Single-Cell Transcriptomics of Endothelial Cells in Upper and Lower Human Esophageal Squamous Cell Carcinoma
Abstract
1. Instruction
2. Methods and Materials
2.1. Sample Collection and Preparation
2.2. Single Cell RNA Sequencing and Sequencing Library Preparation
2.3. Preprocessing of scRNA-Seq Data
2.4. Clustering Analysis and Nonlinear Dimensionality Reduction
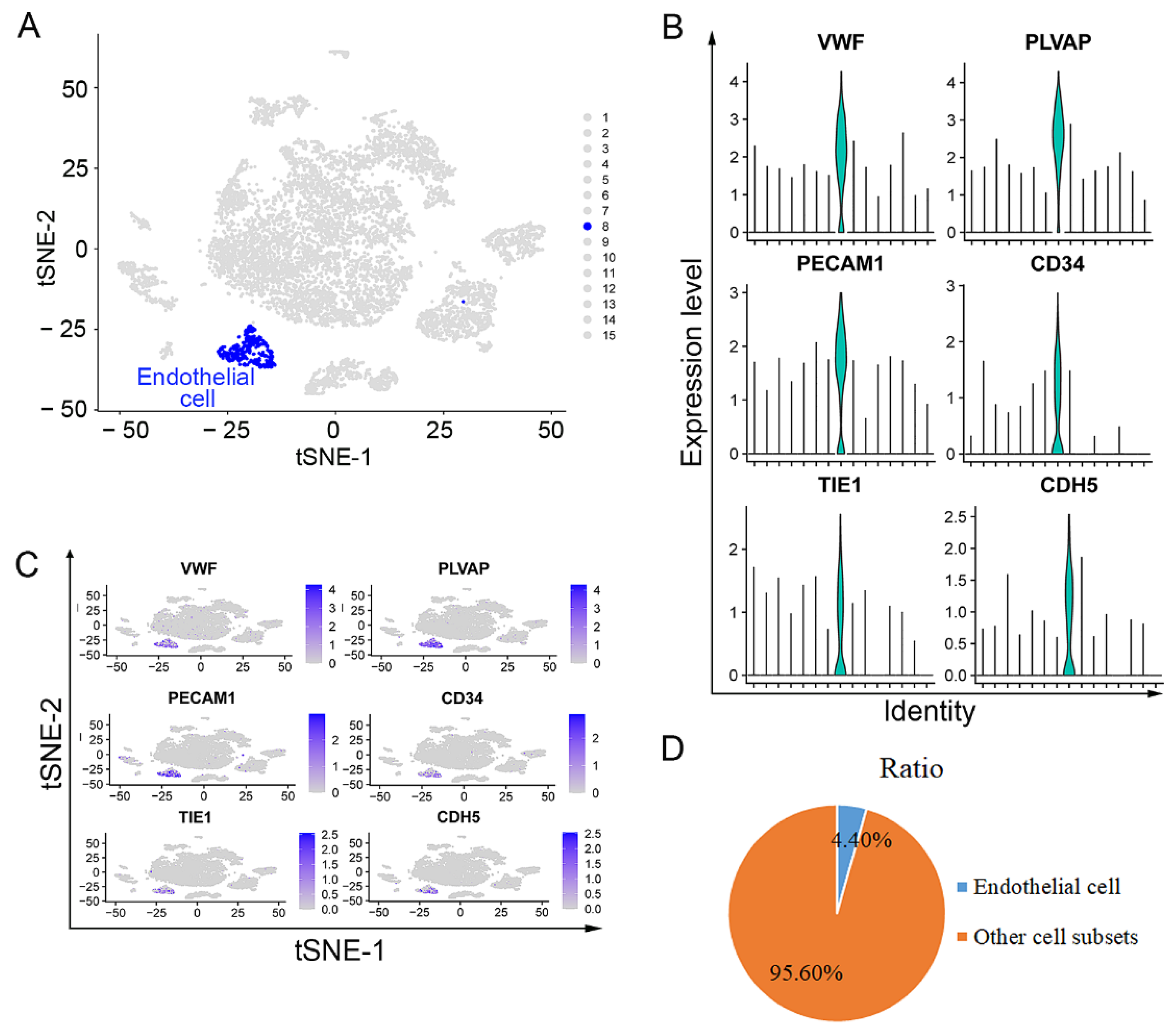
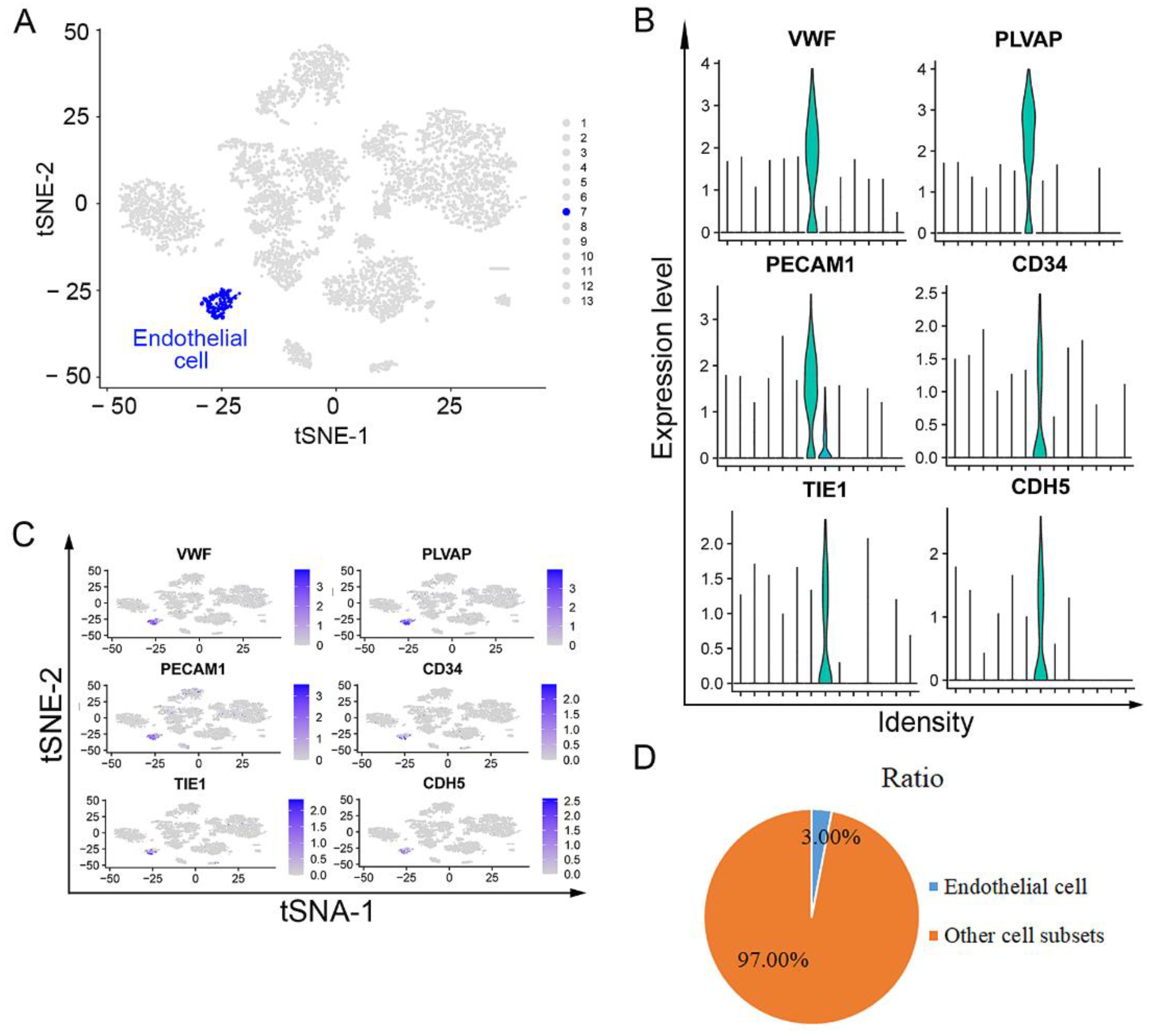
2.5. Identification and Cell Ratio of Endothelial Cells in the Upper and Lower ESCC
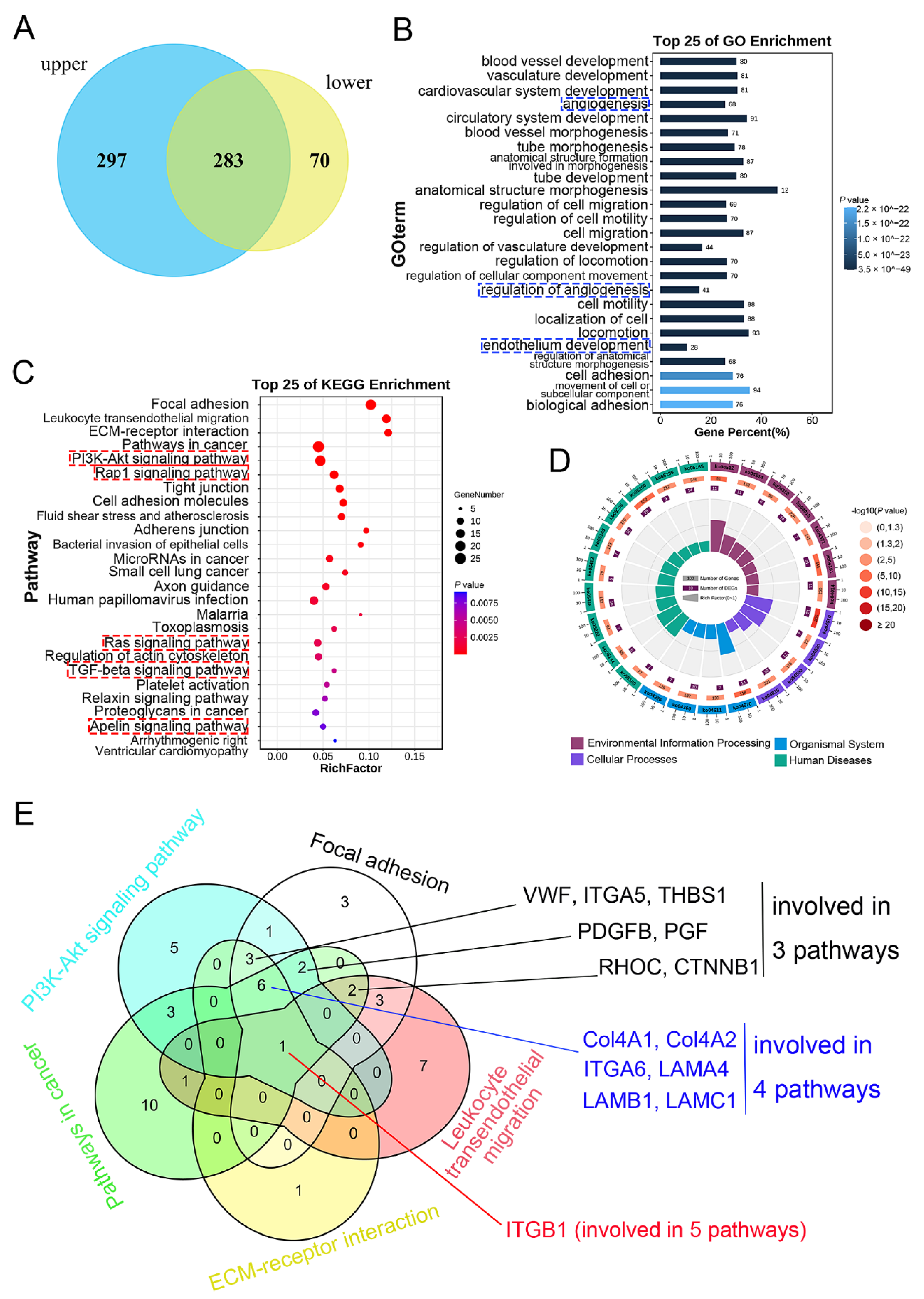
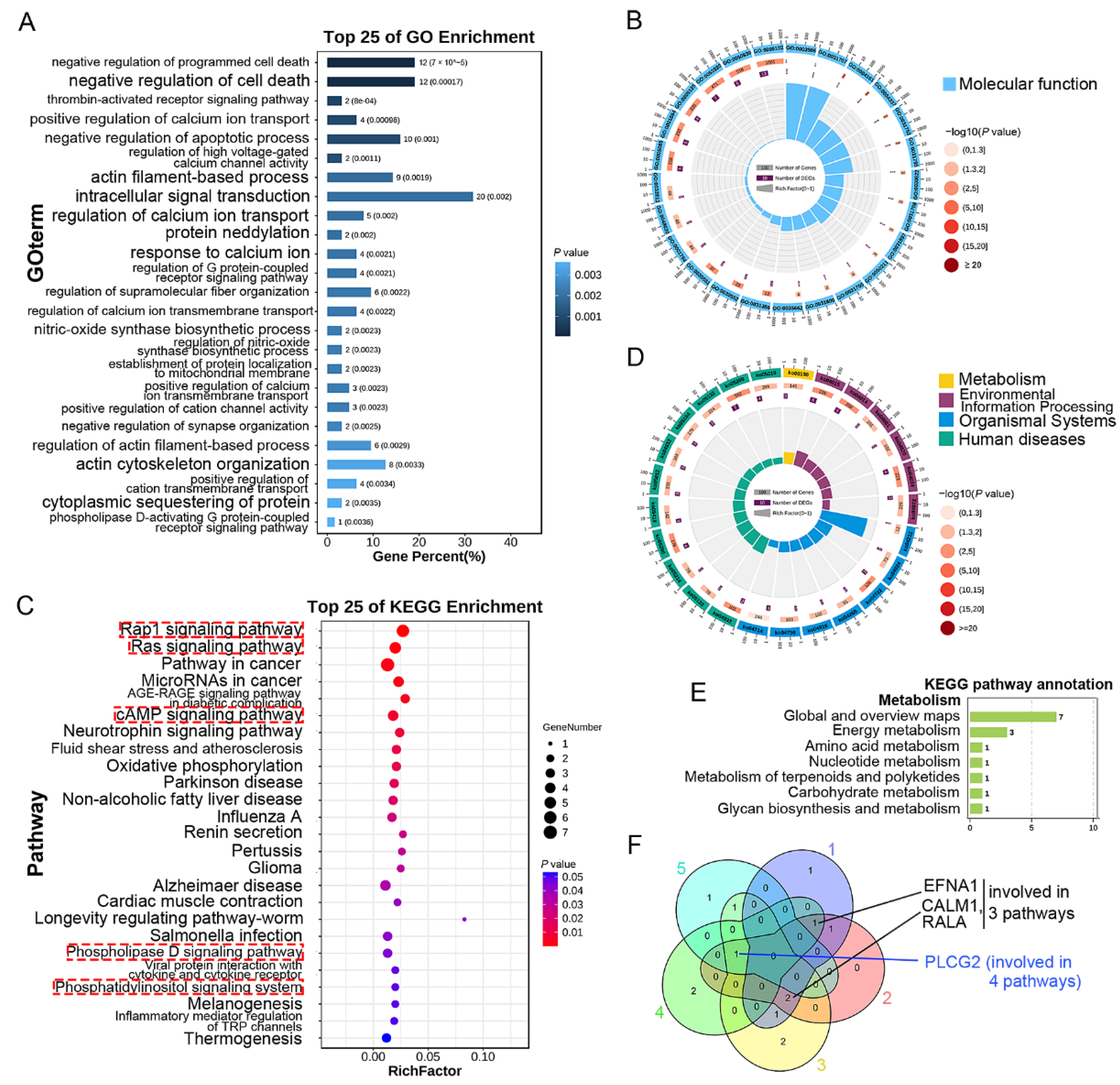
2.6. GO and KEGG Enrichment Analysis
2.7. Screening of Targeted Genes
2.8. Clustering Analysis of the Endothelial Cells and Nonlinear Dimensionality Reduction
2.9. Single-Cell Trajectory Analysis
3. Results
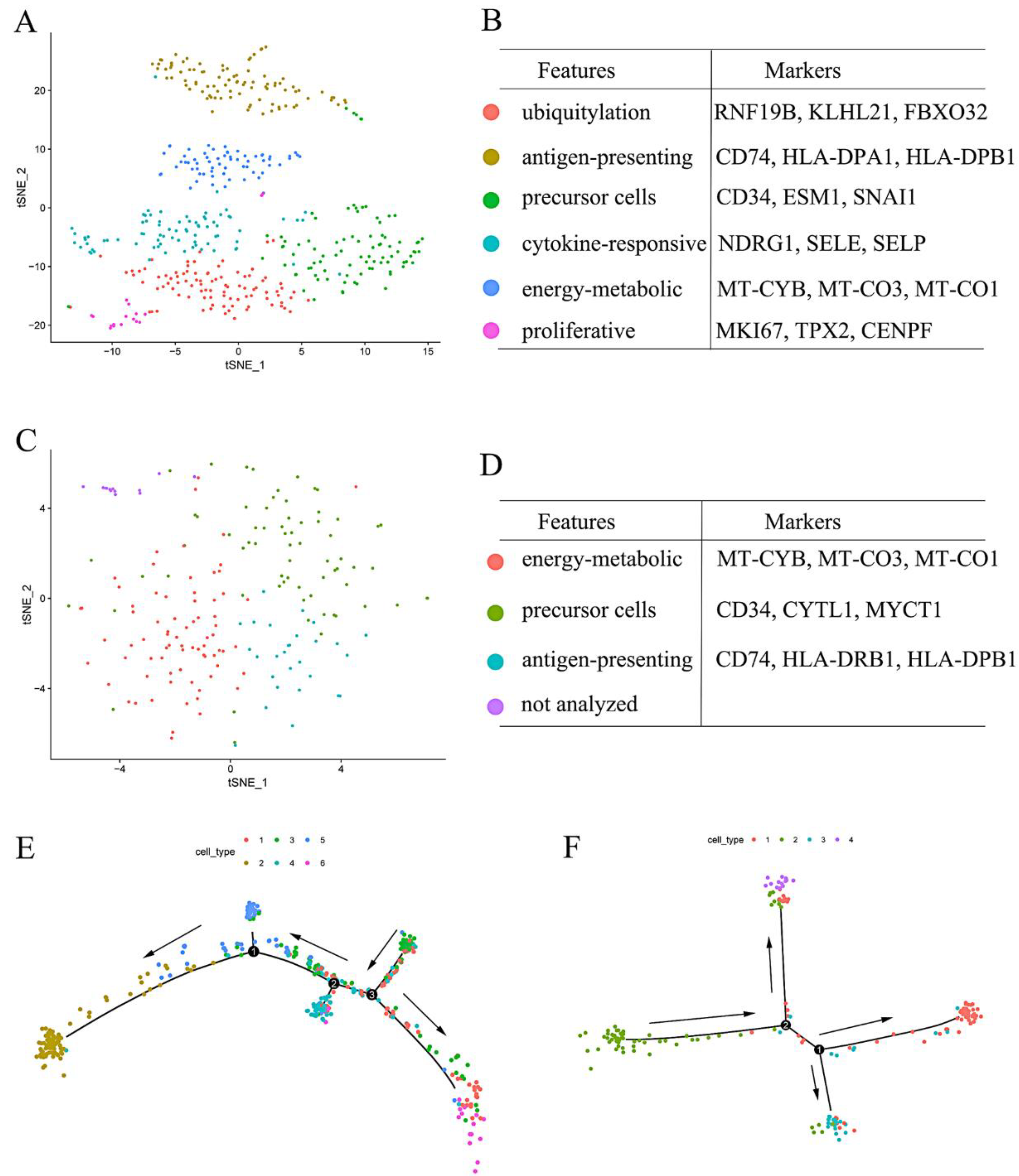
3.1. Endothelial Cells Isolated from Upper ESCC through scRNA Sequencing Analysis
3.2. Endothelial Cells Isolated from Lower ESCC through scRNA Sequencing Analysis
3.3. Detection, GO, and KEGG Enrichment Analysis of Common DEGs
3.4. Optimization of the Targeted Genes
3.5. GO and KEGG Enrichment Analysis of DEGs Only Expressed in Upper Endothelial Cells
3.6. GO and KEGG Enrichment Analysis of DEGs Only Expressed in Lower Endothelial Cells
3.7. Clustering Analysis of the Upper and Lower Endothelial Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhao, T.; Wang, J.; Chen, Y.; Zhang, R.; Lan, X.; Que, J. Etiology, cancer stem cells and potential diagnostic biomarkers for esophageal cancer. Cancer Lett. 2019, 458, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Chen, H.