Removal of Antibiotic Resistance Genes from Animal Wastewater by Ecological Treatment Technology Based on Plant Absorption
Abstract
1. Introduction
2. Pollution of Antibiotic Resistance Genes in Livestock and Poultry Wastewater and Its Impact on the Surrounding Environment
2.1. Generation of Antibiotic Resistance Genes in Livestock and Poultry Wastewater
2.2. Types and Levels of Contamination with Antibiotic Resistance Genes in Livestock and Poultry Wastewater
2.3. Impact of Antibiotic Resistance Genes in Wastewater on the Surrounding Environment
3. Livestock and Poultry Wastewater Treatment Technology
4. Plant Ecological Treatment Technology for Livestock Wastewater
4.1. Effectiveness of Plant Ecological Treatment Technology on the Removal of Antibiotic Resistance Genes
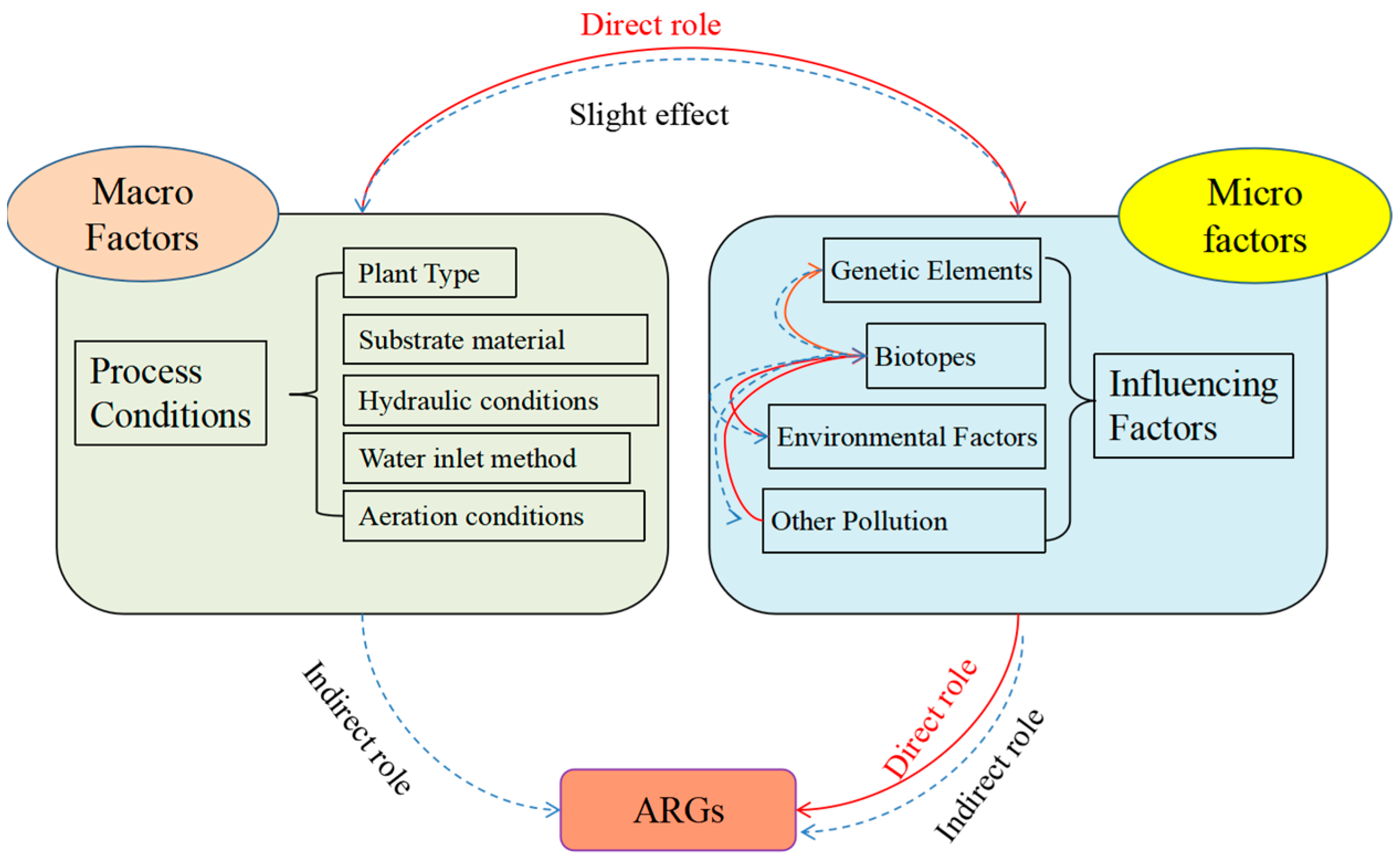
4.2. Drivers of Resistance Gene Elongation in Plant Ecological Treatment Systems
4.3. Transmission Pathways and Distribution Characteristics of Antibiotic Resistance Genes in Plant Tissues
4.4. Mechanism of Removal of Antibiotic Resistance Genes
4.5. Conclusions and Outlook
- (1)
- Continue to clarify the key drivers of ARGs. Since ARG removal is influenced by many factors and there are interactions among the various factors, it is a primary task to sort out the relationships among them, which can help to capture the main factors influencing ARGs. In addition, the current research tools mainly focus on comparative experiments and correlation coefficient analysis methods, but these tools have certain limitations. For example, the correlation coefficient analysis method can only analyze two variables and fluctuates greatly due to the number of data sets, while it is difficult to ensure that other factors remain unchanged under the condition that only one factor can be changed at a time for comparative experiments. Therefore, it is necessary to find a technical tool that can sort out the relationship between various factors, including the direct effect between factors and the indirect effect through other factors.
- (2)
