Mutational Landscape of Patients Referred for Elevated Hemoglobin Level
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects (Patient Selection Criteria)
2.2. Next Generation Sequencing Assay
3. Results
3.1. Patient Cohort
3.2. Tier I/II Variants Identified in JAK2-Positive Patients with Erythrocytosis
3.3. Tier I/II Variants Identified in JAK2-Negative Patients
3.4. Tier III Variants (Unknown Significance)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scott, L.M.; Tong, W.; Levine, R.L.; Scott, M.A.; Beer, P.A.; Stratton, M.R.; Futreal, P.A.; Erber, W.N.; McMullin, M.F.; Harrison, C.N.; et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N. Engl. J. Med. 2007, 356, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Baxter, E.J.; Scott, L.M.; Campbell, P.J.; East, C.; Fourouclas, N.; Swanton, S.; Vassiliou, G.S.; Bench, A.J.; Boyd, E.M.; Curtin, N.; et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005, 365, 1054–1061, Erratum in Lancet 2005, 366, 122. [Google Scholar] [CrossRef]
- Gong, J.Z.; Cook, J.R.; Greiner, T.C.; Hedvat, C.; Hill, C.E.; Lim, M.S.; Longtine, J.A.; Sabath, D.; Wang, Y.L.; Association for Molecular Pathology. Laboratory practice guidelines for detecting and reporting JAK2 and MPL mutations in myeloproliferative neoplasms: A report of the Association for Molecular Pathology. J. Mol. Diagn. 2013, 15, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Link-Lenczowska, D.; Pallisgaard, N.; Cordua, S.; Zawada, M.; Czekalska, S.; Krochmalczyk, D.; Kanduła, Z.; Sacha, T. A comparison of qPCR and ddPCR used for quantification of the JAK2 V617F allele burden in Ph negative MPNs. Ann. Hematol. 2018, 97, 2299–2308. [Google Scholar] [CrossRef]
- Williams, D.M.; Kim, A.H.; Rogers, O.; Spivak, J.L.; Moliterno, A.R. Phenotypic variations and new mutations in JAK2 V617F-negative polycythemia vera, erythrocytosis, and idiopathic myelofibrosis. Exp. Hematol. 2007, 35, 1641–1646. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Xiao, W.; Abdel-Wahab, O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood 2017, 130, 410–423. [Google Scholar] [CrossRef]
- Bhai, P.; Hsia, C.C.; Schenkel, L.C.; Hedley, B.D.; Levy, M.A.; Kerkhof, J.; Santos, S.; Stuart, A.; Lin, H.; Broadbent, R.; et al. Clinical Utility of Implementing a Frontline NGS-Based DNA and RNA Fusion Panel Test for Patients with Suspected Myeloid Malignancies. Mol. Diagn. Ther. 2022, 26, 333–343. [Google Scholar] [CrossRef]
- Ikbal Atli, E.; Gurkan, H.; Atli, E.; Yalcintepe, S.; Demir, S.; Eker, D.; Kalkan, R.; Muzaffer Demir, A. Targeted massively parallel sequencing in the management of cytogenetically normal lymphoid malignancies. J. BU ON 2021, 26, 1540–1548. [Google Scholar]