-D.; Yu, Y.-W.; Li, N.; Chen, W.-Q. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin. Med. J. 2021, 134, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Kumashiro, R.; Kubo, N.; Nakashima, Y.; Yoshida, R.; Yoshinaga, K.; Saeki, H.; Emi, Y.; Kakeji, Y.; Sakaguchi, Y.; et al. Alcohol drinking, cigarette smoking, and the development of squamous cell carcinoma of the esophagus: Epidemiology, clinical findings, and prevention. Int. J. Clin. Oncol. 2010, 15, 126–134. [Google Scholar] [CrossRef]
- Toh, Y.; Oki, E.; Ohgaki, K.; Sakamoto, Y.; Ito, S.; Egashira, A.; Saeki, H.; Kakeji, Y.; Morita, M.; Sakaguchi, Y.; et al. Alcohol drinking, cigarette smoking, and the development of squamous cell carcinoma of the esophagus: Molecular mechanisms of carcinogenesis. Int. J. Clin. Oncol. 2010, 15, 135–144. [Google Scholar]
- Islami, F.; Fedirko, V.; Tramacere, I.; Bagnardi, V.; Jenab, M.; Scotti, L.; Rota, M.; Corrao, G.; Garavello, W.; Schüz, J.; et al. Alcohol drinking and esophageal squamous cell carcinoma with focus on light-drinkers and never-smokers: A systematic review and meta-analysis. Int. J. Cancer 2011, 129, 2473–2484. [Google Scholar] [CrossRef]
- Barzi, A.; Lenz, H.-J. Angiogenesis-related agents in esophageal cancer. Expert Opin. Biol. Ther. 2012, 12, 1335–1345. [Google Scholar] [CrossRef]
- Valverde, C.M.; Macarulla, T.; Casado, E.; Ramos, F.J.; Martinelli, E.; Tabernero, J. Novel targets in gastric and esophageal cancer. Crit. Rev. Oncol. Hematol. 2006, 59, 128–138. [Google Scholar] [CrossRef]
