Epigenetic Alterations Induced by Smoking and Their Intersection with Artificial Intelligence: A Narrative Review
Abstract
1. Introduction
2. Material and Methods
2.1. Selection of Relevant Studies
2.2. Review Approach
2.3. Smoking-Associated Respiratory Diseases Caused by Epigenetic Mechanisms
2.4. DNA Methylation
2.5. Histone Modifications
2.6. Non-Coding RNA Regulation
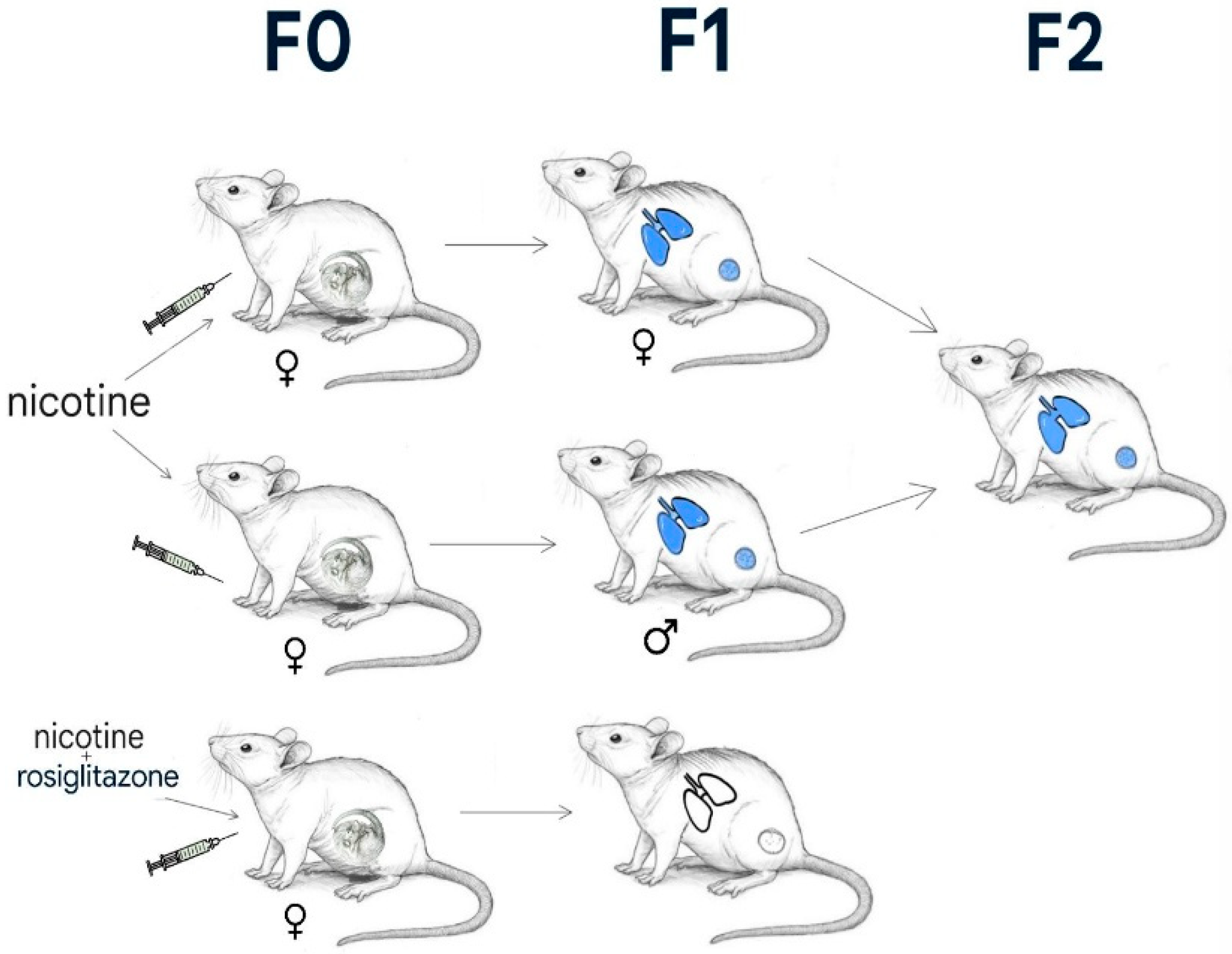
2.7. The Rehan et al. Study
2.8. Paternal Smoking and Epigenetic Transmission
2.9. AI in Epigenetics
2.10. The Molecular Memory of the Genome
2.11. Risk of Bias
3. Disease Prediction
3.1. Respiratory Disease Prediction
3.2. Neurological Disease Prediction
3.3. From Prediction to Prevention
3.4. Challenges and Ethical Considerations
3.5. Future Perspectives
4. Discussion
4.1. The Importance of DNA Methylation
4.2. The HUNT Study
4.3. The Future with the GrimAge Clock
4.4. Past, Present and Future Potential
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADAM33 | A Disintegrin And Metalloprotease 33 |
| AHRR | Aryl-Hydrocarbon Receptor Repressor |
| AI | Artificial Intelligence |
| BAL | Bronchoalveolar Lavage |
| cg05575921 | CpG site 05575921 (in AHRR gene) |
| COPD | Chronic Obstructive Pulmonary Disease |
| DNA | Deoxyribonucleic Acid |
| ECV | Electronic Cigarette Vapour |
| EPIC-Seq | Enhanced Pooled ImmunoCapture Sequencing |
| ETS | Environmental Tobacco Smoke |
| FosB | FBJ Murine Osteosarcoma Viral Oncogene Homolog B |
| H3 | Histone H3 |
| HEP | Human Epigenome Project |
| HGP | Human Genome Project |
| IL13 | Interleukin 13 |
| lncRNA | Long Non-Coding RNA |
| MCS | Maternal Cigarette Smoke |
| miR-21 | MicroRNA 21 |
| miR-223 | MicroRNA 223 |
| miRNA | MicroRNA |
| NAcc | Nucleus Accumbens |
| PAH | Polycyclic Aromatic Hydrocarbons |
| PPAR-γ | Peroxisome Proliferator-Activated Receptor-γ |
| RNA | Ribonucleic Acid |
| SERPINA1 | Serpin Peptidase Inhibitor, Clade A, Member 1 |
References
- WHO. Report on the Global Tobacco Epidemic 2021: Addressing New and Emerging Products; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Skinner, M.K. Environmental epigenomics and disease susceptibility. EMBO Rep. 2011, 12, 620–622. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Reik, W.; Dean, W.; Walter, J. Epigenetic reprogramming in mammalian development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef]
- Breton, C.V.; Byun, H.M.; Wenten, M.; Pan, F.; Yang, A.; Gilliland, F.D. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am. J. Respir. Crit. Care Med. 2009, 180, 462–467. [Google Scholar] [CrossRef]
- Joubert, B.R.; Felix, J.F.; Yousefi, P.; Bakulski, K.M.; Just, A.C.; Breton, C.; Reese, S.E.; Markunas, C.A.; Richmond, R.C.; Xu, C.-J.; et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am. J. Hum. Genet. 2016, 98, 680–696. [Google Scholar] [CrossRef]
