Prevalence of Polymorphism and Post-Training Expression of ACTN3 (R/X) and ACE (I/D) Genes in CrossFit Athletes
Abstract
1. Introduction
2. Materials and Methods
2.1. Maximum Strength Test
2.2. Power Test
2.3. Strength Assessment of the Anteflexion Muscles of the Arm
2.4. VO2max Assessment
2.5. Body Composition Assessment
2.6. RNA and DNA Extraction
2.7. ACTN3 Genotype
2.8. ACE Genotype
2.9. ACTN3 and ACE Expression
2.10. Static Analysis
3. Results
3.1. Results Associated with General Characteristics
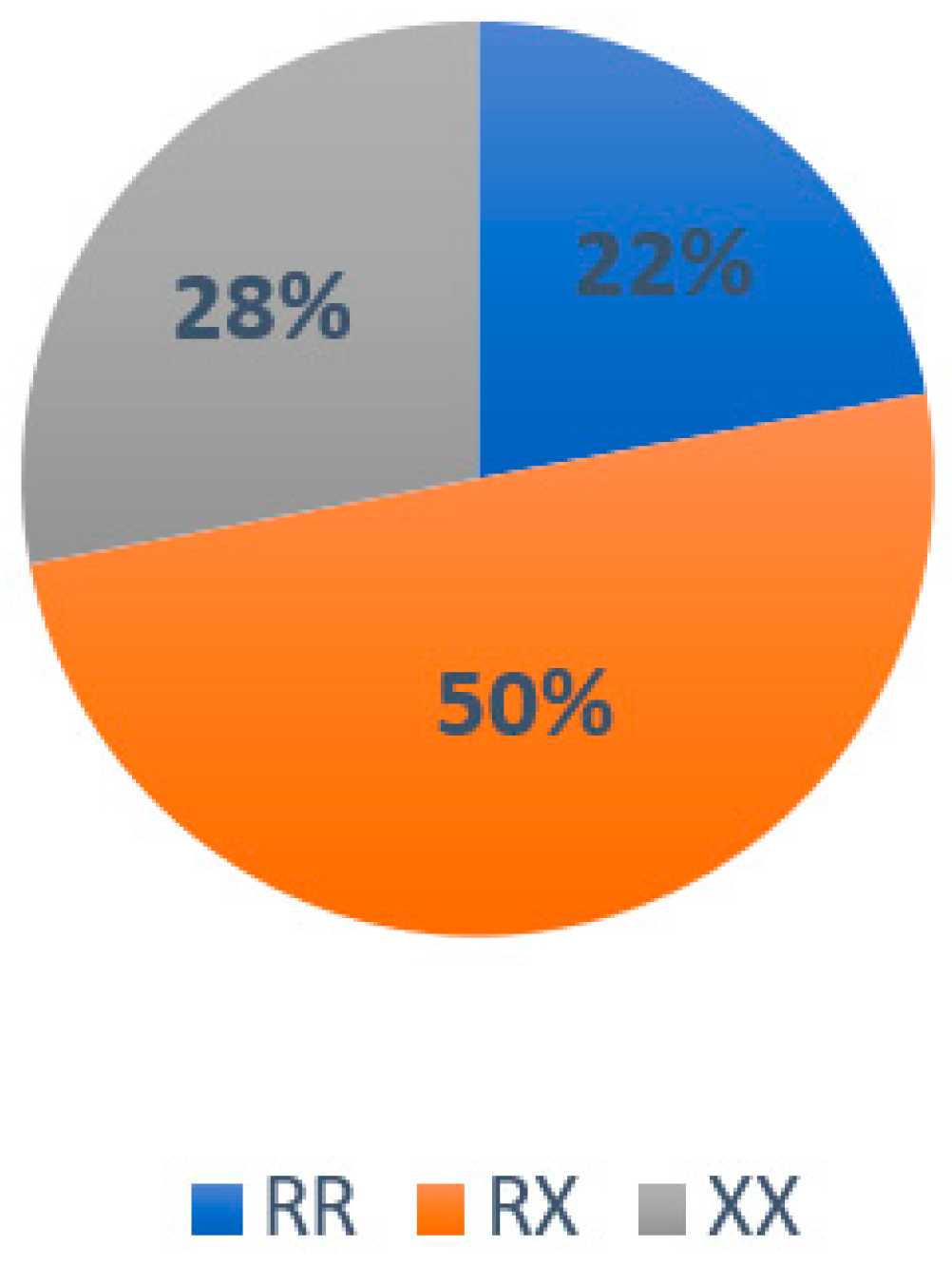
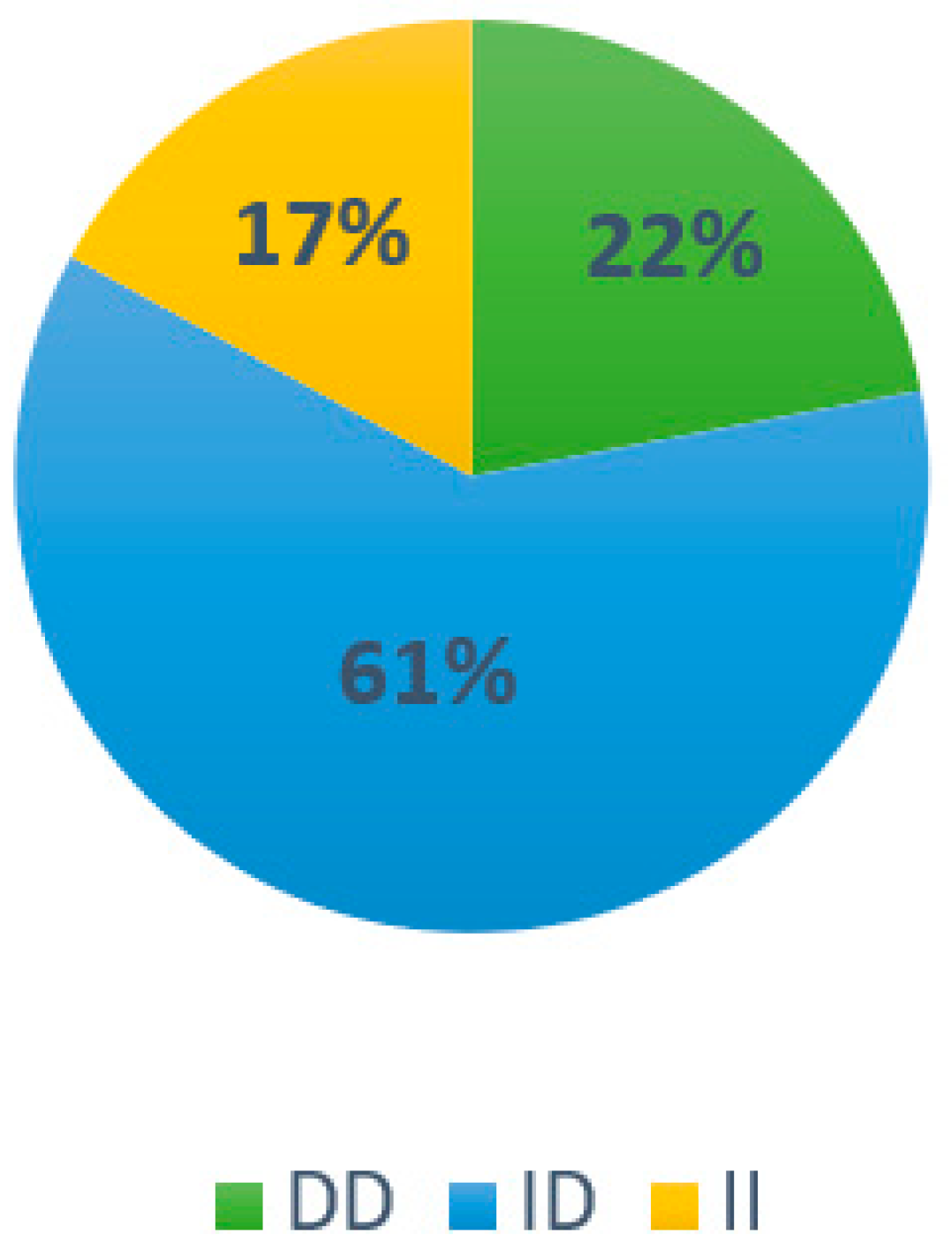
3.2. Results Associated with Genotype Frequency in the Population
3.3. Results Associated with the Effect of Training on Physical Abilities
3.4. Results Related to the Association of ACTN3 and ACE Genotypes with Physical Abilities
3.5. Results Associated with a Correlation of ACTN3 and ACE Expression and Physical Abilities
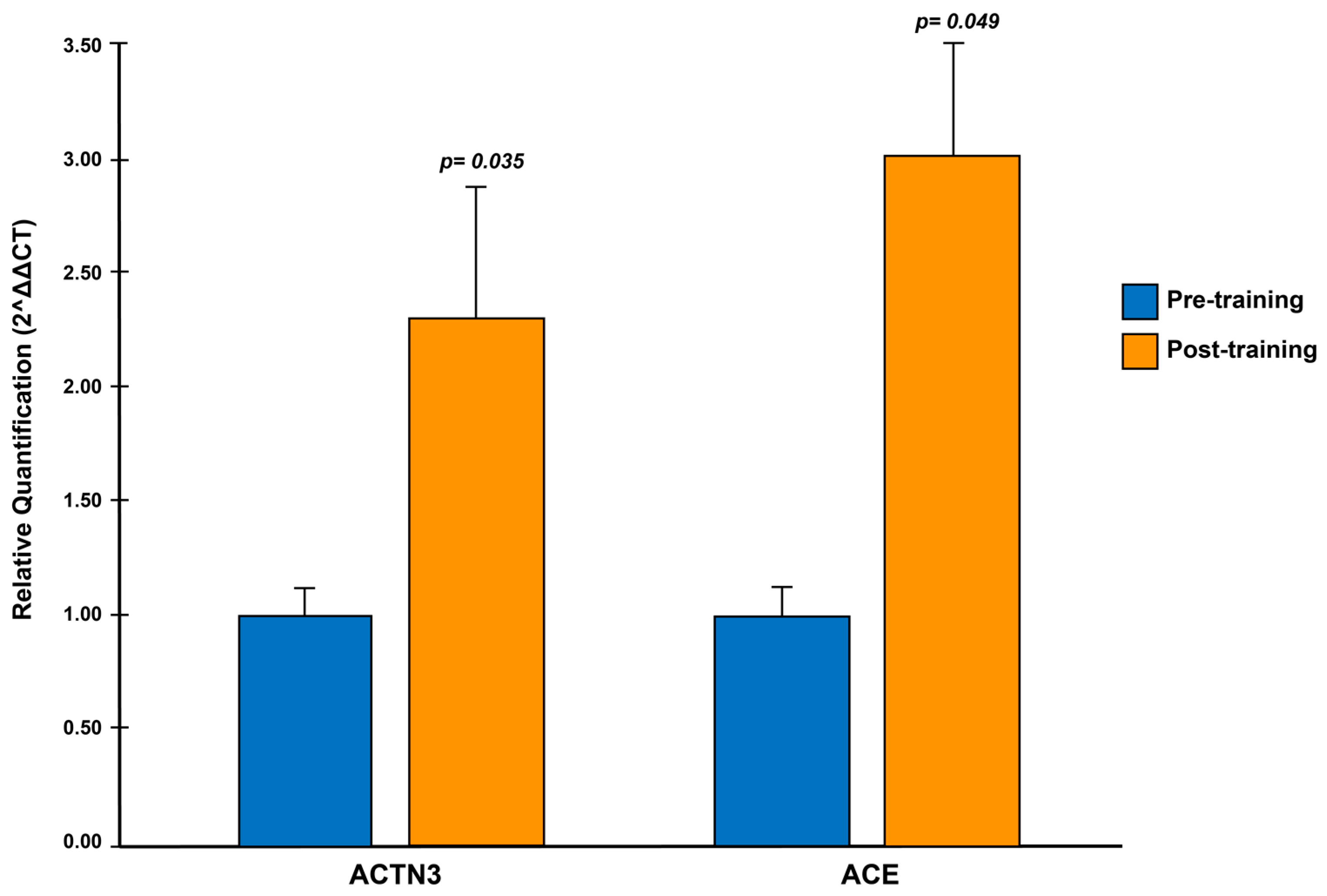
3.6. Results Associated with Gene Expression of the ACTN3 and ACE Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dominski, F.H.; Tibana, R.A.; Andrade, A. “Functional Fitness Training”, CrossFit, HIMT, or HIFT: What Is the Preferable Terminology? Front. Sport. Act. Living 2022, 4, 207. [Google Scholar] [CrossRef]
- Thompson, W.R. Worldwide Survey of Fitness Trends for 2023. ACSM’s Health Fit. J. 2023, 27, 9–18. [Google Scholar] [CrossRef]
- Ahmetov, I.I.; Hall, E.C.R.; Semenova, E.A.; Pranckevičienė, E.; Ginevičienė, V. Advances in sports genomics. Adv. Clin. Chem. 2022, 107, 215–263. [Google Scholar] [CrossRef] [PubMed]
- Pickering, C.; Kiely, J. ACTN3: More than just a gene for speed. Front. Physiol. 2017, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sgourou, A.; Fotopoulos, V.; Kontos, V.; Patrinos, G.P.; Papachatzopoulou, A. Association of genome variations in the renin-angiotensin system with physical performance. Hum. Genom. 2012, 6, 1–7. [Google Scholar] [CrossRef]
- Ma, F.; Yang, Y.; Li, X.; Zhou, F.; Gao, C.; Li, M.; Li, G. The Association of Sport Performance with ACE and ACTN3 Genetic Polymorphisms: A Systematic Review and Meta-Analysis. PLoS ONE 2013, 8, e54685. [Google Scholar] [CrossRef]
