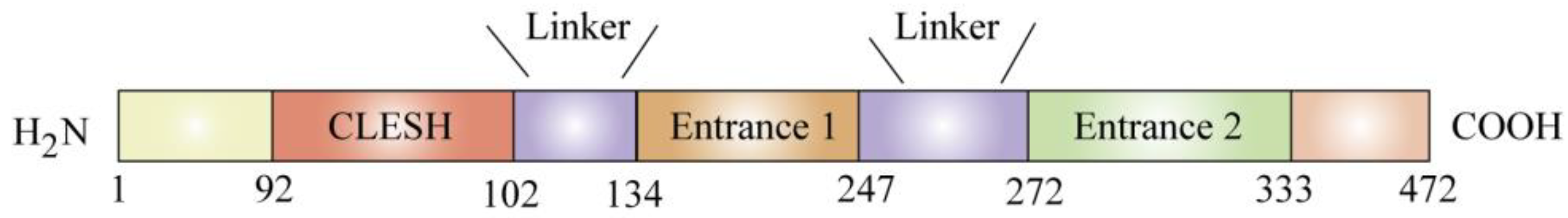
2. Structure and Distribution of CD36
3. CD36 and Tumor-Associated Immune Cells
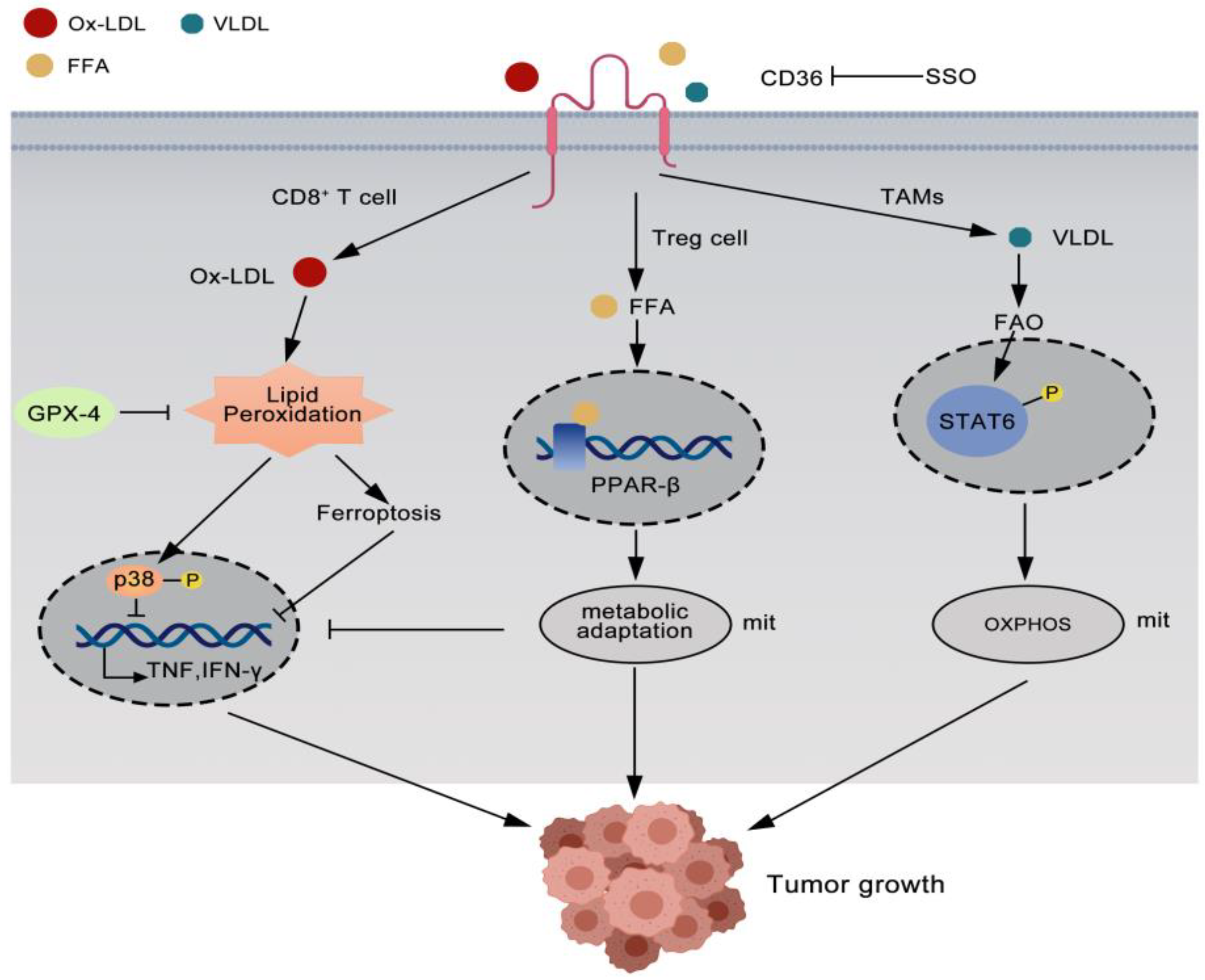
3.1. CD36 Reprogrammed Regulatory Treg Cells
3.2. CD36 Impairs Antitumor Immunity of CD8+T Cells
3.3. CD36 Cooperates with Tumor-Associated Macrophages to Promote Tumor Growth
4. CD36 and CAFs
5. CD36 Activates EMT
6. CD36 and Tumor Angiogenesis
6.1. CD36 Inhibits Tumor Angiogenesis
6.2. CD36 Binds to TSP-1 and TPS-2 to Promote Tumor Metastasis
6.3. CD36 Mediates VM in Tumors
7. Summary and Perspective
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| FAs | fatty acids |
| TSP-1 | thrombospondin-1 |
| TSP-2 | thrombospondin-2 |
| CAFs | cancer-associated fibroblasts |
| EMT | epithelial mesenchymal transition |
| VM | vascular mimicry |
| TME | the tumor microenvironment |
| OvCa | Ovarian cancer |
| SR-B2 | the class B receptor family |
| FAT | the fatty acid transporter |
| LCFA | long chain fatty acids |
| oxLDL | oxidized low density lipoprotein |
| oxPLs | oxidized phospholipids |
| CLESH structural domain | CD36, LIMP-2, Emp sequence homologous |
| TSR | TSP-1 repeat domain 2 |
| PPAR-β | peroxisome proliferator-activated receptor β |
| OXPHOS | oxidative phosphorylation |
| TAMs | tumor-associated macrophages |
References
- Whiteside, T.L. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008, 27, 5904–5912. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Declerck, Y.A. Targeting the tumor microenvironment: From understanding pathways to effective clinical trials. Cancer Res. 2013, 73, 4965–4977. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Geng, F.; Cheng, X.; Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 2018, 38, 27. [Google Scholar] [CrossRef]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.; Liu, R.; Meng, Y.; Xing, D.; Xu, D.; Lu, Z. Lipid metabolism and cancer. J. Exp. Med. 2021, 218, e20201606. [Google Scholar] [CrossRef] [PubMed]
- Ladanyi, A.; Mukherjee, A.; Kenny, H.A.; Johnson, A.; Mitra, A.K.; Sundaresan, S.; Nieman, K.M.; Pascual, G.; Benitah, S.A.; Montag, A.; et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 2018, 37, 2285–2301. [Google Scholar] [CrossRef]
- Pascual, G.; Dominguez, D.; Elosua-Bayes, M.; Beckedorff, F.; Laudanna, C.; Bigas, C.; Douillet, D.; Greco, C.; Symeonidi, A.; Hernandez, I.; et al. Dietary palmitic acid promotes a prometastatic memory via Schwann cells. Nature 2021, 599, 485–490. [Google Scholar] [CrossRef]
- Dawson, D.W.; Pearce, S.F.; Zhong, R.; Silverstein, R.L.; Frazier, W.A.; Bouck, N.P. CD36 mediates the In vitro inhibitory effects of thrombospondin-1 on endothelial cells. J. Cell Biol. 1997, 138, 707–717. [Google Scholar] [CrossRef]
- Martini, C.; DeNichilo, M.; King, D.P.; Cockshell, M.P.; Ebert, B.; Dale, B.; Ebert, L.M.; Woods, A.; Bonder, C.S. CD36 promotes vasculogenic mimicry in melanoma by mediating adhesion to the extracellular matrix. BMC Cancer 2021, 21, 765. [Google Scholar] [CrossRef]