- Zhang, S.; Jin, J.; Xu, B.; Zheng, Q.; Mou, H. The relationship between epigenetic biomarkers and the risk of diabetes and cancer: A machine learning modelling approach. Front. Public Health 2025, 13, 1509458. [Google Scholar] [CrossRef]
- Rauschert, S.; Raubenheimer, K.; Melton, P.E.; Huang, R.C. Machine learning and clinical epigenetics: A review of challenges for diagnosis and classification. Clin. Epigenet. 2020, 12, 51. [Google Scholar] [CrossRef]
- Burton, N.O.; Greer, E.L. Multigenerational epigenetic inheritance: Transmitting information across generations. Semin. Cell Dev. Biol. 2022, 127, 121–132. [Google Scholar] [CrossRef]
- Campagna, M.P.; Xavier, A.; Lechner-Scott, J.; Maltby, V.; Scott, R.J.; Butzkueven, H.; Jokubaitis, V.G.; Lea, R.A. Epigenome-wide association studies: Current knowledge, strategies and recommendations. Clin. Epigenet. 2021, 13, 214. [Google Scholar] [CrossRef] [PubMed]
- Brasil, S.; Neves, C.J.; Rijoff, T.; Falcão, M.; Valadão, G.; Videira, P.A.; Ferreira, V.d.R. Artificial Intelligence in Epigenetic Studies: Shedding Light on Rare Diseases. Front. Mol. Biosci. 2021, 8, 648012. [Google Scholar] [CrossRef]
- Nishitani, S.; Smith, A.K.; Tomoda, A.; Fujisawa, T.X. Data science using the human epigenome for predicting multifactorial diseases and symptoms. Epigenomics 2024, 16, 273–276. [Google Scholar] [CrossRef]
- Vinciguerra, M. The Potential for Artificial Intelligence Applied to Epigenetics. Mayo Clin. Proc. Digit. Health 2023, 1, 476–479. [Google Scholar] [CrossRef]
- Pravettoni, G.; Triberti, S. P5 eHealth: An Agenda for the Health Technologies of the Future; Springer: Cham, Switzerland, 2020. [Google Scholar]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Tobacco Smoking and Involuntary Smoking; World Health Organization: Geneva, Switzerland, 2004. [Google Scholar]
- Keshawarz, A.; Joehanes, R.; Guan, W.; Huan, T.; DeMeo, D.L.; Grove, M.L.; Fornage, M.; Levy, D.; O’cOnnor, G. Longitudinal change in blood DNA epigenetic signature after smoking cessation. Epigenetics 2022, 17, 1098–1109. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Zeilinger, S.; Kühnel, B.; Klopp, N.; Baurecht, H.; Kleinschmidt, A.; Gieger, C.; Weidinger, S.; Lattka, E.; Adamski, J.; Peters, A.; et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE 2013, 8, e63812. [Google Scholar] [CrossRef]
- Suter, M.A.; Abramovici, A.R.; Griffin, E.; Branch, D.W.; Lane, R.H.; Mastrobattista, J.; Rehan, V.K.; Aagaard, K. In utero nicotine exposure epigenetically alters fetal chromatin structure and differentially regulates transcription of the glucocorticoid receptor in a rat model. Birth Defects Res. A Clin. Mol. Teratol. 2015, 103, 583–588. [Google Scholar] [CrossRef]