- Macarthur, D.G.; Seto, J.T.; Chan, S.; Quinlan, K.G.R.; Raftery, J.M.; Turner, N.; Nicholson, M.D.; Kee, A.J.; Hardeman, E.C.; Gunning, P.W.; et al. An Actn3 knockout mouse provides mechanistic insights into the association between α-actinin-3 deficiency and human athletic performance. Hum. Mol. Genet. 2008, 17, 1076–1086. [Google Scholar] [CrossRef]
- Eynon, N.; Banting, L.K.; Ruiz, J.R.; Cieszczyk, P.; Dyatlov, D.A.; Maciejewska-Karlowska, A.; Sawczuk, M.; Pushkarev, V.P.; Kulikov, L.M.; Pushkarev, E.D.; et al. ACTN3 R577X polymorphism and team-sport performance: A study involving three European cohorts. J. Sci. Med. Sport. 2014, 17, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Ivarsson, N.; Westerblad, H. α-Actinin-3: Why Gene Loss Is an Evolutionary Gain. PLoS Genet. 2015, 11, 2–5. [Google Scholar] [CrossRef]
- Broos, S.; Van Leemputte, M.; Deldicque, L. Thomis MA. History-dependent force, angular velocity and muscular endurance in ACTN3 genotypes. Eur. J. Appl. Physiol. 2015, 115, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Cieszczyk, P.; Sawczuk, M.; Maciejewska-Karlowska, A.; Ficek, K. ACTN3 R577X polymorphism in top-level Polish rowers. J. Exerc. Sci. Fitness 2012, 10, 12–15. [Google Scholar] [CrossRef]
- Mikami, E.; Fuku, N.; Murakami, H.; Tsuchie, H.; Takahashi, H.; Ohiwa, N.; Tanaka, H.; Pitsiladis, Y.P.; Higuchi, M.; Miyachi, M.; et al. ACTN3 R577X genotype is associated with sprinting in elite Japanese athletes. Int. J. Sport. Med. 2014, 35, 172–177. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, D.G.; Seto, J.T.; Raftery, J.M.; Quinlan, K.G.; Huttley, G.A.; Hook, J.W.; Lemckert, F.; Kee, A.; Edwards, M.; Berman, Y.; et al. Loss of ACTN3 gene function alters mouse muscle metabolism and shows evidence of positive selection in humans. Nat. Genet. 2007, 39, 1261–1265. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.A.; Irving, R.; Irwin, L.; Morrison, E.; Charlton, V.; Austin, K.; Tladi, D.; Deason, M.; Headley, S.A.; Kolkhorst, F.W.; et al. ACTN3 and ACE genotypes in elite Jamaican and US sprinters. Med. Sci. Sport. Exerc. 2010, 42, 107–112. [Google Scholar] [CrossRef]
- Ahmetov, I.I.; Fedotovskaya, O.N. Current Progress in Sports Genomics, 1st ed.; Advances in Clinical Chemistry; Elsevier Inc.: Amsterdam, The Netherlands, 2015. [Google Scholar] [CrossRef]