- Chen, C.N.; Lin, J.J.; Chen, J.J.; Lee, P.H.; Yang, C.Y.; Kuo, M.L.; Chang, K.J.; Hsieh, F.J. Gene expression profile predicts patient survival of gastric cancer after surgical resection. J. Clin. Oncol. 2005, 23, 7286–7295. [Google Scholar] [CrossRef]
- Feng, W.W.; Wilkins, O.; Bang, S.; Ung, M.; Li, J.; An, J.; Del Genio, C.; Canfield, K.; DiRenzo, J.; Wells, W.; et al. CD36-Mediated Metabolic Rewiring of Breast Cancer Cells Promotes Resistance to HER2-Targeted Therapies. Cell Rep. 2019, 29, 3405–3420.e5. [Google Scholar] [CrossRef] [PubMed]
- Tanase, C.; Gheorghisan-Galateanu, A.A.; Popescu, I.D.; Mihai, S.; Codrici, E.; Albulescu, R.; Hinescu, M.E. CD36 and CD97 in Pancreatic Cancer versus Other Malignancies. Int. J. Mol. Sci. 2020, 21, 5656. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.M. CD36, a scavenger receptor implicated in atherosclerosis. Exp. Mol. Med. 2014, 46, e99. [Google Scholar] [CrossRef] [PubMed]
- Barnwell, J.W.; Asch, A.S.; Nachman, R.L.; Yamaya, M.; Aikawa, M.; Ingravallo, P. A human 88-kD membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum-infected erythrocytes. J. Clin. Investig. 1989, 84, 765–772. [Google Scholar] [CrossRef]
- Yang, X.; Okamura, D.M.; Lu, X.; Chen, Y.; Moorhead, J.; Varghese, Z.; Ruan, X.Z. CD36 in chronic kidney disease: Novel insights and therapeutic opportunities. Nat. Rev. Nephrol. 2017, 13, 769–781. [Google Scholar] [CrossRef]
- Armesilla, A.L.; Vega, M.A. Structural organization of the gene for human CD36 glycoprotein. J. Biol. Chem. 1994, 269, 18985–18991. [Google Scholar] [CrossRef]
- De Fraipont, F.; Nicholson, A.C.; Feige, J.J.; Van Meir, E.G. Thrombospondins and tumor angiogenesis. Trends Mol. Med. 2001, 7, 401–407. [Google Scholar] [CrossRef]
- Silverstein, R.L.; Febbraio, M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009, 2, re3. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y. CD36 tango in cancer: Signaling pathways and functions. Theranostics 2019, 9, 4893–4908. [Google Scholar] [CrossRef]
- Gowda, N.M.; Wu, X.; Kumar, S.; Febbraio, M.; Gowda, D.C. CD36 contributes to malaria parasite-induced pro-inflammatory cytokine production and NK and T cell activation by dendritic cells. PLoS ONE 2013, 8, e77604. [Google Scholar] [CrossRef]
- Pfeiler, S.; Thakur, M.; Grunauer, P.; Megens, R.T.A.; Joshi, U.; Coletti, R.; Samara, V.; Muller-Stoy, G.; Ishikawa-Ankerhold, H.; Stark, K.; et al. CD36-triggered cell invasion and persistent tissue colonization by tumor microvesicles during metastasis. FASEB J. 2019, 33, 1860–1872. [Google Scholar] [CrossRef] [PubMed]
- Rech, A.J.; Mick, R.; Martin, S.; Recio, A.; Aqui, N.A.; Powell, D.J., Jr.; Colligon, T.A.; Trosko, J.A.; Leinbach, L.I.; Pletcher, C.H.; et al. CD25 blockade depletes and selectively reprograms regulatory T cells in concert with immunotherapy in cancer patients. Sci. Transl. Med. 2012, 4, 134ra162. [Google Scholar] [CrossRef] [PubMed]