- Bhattacharya, M. Airway architect Adam33 in asthma. Sci. Transl. Med. 2016, 8, 130. [Google Scholar] [CrossRef]
- Nicodemus-Johnson, J.; Naughton, K.A.; Sudi, J.; Hogarth, K.; Naurekas, E.T.; Nicolae, D.L.; Sperling, A.I.; Solway, J.; White, S.R.; Ober, C. Genome-Wide Methylation Study Identifies an IL-13-induced Epigenetic Signature in Asthmatic Airways. Am. J. Respir. Crit. Care Med. 2016, 193, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Baccarelli, A.; Carey, V.J.; Boutaoui, N.; Bacherman, H.; Klanderman, B.; Rennard, S.; Agusti, A.; Anderson, W.; Lomas, D.A.; et al. Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am. J. Respir. Crit. Care Med. 2012, 185, 373–381. [Google Scholar] [CrossRef]
- Yang, I.V.; Schwartz, D.A. Epigenetic mechanisms and the development of asthma. J. Allergy Clin. Immunol. 2012, 130, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Mahon, G.M.; Koppelman, G.H.; Vonk, J.M. Grandmaternal smoking, asthma and lung function in the offspring: The Lifelines cohort study. Thorax 2021, 76, 441–447. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Sundar, I.K.; Nevid, M.Z.; Friedman, A.E.; Rahman, I. Cigarette smoke induces distinct histone modifications in lung cells: Implications for the pathogenesis of COPD and lung cancer. J. Proteome Res. 2014, 13, 982–996. [Google Scholar] [CrossRef]
- Nilsson, E.E.; Sadler-Riggleman, I.; Skinner, M.K. Environmentally induced epigenetic transgenerational inheritance of disease. Environ. Epigenet. 2018, 4, dvy016. [Google Scholar] [CrossRef] [PubMed]
- Russ, R.; Slack, F.J. Cigarette-Smoke-Induced Dysregulation of MicroRNA Expression and Its Role in Lung Carcinogenesis. Pulm. Med. 2012, 2012, 791234. [Google Scholar] [CrossRef] [PubMed]
- Graff, J.W.; Powers, L.S.; Dickson, A.M.; Kim, J.; Reisetter, A.C.; Hassan, I.H.; Kremens, K.; Gross, T.J.; E Wilson, M.; Monick, M.M. Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PLoS ONE 2012, 7, e44066. [Google Scholar] [CrossRef]
- Tando, Y.; Matsui, Y. Inheritance of environment-induced phenotypic changes through epigenetic mechanisms. Environ. Epigenet. 2023, 9, dvad008. [Google Scholar] [CrossRef]
- Di, H.K.; Gan, Y.; Lu, K.; Wang, C.; Zhu, Y.; Meng, X.; Xia, W.-Q.; Xu, M.-Z.; Feng, J.; Tian, Q.-F.; et al. Maternal smoking status during pregnancy and low birth weight in offspring: Systematic review and meta-analysis of 55 cohort studies published from 1986 to 2020. World J. Pediatr. 2022, 18, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Küpers, L.K.; Xu, X.; Jankipersadsing, S.A.; Vaez, A.; la Bastide-van Gemert, S.; Scholtens, S.; Nolte, I.M.; Richmond, R.C.; Relton, C.L.; Felix, J.F.; et al. DNA methylation mediates the effect of maternal smoking during pregnancy on the birthweight of the offspring. Int. J. Epidemiol. 2015, 44, 1224–1237. [Google Scholar] [CrossRef]