- Tiret, L.; Rigat, B.; Visvikis, S.; Breda, C.; Corvol, P.; Cambien, F.; Soubrier, F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am. J. Hum. Genet. 1992, 51, 197–205. [Google Scholar]
- Papadimitriou, I.D.; Lucia, A.; Pitsiladis, Y.P.; Pushkarev, V.P.; Dyatlov, D.A.; Orekhov, E.F.; Artioli, G.G.; Guilherme, J.P.L.F.; Lancha, A.H.L., Jr.; Ginevičienė, V.; et al. ACTN3 R577X and ACE I/D gene variants influence performance in elite sprinters: A multi-cohort study. BMC Genom. 2016, 17, 285. [Google Scholar] [CrossRef]
- Tsianos, G.; Sanders, J.; Dhamrait, S.; Humphries, S.; Grant, S.; Montgomery, H. The ACE gene insertion/deletion polymorphism and elite endurance swimming. Eur. J. Appl. Physiol. 2004, 92, 360–362. [Google Scholar] [CrossRef]
- Myerson, S.; Hemingway, H.; Budget, R.; Martin, J.; Humphries, S.; Montgomery, H. Human angiotensin I-converting enzyme gene and endurance performance. J. Appl. Physiol. 1999, 87, 1313–1316. [Google Scholar] [CrossRef]
- Collins, M.; Xenophontos, S.L.; Cariolou, M.A.; Mokone, G.G.; Hudson, D.E.; Anastasiades, L.; Noakes, T.D. The ACE gene and endurance performance during the South African Ironman Triathlons. Med. Sci. Sport. Exerc. 2004, 36, 1314–1320. [Google Scholar] [CrossRef]
- Hogarth, M.W.; Garton, F.C.; Houweling, P.J.; Tukiainen, T.; Lek, M.; Macarthur, D.G.; Seto, J.T.; Quinlan, K.G.; Yang, N.; Head, S.I.; et al. Analysis of the ACTN3 heterozygous genotype suggests that α-actinin-3 controls sarcomeric composition and muscle function in a dose-dependent fashion. Hum. Mol. Genet. 2016, 25, 866–877. [Google Scholar] [CrossRef]
- Thomas, K.C.; Zheng, X.F.; Suarez, F.G.; Raftery, J.M.; Quinlan, K.G.R.; Yang, N.; North, K.N.; Houweling, P.J. Evidence based selection of commonly used RT-qPCR reference genes for the analysis of mouse skeletal muscle. PLoS ONE 2014, 9, e88653. [Google Scholar] [CrossRef] [PubMed]