- Sutmuller, R.P.; van Duivenvoorde, L.M.; van Elsas, A.; Schumacher, T.N.; Wildenberg, M.E.; Allison, J.P.; Toes, R.E.; Offringa, R.; Melief, C.J. Synergism of cytotoxic T lymphocyte-associated antigen 4 blockade and depletion of CD25+ regulatory T cells in antitumor therapy reveals alternative pathways for suppression of autoreactive cytotoxic T lymphocyte responses. J. Exp. Med. 2001, 194, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Kohrt, H.; Sagiv-Barfi, I.; Ajami, B.; Axtell, R.C.; Zhou, G.; Rajapaksa, R.; Green, M.R.; Torchia, J.; Brody, J.; et al. Depleting tumor-specific Tregs at a single site eradicates disseminated tumors. J. Clin. Investig. 2013, 123, 2447–2463. [Google Scholar] [CrossRef]
- Walter, S.; Weinschenk, T.; Stenzl, A.; Zdrojowy, R.; Pluzanska, A.; Szczylik, C.; Staehler, M.; Brugger, W.; Dietrich, P.Y.; Mendrzyk, R.; et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat. Med. 2012, 18, 1254–1261. [Google Scholar] [CrossRef]
- Wang, H.; Franco, F.; Tsui, Y.C.; Xie, X.; Trefny, M.P.; Zappasodi, R.; Mohmood, S.R.; Fernandez-Garcia, J.; Tsai, C.H.; Schulze, I.; et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat. Immunol. 2020, 21, 298–308. [Google Scholar] [CrossRef]
- Liu, R.; Pugh, G.H.; Tevonian, E.; Thompson, K.; Lauffenburger, D.A.; Kern, P.A.; Nikolajczyk, B.S. Regulatory T Cells Control Effector T Cell Inflammation in Human Prediabetes. Diabetes 2022, 71, 264–274. [Google Scholar] [CrossRef]
- Perry, J.S.A.; Russler-Germain, E.V.; Zhou, Y.W.; Purtha, W.; Cooper, M.L.; Choi, J.; Schroeder, M.A.; Salazar, V.; Egawa, T.; Lee, B.C.; et al. Transfer of Cell-Surface Antigens by Scavenger Receptor CD36 Promotes Thymic Regulatory T Cell Receptor Repertoire Development and Allo-tolerance. Immunity 2018, 48, 923–936.e4. [Google Scholar] [CrossRef]
- Lee, J.G.; Jaeger, K.E.; Seki, Y.; Wei Lim, Y.; Cunha, C.; Vuchkovska, A.; Nelson, A.J.; Nikolai, A.; Kim, D.; Nishimura, M.; et al. Human CD36(hi) monocytes induce Foxp3+ CD25+ T cells with regulatory functions from CD4 and CD8 subsets. Immunology 2021, 163, 293–309. [Google Scholar] [CrossRef]
- Miao, Y.; Zhang, C.; Yang, L.; Zeng, X.; Hu, Y.; Xue, X.; Dai, Y.; Wei, Z. The activation of PPARgamma enhances Treg responses through up-regulating CD36/CPT1-mediated fatty acid oxidation and subsequent N-glycan branching of TbetaRII/IL-2Ralpha. Cell Commun. Signal. 2022, 20, 48. [Google Scholar] [CrossRef]
- Geys, L.; Vranckx, C.; Lijnen, H.R.; Scroyen, I. CD36 deficiency blunts effects of diet on regulatory T cells in murine gonadal adipose tissue and mesenteric lymph nodes. Cell Immunol. 2015, 298, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Pages, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Farhood, B.; Najafi, M.; Mortezaee, K. CD8+ cytotoxic T lymphocytes in cancer immunotherapy: A review. J. Cell. Physiol. 2019, 234, 8509–8521. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Fearon, D.T. T cell exclusion, immune privilege, and the tumor microenvironment. Science 2015, 348, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Bi, E.; Lu, Y.; Su, P.; Huang, C.; Liu, L.; Wang, Q.; Yang, M.; Kalady, M.F.; Qian, J.; et al. Cholesterol Induces CD8+ T Cell Exhaustion in the Tumor Microenvironment. Cell Metab. 2019, 30, 143–156.e5. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Bai, Y.; Xiong, Y.; Zhang, J.; Chen, S.; Zheng, X.; Meng, X.; Li, L.; Wang, J.; Xu, C.; et al. Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature 2016, 531, 651–655. [Google Scholar] [CrossRef]
- Ayala, A.; Munoz, M.F.; Arguelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Angeli, J.P.F.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascon, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Ma, X.; Xiao, L.; Liu, L.; Ye, L.; Su, P.; Bi, E.; Wang, Q.; Yang, M.; Qian, J.; Yi, Q. CD36-mediated ferroptosis dampens intratumoral CD8+ T cell effector function and impairs their antitumor ability. Cell Metab. 2021, 33, 1001–1012.e5. [Google Scholar] [CrossRef]
- Imai, H.; Matsuoka, M.; Kumagai, T.; Sakamoto, T.; Koumura, T. Lipid Peroxidation-Dependent Cell Death Regulated by GPx4 and Ferroptosis. Curr. Top. Microbiol. Immunol. 2017, 403, 143–170. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Chaudhary, O.; Rodriguez-Morales, P.; Sun, X.; Chen, D.; Zappasodi, R.; Xu, Z.; Pinto, A.F.M.; Williams, A.; Schulze, I.; et al. Uptake of oxidized lipids by the scavenger receptor CD36 promotes lipid peroxidation and dysfunction in CD8+ T cells in tumors. Immunity 2021, 54, 1561–1577.e7. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.H.; Beury, D.W.; Ostrand-Rosenberg, S. Myeloid-Derived Suppressor Cells: Critical Cells Driving Immune Suppression in the Tumor Microenvironment. Adv. Cancer Res. 2015, 128, 95–139. [Google Scholar] [CrossRef]
- Al-Khami, A.A.; Zheng, L.; Del Valle, L.; Hossain, F.; Wyczechowska, D.; Zabaleta, J.; Sanchez, M.D.; Dean, M.J.; Rodriguez, P.C.; Ochoa, A.C. Exogenous lipid uptake induces metabolic and functional reprogramming of tumor-associated myeloid-derived suppressor cells. Oncoimmunology 2017, 6, e1344804. [Google Scholar] [CrossRef] [PubMed]
- Salmaninejad, A.; Valilou, S.F.; Soltani, A.; Ahmadi, S.; Abarghan, Y.J.; Rosengren, R.J.; Sahebkar, A. Tumor-associated macrophages: Role in cancer development and therapeutic implications. Cell. Oncol. 2019, 42, 591–608. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.C.; Everts, B.; Ivanova, Y.; O’Sullivan, D.; Nascimento, M.; Smith, A.M.; Beatty, W.; Love-Gregory, L.; Lam, W.Y.; O’Neill, C.M.; et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 2014, 15, 846–855. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, H.; Li, Y.; Zhang, Y.; Bian, Y.; Zeng, Y.; Yao, X.; Wan, J.; Chen, X.; Li, J.; et al. S100A4 enhances protumor macrophage polarization by control of PPAR-gamma-dependent induction of fatty acid oxidation. J. Immunother. Cancer 2021, 9, e002548. [Google Scholar] [CrossRef]