- Levy, M.A.; Santos, S.; Kerkhof, J.; Stuart, A.; Aref-Eshghi, E.; Guo, F.; Hedley, B.; Wong, H.; Rauh, M.; Feilotter, H.; et al. Implementation of an NGS-based sequencing and gene fusion panel for clinical screening of patients with suspected hematologic malignancies. Eur. J. Haematol. 2019, 103, 178–189. [Google Scholar] [CrossRef]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Zmajkovic, J.; Lundberg, P.; Nienhold, R.; Torgersen, M.L.; Sundan, A.; Waage, A.; Skoda, R.C. A Gain-of-Function Mutation in EPO in Familial Erythrocytosis. N. Engl. J. Med. 2018, 378, 924–930. [Google Scholar] [CrossRef]
- Tavakoli, J.; Ho, G.; Kavecansky, J.; Pai, A.P. A New High Affinity Hemoglobin Variant: Hb San Francisco-KP (HBB: c.104T>C). Hemoglobin 2021, 45, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Negro, A.; Graiani, G.; Nicoli, D.; Farnetti, E.; Casali, B.; Verzicco, I.; Tedeschi, S.; Ghirarduzzi, A.; Cannone, V.; Marco, L.D.; et al. Concurrent heterozygous Von-Hippel–Lindau and transmembrane-protein-127 gene mutation causing an erythropoietin-secreting pheochromocytoma in a normotensive patient with severe erythrocytosis. J. Hypertens. 2020, 38, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Doma, S.A.; Kristan, A.; Debeljak, N.; Zupan, I.P. Congenital erythrocytosis-A condition behind recurrent thromboses: A case report and literature review. Clin. Hemorheol. Micro. 2021, 79, 417–421. [Google Scholar]
- Wouters, H.J.; Mulder, R.; van Zeventer, I.A.; Schuringa, J.J.; van der Klauw, M.M.; van der Harst, P.; Diepstra, A.; Mulder, A.B.; Huls, G. Erythrocytosis in the general population: Clinical characteristics and association with clonal hematopoiesis. Blood Adv. 2020, 4, 6353–6363. [Google Scholar] [CrossRef]
- Núñez-Martínez, P.M.; Taja-Chayeb, L.; Ramírez-Otero, M.A.; Fragoso-Ontiveros, V.; Wegman-Ostrosky, T.; Cruz-Robles, D.; Vidal Millán, S. Familial erythrocytosis 2 and von Hippel-Lindau disease in the same pediatric patient. Boletín Médico Hosp. Infant. México 2021, 78, 341–345. [Google Scholar] [CrossRef]
- Kristan, A.; Gašperšič, J.; Režen, T.; Kunej, T.; Količ, R.; Vuga, A.; Fink, M.; Žula, Š.; Anžej Doma, S.; Preložnik Zupan, I.; et al. Genetic analysis of 39 erythrocytosis and hereditary hemochromatosis-associated genes in the Slovenian family with idiopathic erythrocytosis. J. Clin. Lab. Anal. 2021, 35, e23715. [Google Scholar] [CrossRef]
- Lenglet, M.; Robriquet, F.; Schwarz, K.; Camps, C.; Couturier, A.; Hoogewijs, D.; Buffet, A.; Knight, S.J.; Gad, S.; Couvé, S.; et al. Identification of a new VHL exon and complex splicing alterations in familial erythrocytosis or von Hippel-Lindau disease. Blood 2018, 132, 469–483. [Google Scholar] [CrossRef]
- Kristan, A.; Pajič, T.; Maver, A.; Režen, T.; Kunej, T.; Količ, R.; Vuga, A.; Fink, M.; Žula, Š.; Podgornik, H.; et al. Identification of Variants Associated with Rare Hematological Disorder Erythrocytosis Using Targeted Next-Generation Sequencing Analysis. Front. Genet. 2021, 12, 689868. [Google Scholar] [CrossRef]
- Gangat, N.; Szuber, N.; Pardanani, A.; Tefferi, A. JAK2 unmutated erythrocytosis: Current diagnostic approach and therapeutic views. Leukemia 2021, 35, 2166–2181. [Google Scholar] [CrossRef]