- Rehan, V.K.; Liu, J.; Naeem, E.; Tian, J.; Sakurai, R.; Kwong, K.; Akbari, O.; Torday, J.S. Perinatal nicotine exposure induces asthma in second-generation offspring. BMC Med. 2012, 10, 129. [Google Scholar] [CrossRef]
- Leslie, F.M. Multigenerational epigenetic effects of nicotine on lung function. BMC Med. 2013, 11, 27. [Google Scholar] [CrossRef]
- Bhadsavle, S.S.; Golding, M.C. Paternal epigenetic influences on placental health and their impacts on offspring development and disease. Front. Genet. 2022, 13, 1068408. [Google Scholar] [CrossRef]
- Vlachou, M.; Kyrkou, G.; Georgakopoulou, V.E.; Kapetanaki, A.; Vivilaki, V.; Spandidos, D.A.; Diamanti, A. Smoke signals in the genome: Epigenetic consequences of parental tobacco exposure (Review). Biomed. Rep. 2025, 23, 146. [Google Scholar] [CrossRef] [PubMed]
- Holder, L.B.; Haque, M.M.; Skinner, M.K. Machine learning for epigenetics and future medical applications. Epigenetics 2017, 12, 505–514. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Wang, H.; Yan, J. Application of deep learning in genomics. Sci. China Life Sci. 2020, 63, 1860–1878. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.; Norouzi, M.; Khan, S.S.; Davie, J.R.; Yamanaka, S.; Ashraf, A. Artificial intelligence and deep learning algorithms for epigenetic sequence analysis: A review for epigeneticists and AI experts. Comput. Biol. Med. 2024, 183, 109302. [Google Scholar] [CrossRef]
- Ritzmann, F.; Brand, M.; Bals, R.; Wegmann, M.; Beisswenger, C. Role of Epigenetics in Chronic Lung Disease. Cells 2025, 14, 251. [Google Scholar] [CrossRef]
- Li, D.D.; Chen, T.; Ling, Y.L.; Jiang, Y.; Li, Q.G. A Methylation Diagnostic Model Based on Random Forests and Neural Networks for Asthma Identification. Comput. Math. Methods Med. 2022, 2022, 2679050. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.; Ashley, E.A. Artificial Intelligence in Molecular Medicine. N. Engl. J. Med. 2023, 388, 2456–2465. [Google Scholar] [CrossRef]
- Kitaba, N.T.; Knudsen, G.T.M.; Johannessen, A.; Rezwan, F.I.; Malinovschi, A.; Oudin, A.; Benediktsdottir, B.; Martino, D.; González, F.J.C.; Gómez, L.P.; et al. Fathers’ preconception of smoking and offspring DNA methylation. Clin. Epigenet. 2023, 15, 131. [Google Scholar] [CrossRef]
- Crowley, N. AI Detects Disease Clues from DNA Methyation Data. What Is Epigenetics, 1 November 2023. [Google Scholar]
- Grezenko, H.; Ekhator, C.; Nwabugwu, N.U.; Ganga, H.; Affaf, M.; Abdelaziz, A.M.; Rehman, A.; Shehryar, A.; A Abbasi, F.; Bellegarde, S.B.; et al. Epigenetics in Neurological and Psychiatric Disorders: A Comprehensive Review of Current Understanding and Future Perspectives. Cureus 2023, 15, e43960. [Google Scholar] [CrossRef]