- Domańska-Senderowska, D.; Szmigielska, P.; Snochowska, A.; Jastrzębski, Z.; Jegier, A.; Kiszałkiewicz, J.; Jastrzębska, J.; Pastuszak-Lewandoska, D.; Cięszczyk, P.; Suchanecka, A.; et al. Relationships between the expression of the ACTN3 gene and explosive power of soccer players. J. Hum. Kin. 2019, 69, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Norman, B.; Esbjörnsson, M.; Rundqvist, H.; Österlund, T.; Von Walden, F.; Tesch, P.A. Strength, power, fiber types, and mRNA expression in trained men and women with different ACTN3 R577X genotypes. J. Appl. Physiol. 2009, 106, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Valdivieso, P.; Vaughan, D.; Laczko, E.; Brogioli, M.; Waldron, S.; Rittweger, J.; Flück, M. The metabolic response of skeletal muscle to endurance exercise is modified by the ACE-I/D gene polymorphism and training state. Front. Physiol. 2017, 8, 993. [Google Scholar] [CrossRef] [PubMed]
- Roldán Aguilar, E.E. Bases fisiológicas de los principios del entrenamiento deportivo. Revista Politécnica 2009, 5, 84–93. [Google Scholar]
- García-García, J.A.; Reding-Bernal, A.; López-Alvarenga, J.C. Cálculo del tamaño de la muestra en investigación en educación médica. Investig. Educ. Médica 2013, 2, 217–224. [Google Scholar] [CrossRef]
- García, G.C.; Secchi, J.D. 20 meters shuttle run test with stages of one minute. An original idea that has lasted for 30 years. Apunt. Med. L’Esport 2014, 49, 93–103. [Google Scholar] [CrossRef]
- Ramsbottom, R.; Brewer, J.; Williams, C. A progressive shuttle run test to estimate maximal oxygen uptake. Br. J. Sport. Med. 1988, 22, 141–144. [Google Scholar] [CrossRef]
- Peña, B. VJ y OF. Asociación genética con el rendimiento deportivo en tenistas y voleibolistas de competencia. Rev. Mex. De Investig. Cult. Física Deporte 2016, 3, 79–94. [Google Scholar]
- Güereca-Arvizuo, J.; Ramos-Jiménez, A.; Moreno-Brito, V.; Cervantes-Borunda, M.; Hernández-Torres, R.P. Respuesta De Creatina Quinasa a Un Ejercicio Anaerobio Supramáximo En Genotipos De Actn3. Rev. Int. Med. Cienc. Act. Física Deporte 2020, 20, 381–393. [Google Scholar] [CrossRef]
- Haff, G.G.; Jackson, J.R.; Kawamori, N.; Carlock, J.M.; Hartman, M.J.; Kilgore, J.L.; Morris, R.T.; Ramsey, M.W.; Sands, W.A.; Stone, M.H. Force-time curve characteristics and hormonal alterations during an eleven-week training period in elite women weightlifters. J. Strength Cond. Res. 2008, 22, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Shephard, R.J. Exercise and training in women, Part I: Influence of gender on exercise and training responses. Can. J. Appl. Physiol. 2000, 25, 19–34. [Google Scholar] [CrossRef]