- In-depth exploration of the removal mechanism of ARGs. There are various pathways for ARG removal. Although some studies suggest that microbial degradation plays a major role in ARG removal, the role of plant uptake and substrate particle adsorption cannot be ignored. They provide attachment sites for microorganisms and contaminants. In particular, the root interface is a complex mechanism for ARG removal; therefore, the root interface should be the focus of future research. Therefore, the removal mechanisms of ARGs at the root interface should be studied in depth.
- (3)
- Investigate the distribution characteristics and propagation mechanisms of ARGs in plant tissues. After absorbing nutrients from wastewater, most plants recycle them for resource use. The distribution and propagation of ARGs in plants is the key to whether ARGs can enter the next level of the food chain, and there are few studies focusing on this aspect. Therefore, the distribution characteristics of ARGs in different plant tissues and the mechanisms of their transfer should be further clarified in order to assess the ecological risk posed by ARGs in plant ecological treatment systems.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, S.; Liu, H.; Huang, H.; Lei, Q.; Wang, H.; Zhai, L.; Liu, S.; Zhang, Y.; Hu, Y. Analysis on the amount and utilization of manure in livestock and poultry breeding in China. Strateg. Study CAE 2018, 20, 103–111. (In Chinese) [Google Scholar] [CrossRef]
- Zhao, X.; Yang, C.; Han, Z. Research progress on the resource utilization mode of livestock and poultry feces waste in China. Heilongjiang Anim. Sci. Vet. Med. 2019, 13, 4–7. (In Chinese) [Google Scholar]
- Du, Z.; Liu, Y.; Zhang, R.; Wu, Y. Technologies selection to prevent and control excrement pollution from livestock and poultry farming in Huang-Huai-Hai Region. IOP Conf. Ser. Earth Environ. Sci. 2021, 766, 012104. [Google Scholar]
- Jia, Y.; Zhang, Z.; He, Z.; Zhu, P.; Zhang, Y.; Sun, T. Production Efficiency Prediction of Pig Breeding Industry by Optimized LSTM Computer Algorithm under Environmental Regulation. Sci. Program. 2021, 2021, 3074167. [Google Scholar] [CrossRef]
- UNO. Global Environmemt Outlook, GEO-6; Cambridge University Press: Singapore, 2019. [Google Scholar]
- CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019; p. 148.