- Sinnema, M.; Song, D.; Guan, W.; Janssen, J.W.; van Wijk, R.; Navalsky, B.E.; Peng, K.; Donker, A.E.; Stegmann, A.P.; Lee, F.S. Loss-of-function zinc finger mutation in the EGLN1 gene associated with erythrocytosis. Blood 2018, 132, 1455–1458. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, F.; Marty, C.; Balligand, T.; Verdier, F.; Grosjean, S.; Gryshkova, V.; Raslova, H.; Constantinescu, S.N.; Casadevall, N.; Vainchenker, W.; et al. New pathogenic mechanisms induced by germline erythropoietin receptor mutations in primary erythrocytosis. Haematologica 2018, 103, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Jalowiec, K.A.; Vrotniakaite-Bajerciene, K.; Capraru, A.; Wojtovicova, T.; Joncourt, R.; Rovó, A.; Porret, N.A. NGS Evaluation of a Bernese Cohort of Unexplained Erythrocytosis Patients. Genes 2021, 12, 1951. [Google Scholar] [CrossRef] [PubMed]
- Nabhani, I.A.; Aneke, J.C.; Verhovsek, M.; Eng, B.; Kuo, K.H.; Rudinskas, L.C.; Waye, J.S. Novel High Oxygen Affinity Hemoglobin Variant in a Patient with Polycythemia: Hb Kennisis [β85(F1)Phe→Leu (TTT>TTG); HBB: c.258T>G]. Hemoglobin 2020, 44, 10–12. [Google Scholar] [CrossRef]
- Loganathan, S.E.; Kattaru, S.; Chandrasekhar, C.; Vengamma, B.; Sarma, P.V.G.K. Novel mutations in EPO-R and oxygen-dependent degradation (ODD) domain of EPAS1 genes-a causative reason for Congenital Erythrocytosis. Eur. J. Med. Genet. 2022, 65, 104493. [Google Scholar] [CrossRef]
- Chandrasekhar, C.; Pasupuleti, S.K.; Sarma, P.V.G.K. Novel mutations in the EPO-R, VHL and EPAS1 genes in the Congenital Erythrocytosis patients. Blood Cells Mol. Dis. 2020, 85, 102479. [Google Scholar] [CrossRef]
- Remenyi, G.; Bereczky, Z.; Gindele, R.; Ujfalusi, A.; Illes, A.; Udvardy, M. rs779805 Von Hippel-Lindau Gene Polymorphism Induced/Related Polycythemia Entity, Clinical Features, Cancer Association, and Familiar Characteristics. Pathol. Oncol. Res. 2021, 27, 1609987. [Google Scholar] [CrossRef]
- Ediriwickrema, K.; Wilson, A.J.; O’Nions, J.; Sekhar, M.; Ahmed, S.; Roble, A.; Shambrook, E.L.; Lambert, J.; Alimam, S. Single Centre Analysis of JAK2 V617F and Exon 12 Negative Idiopathic Erythrocytosis. Blood 2021, 138 (Suppl. 1), 4620. [Google Scholar] [CrossRef]
- Mallik, N.; Jamwal, M.; Sharma, R.; Singh, N.; Sharma, P.; Bansal, D.; Trehan, A.; Chhabra, S.; Das, R. Ultra-rare Hb Regina (HBB:c.289C>G) with coinherited β-thalassaemia trait: Solving the puzzle for extreme erythrocytosis. J. Clin. Pathol. 2022, 1–2. [Google Scholar] [CrossRef]
- Lazana, I.; Mohamedali, A.; Smith, F.; Lavallade, H.; McLornan, D.; Raj, K. Uniparental disomy (UPD) of a novel bisphosphoglycerate mutase (BPGM) mutation leading to erythrocytosis. Br. J. Haematol. 2021, 192, 220–223. [Google Scholar] [CrossRef]