- Bhat, A.A.; Nisar, S.; Maacha, S.; Carneiro-Lobo, T.C.; Akhtar, S.; Siveen, K.S.; Wani, N.A.; Rizwan, A.; Bagga, P.; Singh, M.; et al. Cytokine-chemokine network driven metastasis in esophageal cancer; promising avenue for targeted therapy. Mol. Cancer 2021, 20, 2. [Google Scholar] [CrossRef]
- Kuwano, H.; Nakajima, M.; Miyazaki, T.; Kato, H. Distinctive clinicopathological characteristics in esophageal squamous cell carcinoma. Ann. Thorac. Cardiovasc. Surg. 2003, 9, 6–13. [Google Scholar]
- Kuo, H.-Y.; Guo, J.-C.; Hsu, C.-H. Anti-PD-1 immunotherapy in advanced esophageal squamous cell carcinoma: A long-awaited breakthrough finally arrives. J. Formos. Med. Assoc. 2019, 119, 565–568. [Google Scholar] [CrossRef]
- Kato, K.; Cho, B.C.; Takahashi, M.; Okada, M.; Lin, C.Y.; Chin, K.; Kadowaki, S.; Ahn, M.J.; Hamamoto, Y.; Doki, Y.; et al. Nivolumab versus chemotherapy in patients with advanced oesophageal squamous cell carcinoma refractory or intolerant to previous chemotherapy (ATTRACTION-3): A multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019, 20, 1506–1517. [Google Scholar] [CrossRef]
- Sha, Y.Q.; Yang, L.; Lv, Y.G. ERK1/2 and Akt phosphorylation were essential for MGF E peptide regulating cell morphology and mobility but not proangiogenic capacity of BMSCs under severe hypoxia. Cell Biochem. Funct. 2018, 36, 155–165. [Google Scholar] [CrossRef]