- Su, P.; Wang, Q.; Bi, E.; Ma, X.; Liu, L.; Yang, M.; Qian, J.; Yi, Q. Enhanced Lipid Accumulation and Metabolism Are Required for the Differentiation and Activation of Tumor-Associated Macrophages. Cancer Res. 2020, 80, 1438–1450. [Google Scholar] [CrossRef]
- Frank, A.C.; Ebersberger, S.; Fink, A.F.; Lampe, S.; Weigert, A.; Schmid, T.; Ebersberger, I.; Syed, S.N.; Brune, B. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nat. Commun. 2019, 10, 1135. [Google Scholar] [CrossRef]
- Helming, L.; Winter, J.; Gordon, S. The scavenger receptor CD36 plays a role in cytokine-induced macrophage fusion. J. Cell Sci. 2009, 122, 453–459. [Google Scholar] [CrossRef]
- Aguirre, L.A.; Montalban-Hernandez, K.; Avendano-Ortiz, J.; Marin, E.; Lozano, R.; Toledano, V.; Sanchez-Maroto, L.; Terron, V.; Valentin, J.; Pulido, E.; et al. Tumor stem cells fuse with monocytes to form highly invasive tumor-hybrid cells. Oncoimmunology 2020, 9, 1773204. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Pankova, D.; Chen, Y.; Terajima, M.; Schliekelman, M.J.; Baird, B.N.; Fahrenholtz, M.; Sun, L.; Gill, B.J.; Vadakkan, T.J.; Kim, M.P.; et al. Cancer-Associated Fibroblasts Induce a Collagen Cross-link Switch in Tumor Stroma. Mol. Cancer Res. 2016, 14, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Ohlund, D.; Elyada, E.; Tuveson, D. Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 2014, 211, 1503–1523. [Google Scholar] [CrossRef]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef]
- Gascard, P.; Tlsty, T.D. Carcinoma-associated fibroblasts: Orchestrating the composition of malignancy. Genes Dev. 2016, 30, 1002–1019. [Google Scholar] [CrossRef]
- Li, D.; Yu, H.; Guo, Z.; Li, S.; Li, Y.; Guo, Y.; Zhong, H.; Xiong, H.; Liu, Z. SERS analysis of carcinoma-associated fibroblasts in a tumor microenvironment based on targeted 2D nanosheets. Nanoscale 2020, 12, 2133–2141. [Google Scholar] [CrossRef]
- DeFilippis, R.A.; Chang, H.; Dumont, N.; Rabban, J.T.; Chen, Y.Y.; Fontenay, G.V.; Berman, H.K.; Gauthier, M.L.; Zhao, J.; Hu, D.; et al. CD36 repression activates a multicellular stromal program shared by high mammographic density and tumor tissues. Cancer Discov. 2012, 2, 826–839. [Google Scholar] [CrossRef]
- DeFilippis, R.A.; Fordyce, C.; Patten, K.; Chang, H.; Zhao, J.; Fontenay, G.V.; Kerlikowske, K.; Parvin, B.; Tlsty, T.D. Stress signaling from human mammary epithelial cells contributes to phenotypes of mammographic density. Cancer Res. 2014, 74, 5032–5044. [Google Scholar] [CrossRef]
- Jabbari, K.; Winkelmaier, G.; Andersen, C.; Yaswen, P.; Quilici, D.; Furuta, S.; Cheng, Q.; Parvin, B. Protein Ligands in the Secretome of CD36+ Fibroblasts Induce Growth Suppression in a Subset of Breast Cancer Cell Lines. Cancers 2021, 13, 4521. [Google Scholar] [CrossRef]