- Kuehner, J.N.; Bruggeman, E.C.; Wen, Z.; Yao, B. Epigenetic Regulations in Neuropsychiatric Disorders. Front. Genet. 2019, 10, 268. [Google Scholar] [CrossRef] [PubMed]
- Kandel, D.B.; Kandel, E.R. A molecular basis for nicotine as a gateway drug. N. Engl. J. Med. 2014, 371, 2038–2039. [Google Scholar] [CrossRef]
- Dao, J.M.; McQuown, S.C.; Loughlin, S.E.; Belluzzi, J.D.; Leslie, F.M. Nicotine alters limbic function in adolescent rat by a 5-HT1A receptor mechanism. Neuropsychopharmacology 2011, 36, 1319–1331. [Google Scholar] [CrossRef]
- Rauschert, S.; Melton, P.E.; Heiskala, A.; Karhunen, V.; Burdge, G.; Craig, J.M.; Godfrey, K.M.; Lillycrop, K.; Mori, T.A.; Beilin, L.J.; et al. Machine Learning-Based DNA Methylation Score for Fetal Exposure to Maternal Smoking: Development and Validation in Samples Collected from Adolescents and Adults. Environ. Health Perspect. 2020, 128, 97003. [Google Scholar] [CrossRef]
- Zakarya, R.; Adcock, I.; Oliver, B.G. Epigenetic impacts of maternal tobacco and e-vapour exposure on the offspring lung. Clin. Epigenet. 2019, 11, 32. [Google Scholar] [CrossRef]
- Fernando, A.; Kondrup, E.; Cheung, K.; Uberoi, D.; Joly, Y. Still using genetic data? A comparative review of Canadian life insurance application forms before and after the GNDA. FACETS 2024, 9, 1–10. [Google Scholar] [CrossRef]
- Esfahani, M.S.; Hamilton, E.G.; Mehrmohamadi, M.; Nabet, B.Y.; Alig, S.K.; King, D.A.; Steen, C.B.; Macaulay, C.W.; Schultz, A.; Nesselbush, M.C.; et al. Inferring gene expression from cell-free DNA fragmentation profiles. Nat. Biotechnol. 2022, 40, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Leroy, A.; Teh, A.L.; Dondelinger, F.; Alvarez, M.A.; Wang, D. Longitudinal prediction of DNA methylation to forecast epigenetic outcomes. EBioMedicine 2025, 115, 105709. [Google Scholar] [CrossRef]
- Oliva, A.; Kaphle, A.; Reguant, R.; Sng, L.M.F.; Twine, N.A.; Malakar, Y.; Wickramarachchi, A.; Keller, M.; Ranbaduge, T.; Chan, E.K.F.; et al. Future-proofing genomic data and consent management: A comprehensive review of technology innovations. Gigascience 2024, 13, giae021. [Google Scholar] [CrossRef] [PubMed]
- Teare, H.J.A.; Prictor, M.; Kaye, J. Reflections on dynamic consent in biomedical research: The story so far. Eur. J. Hum. Genet. 2021, 29, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Brauneck, A.; Schmalhorst, L.; Weiss, S.; Baumbach, L.; Völker, U.; Ellinghaus, D.; Baumbach, J.; Buchholtz, G. Legal aspects of privacy-enhancing technologies in genome-wide association studies and their impact on performance and feasibility. Genome Biol. 2024, 25, 154. [Google Scholar]
- American Society of Human Genetics. The Genetic Information Nondiscrimination Act (GINA); American Society of Human Genetics: Rockville, MD, USA, 2008. [Google Scholar]