- Sha, Y.; Yang, L.; Lv, Y. MGF E peptide improves anterior cruciate ligament repair by inhibiting hypoxia-induced cell apoptosis and accelerating angiogenesis. J. Cell. Physiol. 2018, 234, 8846–8861. [Google Scholar] [CrossRef]
- Sakurai, T.; Okumura, H.; Matsumoto, M.; Uchikado, Y.; Setoyama, T.; Omoto, I.; Owaki, T.; Maemura, K.; Ishigami, S.; Natsugoe, S. The expression of LC-3 is related to tumor suppression through angiogenesis in esophageal cancer. Med. Oncol. 2013, 30, 701. [Google Scholar] [CrossRef]
- Yamamoto, M.; Ozawa, S.; Ninomiya, Y.; Koyanagi, K.; Oguma, J.; Kazuno, A.; Hara, H.; Yatabe, K.; Kajiwara, H.; Nakamura, N.; et al. Plasma vasohibin-1 and vasohibin-2 are useful biomarkers in patients with esophageal squamous cell carcinoma. Esophagus 2020, 17, 289–297. [Google Scholar] [CrossRef]
- Gan, W.; Wang, W.; Li, T.; Zhang, R.; Hou, Y.; Lv, S.; Zeng, Z.; Yan, Z.; Yang, M. Prognostic Values and Underlying Regulatory Network of Cohesin Subunits in Esophageal Carcinoma. J. Cancer 2022, 13, 1588–1602. [Google Scholar] [CrossRef]
- Chen, Z.C.; Zhao, M.N.; Liang, J.Q.; Hu, Z.Y.; Huang, Y.W.; Li, M.; Pang, Y.R.; Lu, T.; Sui, Q.H.; Zhan, C.; et al. Dissecting the single-cell transcriptome network underlying esophagus non-malignant tissues and esophageal squamous cell carcinoma. EBioMedicine 2021, 69, 103459. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, Z.; Han, Y.; Han, L.; Zou, X.; Zhou, B.; Hu, R.; Hao, J.; Bai, S.; Xiao, H.; et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat. Commun. 2020, 11, 6268. [Google Scholar] [CrossRef]