- Gong, J.; Lin, Y.; Zhang, H.; Liu, C.; Cheng, Z.; Yang, X.; Zhang, J.; Xiao, Y.; Sang, N.; Qian, X.; et al. Reprogramming of lipid metabolism in cancer-associated fibroblasts potentiates migration of colorectal cancer cells. Cell Death Dis. 2020, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- Nan, P.; Dong, X.; Bai, X.; Lu, H.; Liu, F.; Sun, Y.; Zhao, X. Tumor-stroma TGF-beta1-THBS2 feedback circuit drives pancreatic ductal adenocarcinoma progression via integrin alphavbeta3/CD36-mediated activation of the MAPK pathway. Cancer Lett. 2022, 528, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Yelle, N.; Venugopal, C.; Singh, S.K. EMT: Mechanisms and therapeutic implications. Pharmacol. Ther. 2018, 182, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, C.L.; San Juan, B.P.; Lim, E.; Weinberg, R.A. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016, 35, 645–654. [Google Scholar] [CrossRef]
- Gyamfi, J.; Yeo, J.H.; Kwon, D.; Min, B.S.; Cha, Y.J.; Koo, J.S.; Jeong, J.; Lee, J.; Choi, J. Interaction between CD36 and FABP4 modulates adipocyte-induced fatty acid import and metabolism in breast cancer. NPJ Breast Cancer 2021, 7, 129. [Google Scholar] [CrossRef]
- Wang, J.; Wen, T.; Li, Z.; Che, X.; Gong, L.; Jiao, Z.; Qu, X.; Liu, Y. CD36 upregulates DEK transcription and promotes cell migration and invasion via GSK-3beta/beta-catenin-mediated epithelial-to-mesenchymal transition in gastric cancer. Aging 2020, 13, 1883–1897. [Google Scholar] [CrossRef]
- Wang, C.; Yang, Z.; Xu, E.; Shen, X.; Wang, X.; Li, Z.; Yu, H.; Chen, K.; Hu, Q.; Xia, X.; et al. Apolipoprotein C-II induces EMT to promote gastric cancer peritoneal metastasis via PI3K/AKT/mTOR pathway. Clin. Transl. Med. 2021, 11, e522. [Google Scholar] [CrossRef]
- Nath, A.; Li, I.; Roberts, L.R.; Chan, C. Elevated free fatty acid uptake via CD36 promotes epithelial-mesenchymal transition in hepatocellular carcinoma. Sci. Rep. 2015, 5, 14752. [Google Scholar] [CrossRef]
- Deng, M.; Cai, X.; Long, L.; Xie, L.; Ma, H.; Zhou, Y.; Liu, S.; Zeng, C. CD36 promotes the epithelial-mesenchymal transition and metastasis in cervical cancer by interacting with TGF-beta. J. Transl. Med. 2019, 17, 352. [Google Scholar] [CrossRef]
- Hou, Y.; Wu, M.; Wei, J.; Ren, Y.; Du, C.; Wu, H.; Li, Y.; Shi, Y. CD36 is involved in high glucose-induced epithelial to mesenchymal transition in renal tubular epithelial cells. Biochem. Biophys. Res. Commun. 2015, 468, 281–286. [Google Scholar] [CrossRef]
- Folkman, J. Tumor angiogenesis: Therapeutic implications. N. Engl. J. Med. 1971, 285, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Febbraio, M.; Hajjar, D.P.; Silverstein, R.L. CD36: A class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Investig. 2001, 108, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Osz, K.; Ross, M.; Petrik, J. The thrombospondin-1 receptor CD36 is an important mediator of ovarian angiogenesis and folliculogenesis. Reprod. Biol. Endocrinol. 2014, 12, 21. [Google Scholar] [CrossRef]