- Mao, D.; Yu, S.; Rysz, M.; Luo, Y.; Yang, F.; Li, F.; Hou, J.; Mu, Q.; Alvarez, P.J.J. Prevalence and proliferation of antibiotic resistance genes in two municipal wastewater treatment plants. Water Res. 2015, 85, 458–466. [Google Scholar] [CrossRef]
- Li, S.; Hu, J. Photolytic and photocatalytic degradation of tetracycline: Effect of humic acid on degradation kinetics and mechanisms. J. Hazard. Mater. 2016, 318, 134–144. [Google Scholar] [CrossRef]
- Sui, Q.; Chen, Y.; Yu, D.; Wang, T.; Hai, Y.; Zhang, J.; Chen, M.; Wei, Y. Fates of intracellular and extracellular antibiotic resistance genes and microbial community structures in typical swine wastewater treatment processes. Environ. Int. 2019, 133, 105183. [Google Scholar] [CrossRef]
- Rysz, M.; Alvarez, P. Amplification and attenuation of tetracycline resistance in soil bacteria: Aquifer column experiments. Water Res. 2004, 38, 3705–3712. [Google Scholar] [CrossRef]
- Zhou, L.J.; Ying, G.G.; Liu, S.; Zhang, R.Q.; Lai, H.J.; Chen, Z.F.; Pan, C.G. Excretion masses and environmental occurrence of antibiotics in typical swine and dairy cattle farms in China. Sci. Total Environ. 2013, 444, 183–195. [Google Scholar] [CrossRef]
- Zhi, S.; Zhou, J.; Yang, F.; Tian, L.; Zhang, K. Systematic analysis of occurrence and variation tendency about 58 typical veterinary antibiotics during animal wastewater disposal processes in Tianjin, China. Ecotoxicol. Environ. Saf. 2018, 165, 376–385. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarden, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Van, H.A.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J. Acquired antibiotic resistance genes: An overview. Front. Microbiol. 2011, 2, 203. [Google Scholar]
- Gu, Y.; Han, B.; Huang, J.; Yang, F.; Zhang, K. Occurrence characteristics and risk assessment of resistance genes in live-stock waste from family farms in Tianjin City, China. J. Agro-Environ. Sci. 2020, 39, 394–402. (In Chinese) [Google Scholar]
- Ji, X.; Shen, Q.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.; Wu, M. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai: China. J. Hazard. Mater. 2012, 235, 178–185. [Google Scholar] [CrossRef] [PubMed]
- McKinney, C.W.; Loftin, K.A.; Meyer, M.T.; Davis, J.G.; Pruden, A. Tet and sul antibiotic resistance genes in livestock lagoons of various operation type, configuration, and antibiotic occurrence. Environ. Sci. Technol. 2010, 44, 6102–6109. [Google Scholar] [CrossRef]
- Yuan, Q.B.; Zhai, Y.F.; Mao, B.Y.; Hu, N. Antibiotic Resistance Genes and, IntI1 Prevalence in a Swine Wastewater Treatment Plant and Correlation with Metal Resistance, Bacterial Community and Wastewater Parameters. Ecotoxicol. Env. Ment. Saf. 2018, 161, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Tamminen, M.; Karkman, A.; Lõhmus, A.; Muziasari, W.I.; Takasu, H.; Wada, S.; Suzuki, S.; Virta, M. Tetracycline Resistance Genes Persist at Aquaculture Farms in the Absence of Selection Pressure. Environ. Sci. 2010, 45, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, K.; Zhi, S.; Tian, X.; Gu, Y.; Zhou, J. High prevalence and dissemination of β-lactamase genes in swine farms in northern China. Sci. Total Environ. 2019, 651, 2507–2513. [Google Scholar] [CrossRef]
- Yang, F.; Gu, Y.; Zhou, J.; Zhang, K. Swine waste: A reservoir of high-risk blaNDM and mcr-1. Sci. Total Environ. 2019, 683, 308–316. [Google Scholar] [CrossRef]