- Chang, J.; Tan, W.L.; Ling, Z.Q.; Xi, R.B.; Shao, M.M.; Chen, M.J.; Luo, Y.Y.; Zhao, Y.J.; Liu, Y.; Huang, X.C.; et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol-drinking-related mutation signature and genomic alterations. Nat. Commun. 2017, 26, 15290. [Google Scholar] [CrossRef]
- Kim, J.; Bowlby, R.; Mungall, A.J.; Gordon Robertson, A.; Odze, R.D.; Cherniack, A.D.; Shih, J.; Pedamallu, C.S.; Cibulskis, C.; Dunford, A.; et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175. [Google Scholar]
- Rice, T.W.; Ishwaran, H.; Hofstetter, W.L.; Kelsen, D.P.; Apperson-Hansen, C.; Blackstone, E.H. Worldwide Esophageal Cancer Collaboration Investigators, Recommendations for pathologic staging (pTNM) of cancer of the esophageal and esophagogastric junction for the 8th edition AJCC/UICC staging manuals. Dis. Esophagus 2016, 29, 897–905. [Google Scholar] [CrossRef]
- Gao, C.; Zhang, M.; Chen, L. The Comparison of Two Single-cell Sequencing Platforms: BD Rhapsody and 10x Genomics Chromium. Curr. Genom. 2020, 21, 602–609. [Google Scholar] [CrossRef]
- Zhang, X.; Peng, L.; Luo, Y.; Zhang, S.; Pu, Y.; Chen, Y.; Guo, W.; Yao, J.; Shao, M.; Fan, W.; et al. Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat. Commun. 2021, 12, 5291. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.-R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef]
- Sha, Y.; Zhang, B.; Chen, L.; Wang, C.; Sun, T. Dehydrocorydaline Accelerates Cell Proliferation and Extracellular Matrix Synthesis of TNFα-Treated Human Chondrocytes by Targeting Cox2 through JAK1-STAT3 Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 7268. [Google Scholar] [CrossRef]