- Otto III, W.H.; Coburn, J.W.; Brown, L.E.; Spiering, B.A. Effects of weightlifting vs. kettlebell training on vertical jump, strength, and body composition. J. Strength Cond. Res. 2012, 26, 1199–1202. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zaken, S.; Eliakim, A.; Nemet, D.; Meckel, Y. Genetic variability among power athletes: The stronger vs. the faster. J. Strength Cond. Res. 2019, 33, 1505–1511. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, N.; Miyamoto-Mikami, E.; Murakami, H.; Nakamura, T.; Min, S.K.; Mizuno, M.; Naito, H.; Miyachi, M.; Nakazato, K.; Fuku, N. ACTN3 R577X genotype and athletic performance in a large cohort of Japanese athletes. Eur. J. Sport Sci. 2016, 16, 694–701. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.Q.; Yi, L.; Liu, J.; Hu, Y.; Lu, Y.; Wang, M. ACTN3 R577X genotype and performance of elite middle-long distance swimmers in China. Biol. Sport. 2017, 34, 39–43. [Google Scholar] [CrossRef]
- Papadimitriou, I.D.; Papadopoulos, C.; Kouvatsi, A.; Triantaphyllidis, C. The ACE I/D polymorphism in elite Greek track and field athletes. J. Sport. Med. Phys. Fit. 2009, 49, 459–463. [Google Scholar]
- Peplonska, B.; Adamczyk, J.G.; Siewierski, M.; Safranow, K.; Maruszak, A.; Sozanski, H.; Gajewski, A.K.; Zekanowski, C. Genetic variants associated with physical and mental characteristics of the elite athletes in the Polish population. Scand. J. Med. Sci. Sports 2017, 27, 788–800. [Google Scholar] [CrossRef]
- Wang, G.; Mikami, E.; Chiu, L.L.; De Perini, A.; Deason, M.; Fuku, N.; Miyachi, M.; Kaneoka, K.; Murakami, H.; Tanaka, M.; et al. Association analysis of ACE and ACTN3 in Elite Caucasian and East Asian Swimmers. Med. Sci. Sport. Exerc. 2013, 45, 892–900. [Google Scholar] [CrossRef]
- Orysiak, J.; Busko, K.; Michalski, R.; Mazur-Rózycka, J.; Gajewski, J.; Malczewska-Lenczowska, J.; Sitkowski, D.; Pokrywka, A. Relationship between ACTN3 R577x polymorphism and maximal power output in elite polish athletes. Medicina 2014, 50, 303–308. [Google Scholar] [CrossRef]