- van Bekkum, M.; Borgesius, F.Z.; Heskes, T. AI, insurance, discrimination and unfair differentiation: An overview and research agenda. Law Innov. Technol. 2025, 17, 177–204. [Google Scholar] [CrossRef]
- Gorzynski, J.E.; Goenka, S.D.; Shafin, K.; Jensen, T.D.; Fisk, D.G.; Grove, M.E.; Spiteri, E.; Pesout, T.; Monlong, J.; Baid, G.; et al. Ultrarapid Nanopore Genome Sequencing in a Critical Care Setting. N. Engl. J. Med. 2022, 386, 700–702. [Google Scholar] [CrossRef]
- Alsaedi, S.; Ogasawara, M.; Alarawi, M.; Gao, X.; Gojobori, T. AI-powered precision medicine: Utilizing genetic risk factor optimization to revolutionize healthcare. NAR Genom. Bioinform. 2025, 7, lqaf038. [Google Scholar] [CrossRef]
- Jacobsen, K.K.; Schnohr, P.; Jensen, G.B.; Bojesen, S.E. AHRR (cg05575921) Methylation Safely Improves Specificity of Lung Cancer Screening Eligibility Criteria: A Cohort Study. Cancer Epidemiol. Biomark. Prev. 2022, 31, 758–765. [Google Scholar] [CrossRef]
- Arefeen, M.A.; Nimi, S.T.; Rahman, M.S.; Arshad, S.H.; Holloway, J.W.; Rezwan, F.I. Prediction of Lung Function in Adolescence Using Epigenetic Aging: A Machine Learning Approach. Methods Protoc. 2020, 3, 77. [Google Scholar] [CrossRef]
- Krauss-Etschmann, S.; Meyer, K.F.; Dehmel, S.; Hylkema, M.N. Inter- and transgenerational epigenetic inheritance: Evidence in asthma and COPD? Clin Epigenet. 2015, 7, 53. [Google Scholar] [CrossRef]
- Tsai, P.C.; Glastonbury, C.A.; Eliot, M.N.; Bollepalli, S.; Yet, I.; Castillo-Fernandez, J.E.; Carnero-Montoro, E.; Hardiman, T.; Martin, T.C.; Vickers, A.; et al. Smoking induces coordinated DNA methylation and gene expression changes in adipose tissue with consequences for metabolic health. Clin. Epigenet. 2018, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Stueve, T.R.; Li, W.Q.; Shi, J.; Marconett, C.N.; Zhang, T.; Yang, C.; Mullen, D.; Yan, C.; Wheeler, W.; Hua, X.; et al. Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Hum. Mol. Genet. 2017, 26, 3014–3027. [Google Scholar] [CrossRef]
- Li, J.L.; Jain, N.; Tamayo, L.I.; Tong, L.; Jasmine, F.; Kibriya, M.G.; Demanelis, K.; Oliva, M.; Chen, L.S.; Pierce, B.L. The association of cigarette smoking with DNA methylation and gene expression in human tissue samples. Am. J. Hum. Genet. 2024, 111, 636–653. [Google Scholar] [CrossRef]
- Chen, Q.; Nwozor, K.O.; van den Berge, M.; Slebos, D.J.; Faiz, A.; Jonker, M.R.; Boezen, H.M.; Heijink, I.H.; de Vries, M. From Differential DNA Methylation in COPD to Mitochondria: Regulation of AHRR Expression Affects Airway Epithelial Response to Cigarette Smoke. Cells 2022, 11, 3423. [Google Scholar] [CrossRef]