- Augustin, H.G.; Koh, G.Y. Antiangiogenesis: Vessel Regression, Vessel Normalization, or Both? Cancer Res. 2022, 82, 15–17. [Google Scholar] [CrossRef]
- Zhu, H.; Yang, X.; Ding, Y.; Liu, J.; Lu, J.; Zhan, L.; Qin, Q.; Zhang, H.; Chen, X.; Yang, Y.; et al. Recombinant human endostatin enhances the radioresponse in esophageal squamous cell carcinoma by normalizing tumor vasculature and reducing hypoxia. Sci. Rep. 2015, 5, 14503. [Google Scholar] [CrossRef][Green Version]
- Ma, S.; Lu, C.-C.; Yang, L.-Y.; Wang, J.-J.; Wang, B.-S.; Cai, H.-Q.; Hao, J.-J.; Xu, X.; Cai, Y.; Zhang, Y.; et al. ANXA2 promotes esophageal cancer progression by activating MYC-HIF1A-VEGF axis. J. Exp. Clin. Cancer Res. 2018, 37, 183. [Google Scholar] [CrossRef]
- Zhao, Y.; Adjei, A.A. Targeting Angiogenesis in Cancer Therapy: Moving Beyond Vascular Endothelial Growth Factor. Oncologist 2015, 20, 660–673. [Google Scholar] [CrossRef]
- Sha, Y.Q.; Afandi, R.; Zhang, B.B.; Yang, L.; Lv, G. MGF E peptide pretreatment improves collagen synthesis and cell proliferation of injured human ACL fibroblasts via MEK-ERK1/2 signaling pathway. Growth Factors 2017, 35, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Duncan, E.D.; Han, K.-J.; Trout, M.A.; Prekeris, R. Ubiquitylation by Rab40b/Cul5 regulates Rap2 localization and activity during cell migration. J. Cell Biol. 2022, 221, e202107114. [Google Scholar] [CrossRef] [PubMed]