- Boraita, A.; de la Rosa, A.; Heras, M.E.; de la Torre, A.I.; Canda, A.; Rabadán, M.; Díaz, E.; González, C.; López, M.; Hernández, M. Cardiovascular Adaptation, Functional Capacity, and Angiotensin-Converting Enzyme I/D Polymorphism in Elite Athletes. Rev. Española Cardiol. 2010, 63, 810–819. [Google Scholar] [CrossRef]
- Tobina, T.; Michishita, R.; Yamasawa, F.; Zhang, B.; Sasaki, H.; Tanaka, H.; Saku, K.; Kiyonaga, A. Association between the angiotensin I-converting enzyme gene insertion/deletion polymorphism and endurance running speed in Japanese runners. J. Physiol. Sci. 2010, 60, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Znazen, H.; Mejri, A.; Touhami, I.; Chtara, M.; Siala, H.; Ahmetov, I.I.; Messaoud, T.; Chamari, K.; Soussi, N. Genetic advantageous predisposition of angiotensin converting enzyme id polymorphism in Tunisian athletes. J. Sport. Med. Phys. Fit. 2015, 56, 724–730. [Google Scholar] [CrossRef]
- Yusof, H.A.; Singh, R.; Zainuddin, Z.; Rooney, K.; Muhamed, A.M.C. Alpha-Actinin-3 (ACTN3) R/X Gene Polymorphism and Physical Performance of Multi-Ethnic Malaysian Population. Int. J. Appl. Exerc. Physiol. 2016, 5, 18–30. [Google Scholar]
- Ahmetov, I.I.; Vinogradova, O.L.; Williams, A.G. Gene polymorphisms and fiber-type composition of human skeletal muscle. Int. J. Sport Nutr. Exerc. Metabol. 2012, 22, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Ginszt, M.; Michalak-Wojnowska, M.; Gawda, P.; Wojcierowska-Litwin, M.; Korszén-Pilecka, I.; Kusztelak, M.; Muda, R.; Filip, A.A.; Majcher, P. ACTN3 genotype in professional sport climbers. J. Strength Cond. Res. 2018, 32, 1311–1315. [Google Scholar] [CrossRef]
- Seip, R.L.; Mair, K.; Cole, T.G.; Semenkovich, C.F. Induction of human skeletal muscle lipoprotein lipase gene expression by short-term exercise is transient. Am. J. Physiol. Endocrinol. Metab. 1997, 272, 255–261. [Google Scholar] [CrossRef]
- Egan, B.; O’Connor, P.L.; Zierath, J.R.; O’Gorman, D.J. Time Course Analysis Reveals Gene-Specific Transcript and Protein Kinetics of Adaptation to Short-Term Aerobic Exercise Training in Human Skeletal Muscle. PLoS ONE 2013, 8, e74098. [Google Scholar] [CrossRef]
| Variable | Women | Men | ||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| Age (years) | 27.1 | ±4.8 | 28.2 | ±5.6 |
| Weight (kg) | 60.5 | ±7.1 | 81.82 | ±11.3 |