- Chin-Yee, B.; Cheong, I.; Matyashin, M.; Lazo-Langner, A.; Chin-Yee, I.; Bhayana, V.; Bhai, P.; Lin, H.; Sadikovic, B.; Hsia, C.C. Serum erythropoietin levels in 696 patients investigated for erythrocytosis with JAK2 mutation analysis. Am. J. Hematol. 2022, 97, E150–E153. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Lasho, T.L.; Guglielmelli, P.; Finke, C.M.; Rotunno, G.; Elala, Y.; Pacilli, A.; Hanson, C.A.; Pancrazzi, A.; Ketterling, R.P.; et al. Targeted deep sequencing in polycythemia vera and essential thrombocythemia. Blood Adv. 2016, 1, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Andréasson, B.; Pettersson, H.; Wasslavik, C.; Johansson, P.; Palmqvist, L.; Asp, J. ASXL1 mutations, previous vascular complications and age at diagnosis predict survival in 85 WHO-defined polycythaemia vera patients. Br. J. Haematol. 2020, 189, 913–919. [Google Scholar] [CrossRef]
- Tefferi, A.; Guglielmelli, P.; Lasho, T.L.; Coltro, G.; Finke, C.M.; Loscocco, G.G.; Sordi, B.; Szuber, N.; Rotunno, G.; Pacilli, A.; et al. Mutation-enhanced international prognostic systems for essential thrombocythaemia and polycythaemia vera. Br. J. Haematol. 2020, 189, 291–302. [Google Scholar] [CrossRef]
- McMullin, M.F.; Reilly, J.T.; Campbell, P.; Bareford, D.; Green, A.R.; Harrison, C.N.; Conneally, E.; National Cancer Research Institute, Myeloproliferative Disorder Subgroup; Ryan, K.; British Committee for Standards in Haematology. Amendment to the guideline for diagnosis and investigation of polycythaemia/erythrocytosis. Br. J. Haematol. 2007, 138, 821–822. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, G.A.; Stella, S.; Pennisi, M.S.; Pirosa, C.; Fermo, E.; Fabris, S.; Cattaneo, D.; Iurlo, A. The Role of New Technologies in Myeloproliferative Neoplasms. Front. Oncol. 2019, 9, 321. [Google Scholar] [CrossRef]
- Loscocco, G.G.; Guglielmelli, P.; Vannucchi, A.M. Impact of Mutational Profile on the Management of Myeloproliferative Neoplasms: A Short Review of the Emerging Data. OncoTargets Ther. 2020, 13, 12367–12382. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Maslah, N.; Cassinat, B.; Verger, E.; Kiladjian, J.J.; Velazquez, L. The role of LNK/SH2B3 genetic alterations in myeloproliferative neoplasms and other hematological disorders. Leukemia 2017, 31, 1661–1670. [Google Scholar] [CrossRef]
| Total Patients | Male | Females | |
|---|---|---|---|
| Total Patients, number (%) | 529 (100) | 390 (73.7) | 139 (26.3) |
| Age; median (range) years | 60.6 (18–95) | 60.4 (18–95) | 61.6 (24–85) |
| * Height; median (range) cm | 173 (125–198) | 177 (125–198) | 160.5 (141–180) |
| ° Weight; median (range) kg | 91.4 (42–227.7) | 96.1(52.7–227.7) | 79.1 (42–150.3) |
| ** BMI; median (range) kg/m2 | 30.5 (14.9–68.7) | 30.9 (17.9–68.7) | 30.4 (14.9–53.8) |
| Hemoglobin; median (range) g/L | 175 (160–240) | 178 (165–240) | 167 (160–217) |
| Leukocytes; median (range) × 109/L | 8.3 (2.2–43.6) | 8 (2.2–43.6) | 8.8 (4.4–31.7) |
| Platelet count; median (range) × 109/L | 232.5 (31–1498) | 220 (31–1161) | 278 (50–1498) |