- Elangovan, M.; Ka, J.; Pak, B.; Choi, W.; Oh, S.; Jin, S.; Yoo, Y.J. Ubiquitin-conjugating enzyme V variant 1 enables cellular responses toward fibroblast growth factor signaling in endothelium. FASEB J. 2021, 36, e22103. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Sun, X.J.; Niu, S.S.; Yang, C.Y.; Hao, Y.P.; Kou, J.T.; Li, X.Z.; Wang, X.X. Overexpression of IQGAP1 promotes the angiogenesis of esophageal squamous cell carcinoma through the AKT and ERK-mediated VEGF-VEGFR2 signaling pathway. Oncol. Rep. 2018, 40, 1795–1802. [Google Scholar] [CrossRef]
- Zheng, Z.-Y.; Chu, M.-Y.; Lin, W.; Zheng, Y.-Q.; Xu, X.-E.; Chen, Y.; Liao, L.-D.; Wu, Z.-Y.; Wang, S.-H.; Li, E.-M.; et al. Blocking STAT3 signaling augments MEK/ERK inhibitor efficacy in esophageal squamous cell carcinoma. Cell Death Dis. 2022, 13, 496. [Google Scholar] [CrossRef]
- Fang, J.; Chu, L.; Li, C.; Chen, Y.; Hu, F.; Zhang, X.; Zhao, H.; Liu, Z.; Xu, Q. JAK2 inhibitor blocks the inflammation and growth of esophageal squamous cell carcinoma in vitro through the JAK/STAT3 pathway. Oncol. Rep. 2014, 33, 494–502. [Google Scholar] [CrossRef]
- Thomas, P.; Pranatharthi, A.; Ross, C.; Srivastava, S. Rhoc: A fascinating journey from a cytoskeletal organizer to a Cancer stem cell therapeutic target. J. Exp. Clin. Cancer Res. 2019, 38, 328. [Google Scholar] [CrossRef]


| Metabolism | Genes |
|---|---|
| Lipid metabolism | PLPP1, PLPP3, PLA2G16, SPHK1, DGKZ, GPX1 |
| Glycan biosynthesis and metabolism | GNS, CSGALNACT1, NDST1, GALNT18 |
| Metabolism of cofactors and vitamin | HMOX1, NNMT, NT5E |