| Size (m) | 1.6 | ±0.05 | 1.7 | ±0.06 |
| Fat (kg) | 13.3 | ±6.1 | 15.2 | ±7.0 |
| PBF (%) | 20.2 | ±4.1 | 18.3 | ±6.2 |
| Lean mass (kg) | 26.6 | ±2.5 | 38 | ±5.0 |
| Variable | Women | ||||||
|---|---|---|---|---|---|---|---|
| Pre | Post | ||||||
| Mean | SD | Mean | SD | p | |||
| Left dynamo (N) | 30.00 | ± | 6.116 | 32.70 | ± | 4.999 | 0.061 |
| Right dynamo (N) | 32.06 | ± | 5.485 | 36.53 | ± | 5.781 | 0.019 * |
| RM Front SQ (lbs) | 166.87 | ± | 32.175 | 176.87 | ± | 30.698 | 0.010 * |
| RM Shoulder P (lbs) | 90.00 | ± | 7.559 | 95.00 | ± | 10.351 | 0.050 * |
| VO2max (mL/kg/min) | 38.18 | ± | 4.826 | 38.53 | ± | 4.537 | 0.811 |
| PPower35 (N) | 1229.16 | ± | 242.562 | 1307.5 | ± | 299.983 | 0.025 * |
| PPower55 (N) | 1676.00 | ± | 446.957 | 1612.05 | ± | 421.628 | 0.281 |
| PPower75 (N) | 1957.73 | ± | 463.770 | 1895.91 | ± | 391.960 | 0.484 |
| PSplitj35 (N) | 738.88 | ± | 203.122 | 835.37 | ± | 317.444 | 0.196 |
| PSplitj55 (N) | 1676.00 | ± | 446.957 | 1612.05 | ± | 421.628 | 0.281 |
| PSplitj75 (N) | 1957.73 | ± | 463.770 | 1895.91 | ± | 391.960 | 0.484 |
| Variable | Men | ||||||
|---|---|---|---|---|---|---|---|
| Pre | Post | ||||||
| Mean | SD | Mean | SD | p | |||
| Left dynamo (N) | 49.15 | ± | 3.915 | 50.88 | ± | 3.672 | 0.072 |
| Right dynamo (N) | 51.44 | ± | 3.482 | 52.54 | ± | 4.263 | 0.160 |
| RM Front SQ (lbs) | 287.00 | ± | 35.683 | 301.75 | ± | 31.314 | 0.007 * |
| RM Shoulder P (lbs) | 159.50 | ± | 14.424 | 165.00 | ± | 14.529 | 0.003 * |
| VO2max (mL/kg/min) | 43.24 | ± | 4.538 | 45.00 | ± | 3.924 | 0.178 |
| PPower35 (N) | 2314.00 | ± | 447.057 | 2085.85 | ± | 364.059 | 0.051 |
| PPower55 (N) | 2905.73 | ± | 433.930 | 2440.71 | ± | 520.308 | 0.003 * |
| PPower75 (N) | 3022.76 | ± | 450.317 | 2544.44 | ± | 485.093 | 0.010 * |
| PSplitj35 (N) | 1317.68 | ± | 385.735 | 1320.23 | ± | 319.553 | 0.980 |
| PSplitj55 (N) | 2905.73 | ± | 433.930 | 2440.71 | ± | 520.308 | 0.003 * |
| PSplitj75 (N) | 3022.76 | ± | 450.317 | 2544.44 | ± | 485.093 | 0.010 * |
| Variable | Correlation with ACTN3 Expression | Correlation with ACE Expression | ||
|---|---|---|---|---|
| Pearson | p | Pearson | p | |
| Left dynamo (N) | 0.344 | 0.162 | 0.400 | 0.100 |