- Sun, Y.Q.; Richmond, R.C.; Suderman, M.; Min, J.L.; Battram, T.; Flatberg, A.; Beisvag, V.; Nøst, T.H.; Guida, F.; Jiang, L.; et al. Assessing the role of genome-wide DNA methylation between smoking and risk of lung cancer using repeated measurements: The HUNT study. Int. J. Epidemiol. 2021, 50, 1482–1497. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.T.; Quach, A.; Wilson, J.G.; Reiner, A.P.; Aviv, A.; Raj, K.; Hou, L.; Baccarelli, A.A.; Li, Y.; Stewart, J.D.; et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 2019, 11, 303–327. [Google Scholar] [CrossRef]
- Lu, A.T.; Binder, A.M.; Zhang, J.; Yan, Q.; Reiner, A.P.; Cox, S.R.; Corley, J.; Harris, S.E.; Kuo, P.-L.; Moore, A.Z.; et al. DNA methylation GrimAge version 2. Aging 2022, 14, 9484–9549. [Google Scholar] [CrossRef]
- Zhang, Q. An interpretable biological age. Lancet Healthy Longev. 2023, 4, e662–e663. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Chen, H.; Kaeberlein, M.; Lee, S.I. ExplaiNAble BioLogical Age (ENABL Age): An artificial intelligence framework for interpretable biological age. Lancet Healthy Longev. 2023, 4, e711–e723. [Google Scholar] [CrossRef] [PubMed]
- Song, M.A.; Mori, K.M.; McElroy, J.P.; Freudenheim, J.L.; Weng, D.Y.; Reisinger, S.A.; Brasky, T.M.; Wewers, M.D.; Shields, P.G. Accelerated epigenetic age, inflammation, and gene expression in lung: Comparisons of smokers and vapers with non-smokers. Clin Epigenet. 2023, 15, 160. [Google Scholar] [CrossRef]
- Klose, D.; Needhamsen, M.; Ringh, M.V.; Hagemann-Jensen, M.; Jagodic, M.; Kular, L. Smoking affects epigenetic ageing of lung bronchoalveolar lavage cells in Multiple Sclerosis. Mult. Scler. Relat. Disord. 2023, 79, 104991. [Google Scholar] [CrossRef]
- National Human Genome Research Institute (NHGRI). Human Genome Project: The Most Important Biomedical Research Undertaking of the 20th Century; National Human Genome Research Institute: Bethesda, MD, USA, 2024. [Google Scholar]
- Hood, L.; Rowen, L. The Human Genome Project: Big science transforms biology and medicine. Genome Med. 2013, 5, 79. [Google Scholar] [CrossRef] [PubMed]
- Kabata, F.; Thaldar, D. The human genome as the common heritage of humanity. Front. Genet. 2023, 14, 1282515. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, J. Human epigenome project–up and running. PLoS Biol. 2003, 1, E82. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, F.; Beck, S.; Gut, I.G.; Berlin, K. Future potential of the Human Epigenome Project. Expert Rev. Mol. Diagn. 2004, 4, 609–618. [Google Scholar] [CrossRef]
| Gene/Element | Epigenetic Change Induced by Smoking | Related Disease Risk/Effect |
|---|---|---|
| AHRR | DNA hypomethylation at cg05575921; | Biomarker of smoking exposure; associated with lung cancer, COPD, altered lung function |
| ADAM33 | DNA methylation changes in airway epithelial cells | Asthma susceptibility, airway remodelling, and reduced lung function |