| Amino acid metabolism | SAT1, KMT2E |
| Nucleotide metabolism | NT5E, GUK1 |
| Energy metabolism | MT-ATP6, MT-ND5 |
| Carbohydrate metabolism | CYB5R3, PLCB1 |
| Metabolism of other amino acid | GPX1 |
| Metabolism | Genes |
|---|---|
| Energy metabolism | UQCR11, NDUFB2, COX7A2 |
| Amino acid metabolism | COLGALT1 |
| Nucleotide metabolism | PDE4B |
| Metabolism of terpenoids and polyketides | FDPS |
| Carbohydrate metabolism | PLCG2 |
| Glycan biosynthesis and metabolism | COLGALT1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sha, Y.; Hong, H.; Cai, W.; Sun, T. Single-Cell Transcriptomics of Endothelial Cells in Upper and Lower Human Esophageal Squamous Cell Carcinoma. Curr. Oncol. 2022, 29, 7680-7694. https://doi.org/10.3390/curroncol29100607
Sha Y, Hong H, Cai W, Sun T. Single-Cell Transcriptomics of Endothelial Cells in Upper and Lower Human Esophageal Squamous Cell Carcinoma. Current Oncology. 2022; 29(10):7680-7694. https://doi.org/10.3390/curroncol29100607
Chicago/Turabian StyleSha, Yongqiang, Huhai Hong, Wenjie Cai, and Tao Sun. 2022. "Single-Cell Transcriptomics of Endothelial Cells in Upper and Lower Human Esophageal Squamous Cell Carcinoma" Current Oncology 29, no. 10: 7680-7694. https://doi.org/10.3390/curroncol29100607
APA StyleSha, Y., Hong, H., Cai, W., & Sun, T. (2022). Single-Cell Transcriptomics of Endothelial Cells in Upper and Lower Human Esophageal Squamous Cell Carcinoma. Current Oncology, 29(10), 7680-7694. https://doi.org/10.3390/curroncol29100607