- Jimenez, B.; Volpert, O.V.; Crawford, S.E.; Febbraio, M.; Silverstein, R.L.; Bouck, N. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nat. Med. 2000, 6, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lawler, J. Thrombospondin-based antiangiogenic therapy. Microvasc. Res. 2007, 74, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Lawler, P.R.; Lawler, J. Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2. Cold Spring Harb. Perspect. Med. 2012, 2, a006627. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Cork, S.M.; Sandberg, E.M.; Devi, N.S.; Zhang, Z.; Klenotic, P.A.; Febbraio, M.; Shim, H.; Mao, H.; Tucker-Burden, C.; et al. Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer Res. 2009, 69, 1212–1220. [Google Scholar] [CrossRef]
- Klenotic, P.A.; Huang, P.; Palomo, J.; Kaur, B.; Van Meir, E.G.; Vogelbaum, M.A.; Febbraio, M.; Gladson, C.L.; Silverstein, R.L. Histidine-rich glycoprotein modulates the anti-angiogenic effects of vasculostatin. Am. J. Pathol. 2010, 176, 2039–2050. [Google Scholar] [CrossRef]
- Dong, L.; Yuan, Y.; Opansky, C.; Chen, Y.; Aguilera-Barrantes, I.; Wu, S.; Yuan, R.; Cao, Q.; Cheng, Y.C.; Sahoo, D.; et al. Diet-induced obesity links to ER positive breast cancer progression via LPA/PKD-1-CD36 signaling-mediated microvascular remodeling. Oncotarget 2017, 8, 22550–22562. [Google Scholar] [CrossRef]
- Koch, M.; Hussein, F.; Woeste, A.; Grundker, C.; Frontzek, K.; Emons, G.; Hawighorst, T. CD36-mediated activation of endothelial cell apoptosis by an N-terminal recombinant fragment of thrombospondin-2 inhibits breast cancer growth and metastasis in vivo. Breast Cancer Res. Treat. 2011, 128, 337–346. [Google Scholar] [CrossRef]
- Russell, S.; Duquette, M.; Liu, J.; Drapkin, R.; Lawler, J.; Petrik, J. Combined therapy with thrombospondin-1 type I repeats (3TSR) and chemotherapy induces regression and significantly improves survival in a preclinical model of advanced stage epithelial ovarian cancer. FASEB J. 2015, 29, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Jeanne, A.; Schneider, C.; Martiny, L.; Dedieu, S. Original insights on thrombospondin-1-related antireceptor strategies in cancer. Front. Pharmacol. 2015, 6, 252. [Google Scholar] [CrossRef] [PubMed]
- Kyriakides, T.R.; Maclauchlan, S. The role of thrombospondins in wound healing, ischemia, and the foreign body reaction. J. Cell Commun. Signal. 2009, 3, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Kunstfeld, R.; Hawighorst, T.; Streit, M.; Hong, Y.K.; Nguyen, L.; Brown, L.F.; Detmar, M. Thrombospondin-2 overexpression in the skin of transgenic mice reduces the susceptibility to chemically induced multistep skin carcinogenesis. J. Dermatol. Sci. 2014, 74, 106–115. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, P.C.; Tang, C.H.; Lin, L.W.; Tsai, C.H.; Chu, C.Y.; Lin, T.H.; Huang, Y.L. Thrombospondin-2 promotes prostate cancer bone metastasis by the up-regulation of matrix metalloproteinase-2 through down-regulating miR-376c expression. J. Hematol. Oncol. 2017, 10, 33. [Google Scholar] [CrossRef]