| Hematocrit; median (range) L/L | 0.52 (0.45–0.73) | 0.53 (0.46–0.73) | 0.51 (0.45–0.67) |
| # EPO levels; median (range) mIU/mL | 8 (1.1–456.1) | 8.1 (1.1–456.1) | 8 (1.2–111.7) |
| . | Total Patients n = 529 (%) | JAK2 Positive Cohort n = 58 (%) | JAK2 Negative Cohort n = 471(%) |
| Hemoglobin; median (range) g/L | 175 (160–240) | 174.5(161–217) | 175(160–240) |
| Leukocytes; median (range) × 109/L | 8.3(2.2–43.6) | 10.9(4.3–31.7) | 8(2.2–43.6) |
| Platelet count; median (range) × 109/L | 232.5(31–1498) | 602.5(50–1498) | 224(31–566) |
| Hematocrit; median (range) L/L | 0.52(0.45–0.73) | 0.54(0.48–0.67) | 0.52(0.45–0.73) |
| EPO levels; median (range) mIU/mL | 8(1.1–456.1) | 1.7(1.1–6.5) | 8.6(1.3–456.1) |
| Positive for Tier I/II variants by NGS | 91 (17.2) | 58 | 33 (7) |
| JAK2 V617F | 57 (10.7) | 57 (98.3) | 0 |
| JAK2 exon 12 mutation | 1 (0.2) | 1 (1.7) | 0 |
| Clinical Diagnosis | |||
| Diagnosis of PV * | 60 | 58 (100) | 2 $ (0.4) |
| Diagnosis of CML | 3 | 0 | 3 (0.6) |
| Secondary erythrocytosis | 466 | 0 | 466 (99.0) |
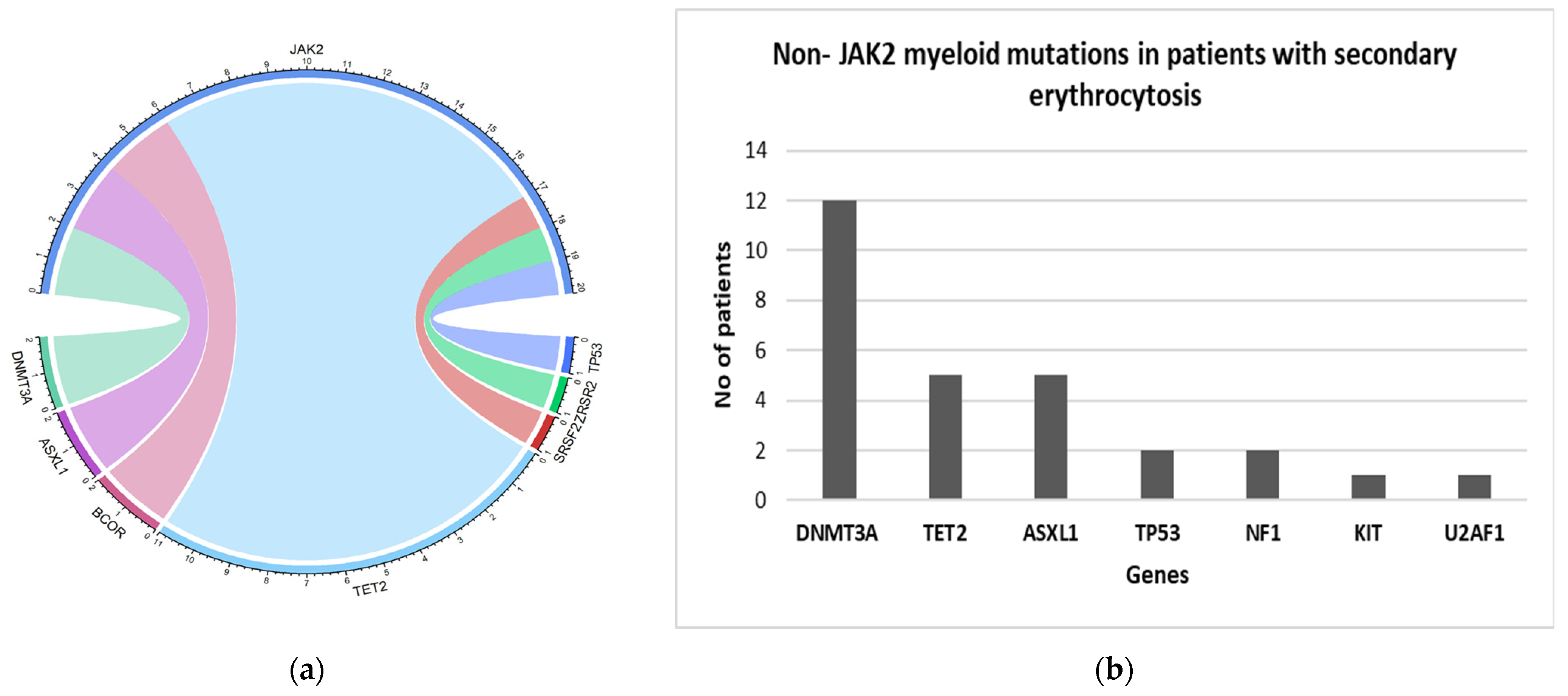
| Non-JAK2 mutations | Total Patients n = 529 (%) | JAK2 positive cohort n = 58 (%) | JAK2 negative with secondary erythrocytosis n = 466 (%) |
| TET2 | 16 (3) | 11 (19.0) | 5 (1.1) |
| DNMT3A | 14 (2.6) | 2 (3.4) | 12 (2.5) |
| ASXL1 | 7 (1.3) | 2 (3.4) | 5 (1.1) |
| BCOR | 2 (0.3) | 2 (1.7) | 0 |
| SRSF2 | 1 (0.2) | 1 (1.7) | 0 |
| TP53 | 1(0.2) | 1 (1.7) | 2 (0.4) |
| ZRSR2 | 1(0.2) | 1 | 0 |
| NF1 | 2 (0.3) | 0 | 2 (0.4) |
| KIT | 1 (0.2) | 0 | 1 (0.2) |
| U2AF1 | 1(0.2) | 0 | 1 (0.2) |
| Total non-JAK2 mutations | 48 (9) | 20 (34.5) | 28 (6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhai, P.; Chin-Yee, B.; Pope, V.; Cheong, I.; Matyashin, M.; Levy, M.A.; Foroutan, A.; Stuart, A.; Hsia, C.C.; Lin, H.; et al. Mutational Landscape of Patients Referred for Elevated Hemoglobin Level. Curr. Oncol. 2022, 29, 7209-7217. https://doi.org/10.3390/curroncol29100568
Bhai P, Chin-Yee B, Pope V, Cheong I, Matyashin M, Levy MA, Foroutan A, Stuart A, Hsia CC, Lin H, et al. Mutational Landscape of Patients Referred for Elevated Hemoglobin Level. Current Oncology. 2022; 29(10):7209-7217. https://doi.org/10.3390/curroncol29100568
Chicago/Turabian StyleBhai, Pratibha, Benjamin Chin-Yee, Victor Pope, Ian Cheong, Maxim Matyashin, Michael A. Levy, Aidin Foroutan, Alan Stuart, Cyrus C. Hsia, Hanxin Lin, and et al. 2022. "Mutational Landscape of Patients Referred for Elevated Hemoglobin Level" Current Oncology 29, no. 10: 7209-7217. https://doi.org/10.3390/curroncol29100568
APA StyleBhai, P., Chin-Yee, B., Pope, V., Cheong, I., Matyashin, M., Levy, M. A., Foroutan, A., Stuart, A., Hsia, C. C., Lin, H., Sadikovic, B., & Chin-Yee, I. (2022). Mutational Landscape of Patients Referred for Elevated Hemoglobin Level. Current Oncology, 29(10), 7209-7217. https://doi.org/10.3390/curroncol29100568