| IL13 | DNA methylation changes | Asthma, allergic airway inflammation |
| SERPINA1 | DNA methylation changes | COPD, impaired lung function |
| PPAR-γ | Histone modification; suppressed expression after smoke exposure | Impaired alveolar development, intergenerational lung pathology |
| Histone H3 | Acetylation induced by tobacco smoke | Activation of pro-inflammatory genes, chronic lung inflammation |
| miR-21, miR-223 | Dysregulated expression due to smoking | Airway inflammation, fibrosis, altered airway responsiveness, transgenerational effects |
| FosB | Histone acetylation in reward-related brain regions | Altered neuronal plasticity, increased susceptibility to nicotine addiction |
| Application | Input Data | AI Approach | Predictive Outcome |
|---|---|---|---|
| Lung disease risk | DNA methylation, histone marks, ncRNA | Machine learning classifiers | Early identification of asthma, COPD, lung cancer risk |
| Biological ageing | DNA methylation | Epigenetic clocks (GrimAge, GrimAge2) | Estimation of biological age, prediction of age-related disease |
| Epigenetic biomarker discovery | Multi-omic datasets | Deep learning models | Identification of novel predictive markers for respiratory and neurological diseases |
| Prenatal exposure effects | DNA methylation in fetal tissues | Predictive modeling | Detection of offspring at risk from maternal smoking exposure |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ianosi, E.S.; Tomoroga, D.M.; Văsieșiu, A.M.; Grigorescu, B.L.; Vultur, M.; Ianosi, M.B. Epigenetic Alterations Induced by Smoking and Their Intersection with Artificial Intelligence: A Narrative Review. Int. J. Environ. Res. Public Health 2025, 22, 1622. https://doi.org/10.3390/ijerph22111622
Ianosi ES, Tomoroga DM, Văsieșiu AM, Grigorescu BL, Vultur M, Ianosi MB. Epigenetic Alterations Induced by Smoking and Their Intersection with Artificial Intelligence: A Narrative Review. International Journal of Environmental Research and Public Health. 2025; 22(11):1622. https://doi.org/10.3390/ijerph22111622
Chicago/Turabian StyleIanosi, Edith Simona, Daria Maria Tomoroga, Anca Meda Văsieșiu, Bianca Liana Grigorescu, Mara Vultur, and Maria Beatrice Ianosi. 2025. "Epigenetic Alterations Induced by Smoking and Their Intersection with Artificial Intelligence: A Narrative Review" International Journal of Environmental Research and Public Health 22, no. 11: 1622. https://doi.org/10.3390/ijerph22111622
APA StyleIanosi, E. S., Tomoroga, D. M., Văsieșiu, A. M., Grigorescu, B. L., Vultur, M., & Ianosi, M. B. (2025). Epigenetic Alterations Induced by Smoking and Their Intersection with Artificial Intelligence: A Narrative Review. International Journal of Environmental Research and Public Health, 22(11), 1622. https://doi.org/10.3390/ijerph22111622