| Right dynamo (N) | 0.252 | 0.312 | 0.278 | 0.264 |
| RM Front SQ (lbs) | 0.434 | 0.072 | 0.353 | 0.151 |
| RM Shoulder P (lbs) | 0.431 | 0.074 | 0.385 | 0.115 |
| VO2max (ml/kg/min) | 0.041 | 0.871 | −0.106 | 0.675 |
| PPower35 (N) | 0.489 | 0.040 * | 0.512 | 0.030 * |
| PPower55 (N) | 0.421 | 0.082 | 0.433 | 0.073 |
| PPower75 (N) | 0.412 | 0.089 | 0.425 | 0.079 |
| PSplitj35 (N) | 0.218 | 0.385 | 0.336 | 0.173 |
| PSplitj55 (N) | 0.376 | 0.124 | 0.407 | 0.094 |
| PSplitj75 (N) | 0.350 | 0.155 | 0.432 | 0.074 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peña-Vázquez, O.; Enriquez-del Castillo, L.A.; González-Chávez, S.A.; Güereca-Arvizuo, J.; Candia Lujan, R.; Carrasco Legleu, C.E.; Cervantes Hernández, N.; Pacheco-Tena, C. Prevalence of Polymorphism and Post-Training Expression of ACTN3 (R/X) and ACE (I/D) Genes in CrossFit Athletes. Int. J. Environ. Res. Public Health 2023, 20, 4404. https://doi.org/10.3390/ijerph20054404
Peña-Vázquez O, Enriquez-del Castillo LA, González-Chávez SA, Güereca-Arvizuo J, Candia Lujan R, Carrasco Legleu CE, Cervantes Hernández N, Pacheco-Tena C. Prevalence of Polymorphism and Post-Training Expression of ACTN3 (R/X) and ACE (I/D) Genes in CrossFit Athletes. International Journal of Environmental Research and Public Health. 2023; 20(5):4404. https://doi.org/10.3390/ijerph20054404
Chicago/Turabian StylePeña-Vázquez, Omar, Liliana Aracely Enriquez-del Castillo, Susana Aideé González-Chávez, Jaime Güereca-Arvizuo, Ramon Candia Lujan, Claudia Esther Carrasco Legleu, Natanael Cervantes Hernández, and César Pacheco-Tena. 2023. "Prevalence of Polymorphism and Post-Training Expression of ACTN3 (R/X) and ACE (I/D) Genes in CrossFit Athletes" International Journal of Environmental Research and Public Health 20, no. 5: 4404. https://doi.org/10.3390/ijerph20054404
APA StylePeña-Vázquez, O., Enriquez-del Castillo, L. A., González-Chávez, S. A., Güereca-Arvizuo, J., Candia Lujan, R., Carrasco Legleu, C. E., Cervantes Hernández, N., & Pacheco-Tena, C. (2023). Prevalence of Polymorphism and Post-Training Expression of ACTN3 (R/X) and ACE (I/D) Genes in CrossFit Athletes. International Journal of Environmental Research and Public Health, 20(5), 4404. https://doi.org/10.3390/ijerph20054404