- Harmsen, M.J.; Wong, C.F.C.; Mijatovic, V.; Griffioen, A.W.; Groenman, F.; Hehenkamp, W.J.K.; Huirne, J.A.F. Role of angiogenesis in adenomyosis-associated abnormal uterine bleeding and subfertility: A systematic review. Hum. Reprod. Update 2019, 25, 647–671. [Google Scholar] [CrossRef]
- Liu, Y.; Li, F.; Yang, Y.T.; Xu, X.D.; Chen, J.S.; Chen, T.L.; Chen, H.J.; Zhu, Y.B.; Lin, J.Y.; Li, Y.; et al. IGFBP2 promotes vasculogenic mimicry formation via regulating CD144 and MMP2 expression in glioma. Oncogene 2019, 38, 1815–1831. [Google Scholar] [CrossRef]
- Firlej, V.; Mathieu, J.R.; Gilbert, C.; Lemonnier, L.; Nakhle, J.; Gallou-Kabani, C.; Guarmit, B.; Morin, A.; Prevarskaya, N.; Delongchamps, N.B.; et al. Thrombospondin-1 triggers cell migration and development of advanced prostate tumors. Cancer Res. 2011, 71, 7649–7658. [Google Scholar] [CrossRef]
- Hendrix, M.J.; Seftor, E.A.; Hess, A.R.; Seftor, R.E. Vasculogenic mimicry and tumour-cell plasticity: Lessons from melanoma. Nat. Rev. Cancer 2003, 3, 411–421. [Google Scholar] [CrossRef]
- Seftor, R.E.; Hess, A.R.; Seftor, E.A.; Kirschmann, D.A.; Hardy, K.M.; Margaryan, N.V.; Hendrix, M.J. Tumor cell vasculogenic mimicry: From controversy to therapeutic promise. Am. J. Pathol. 2012, 181, 1115–1125. [Google Scholar] [CrossRef]
- Mwaikambo, B.R.; Yang, C.; Chemtob, S.; Hardy, P. Hypoxia up-regulates CD36 expression and function via hypoxia-inducible factor-1- and phosphatidylinositol 3-kinase-dependent mechanisms. J. Biol. Chem. 2009, 284, 26695–26707. [Google Scholar] [CrossRef] [PubMed]
- Bou Khzam, L.; Son, N.H.; Mullick, A.E.; Abumrad, N.A.; Goldberg, I.J. Endothelial cell CD36 deficiency prevents normal angiogenesis and vascular repair. Am. J. Transl. Res. 2020, 12, 7737–7761. [Google Scholar] [PubMed]
- Nolan, D.J.; Ginsberg, M.; Israely, E.; Palikuqi, B.; Poulos, M.G.; James, D.; Ding, B.S.; Schachterle, W.; Liu, Y.; Rosenwaks, Z.; et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell 2013, 26, 204–219. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, X.; Yan, S.; Li, J.; Jiang, C.; Huang, S.; Liu, S.; Zou, X.; Zhang, G.; Zou, J.; Liu, Q. CD36 and Its Role in Regulating the Tumor Microenvironment. Curr. Oncol. 2022, 29, 8133-8145. https://doi.org/10.3390/curroncol29110642
Liao X, Yan S, Li J, Jiang C, Huang S, Liu S, Zou X, Zhang G, Zou J, Liu Q. CD36 and Its Role in Regulating the Tumor Microenvironment. Current Oncology. 2022; 29(11):8133-8145. https://doi.org/10.3390/curroncol29110642
Chicago/Turabian StyleLiao, Xinzhi, Sheng Yan, Jialin Li, Chengming Jiang, Sigen Huang, Shengyin Liu, Xiaofeng Zou, Guoxi Zhang, Junrong Zou, and Quanliang Liu. 2022. "CD36 and Its Role in Regulating the Tumor Microenvironment" Current Oncology 29, no. 11: 8133-8145. https://doi.org/10.3390/curroncol29110642
APA StyleLiao, X., Yan, S., Li, J., Jiang, C., Huang, S., Liu, S., Zou, X., Zhang, G., Zou, J., & Liu, Q. (2022). CD36 and Its Role in Regulating the Tumor Microenvironment. Current Oncology, 29(11), 8133-8145. https://doi.org/10.3390/curroncol29110642