- Wu, N.; Qiao, M. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Sci. Total Environ. 2010, 44, 6933–6939. [Google Scholar] [CrossRef]
- Lu, Y.; Li, J.; Meng, J.; Zhang, J.; Zhuang, H.; Zheng, G.; Xie, W.; Ping, L.; Shan, S. Long-term biogas slurry application increased antibiotics accumulation and antibiotic resistance genes (ARGs) spread in agricultural soils with different properties. Sci. Total Environ. 2021, 759, 143473. [Google Scholar] [CrossRef] [PubMed]
- Dungan, R.S.; McKinney, C.W.; Leytem, A.B. Tracking antibiotic resistance genes in soil irrigated with dairy wastewater. Sci. Total Environ. 2018, 635, 1477–1483. [Google Scholar] [CrossRef] [PubMed]
- Kampouris, I.D.; Agrawal, S.; Orschler, L.; Cacace, D.; Kunze, S.; Berendonk, T.U.; Klümper, U. Antibiotic resistance gene load and irrigation intensity determine the impact of wastewater irrigation on antimicrobial resistance in the soil microbiome. Water Res. 2021, 193, 116818. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- GBORTONE. Integrated Anaerobic/Aerobic Biological Treatment for Intensive Swine Production. Bioresour. Technol. 2009, 100, 5424–5430. [Google Scholar] [CrossRef]
- Yang, F.; Tian, X.; Han, B.; Zhao, R.; Li, J.; Zhang, K. Tracking high-risk β-lactamase gene (bla gene) transfers in two Chinese intensive dairy farms. Environ. Pollut. 2021, 274, 116593. [Google Scholar] [CrossRef]
- Ting WH, T.; Tan IA, W.; Salleh, S.F.; Wahab, N.A. Application of water hyacinth (Eichhornia crassipes) for phytoremediation of ammoniacal nitrogen: A review. J. Water Process Eng. 2018, 22, 239–249. [Google Scholar] [CrossRef]
- Chen, J.; Deng, W.J.; Liu, Y.S.; Hu, L.X.; He, L.Y.; Zhao, J.L.; Wang, T.T.; Ying, G.G. Fate and removal of antibiotics and antibiotic resistance genes in hybrid constructed wetlands. Environ. Pollut. 2019, 249, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Guo, X.; Liu, Y.; Lu, S.; Xi, B.; Zhang, J.; Wang, Z.; Bi, B. A review on removing antibiotics and antibiotic resistance genes from wastewater by constructed wetlands: Performance and microbial response. Environ. Pollut. 2019, 254, 112996. [Google Scholar] [CrossRef]
- Ávila, C.; García-Galán, M.J.; Borrego, C.M.; Rodríguez-Mozaz, S.; García, J.; Barceló, D. New insights on the combined removal of antibiotics and ARGs in urban wastewater through the use of two configurations of vertical subsurface flow constructed wetlands. Sci. Total Environ. 2021, 755, 142554. [Google Scholar] [CrossRef]
- Chen, Y.; Vymazal, J.; Březinová, T.; Koželuh, M.; Kule, L.; Huang, J.; Chen, Z. Occurrence, removal and environmental risk assessment of pharmaceuticals and personal care products in rural wastewater treatment wetlands. Sci. Total Environ. 2016, 566, 1660–1669. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gersberg, R.M.; Ng, W.J.; Tan, S.K. Removal of pharmaceuticals and personal care products in aquatic plant-based systems: A review. Environ. Pollut. 2014, 184, 620–639. [Google Scholar] [CrossRef] [PubMed]
- García, J.; García-Galán, M.J.; Day, J.W.; Boopathy, R.; White, J.R.; Wallace, S.; Hunter, R.G. A review of emerging organic contaminants (EOCs), antibiotic resistant bacteria (ARB), and antibiotic resistance genes (ARGs) in the environment: Increasing removal with wetlands and reducing environmental impacts. Bioresour. Technol. 2020, 307, 123228. [Google Scholar] [CrossRef]
- Chen, J.; Ying, G.G.; Wei, X.D.; Liu, Y.S.; Liu, S.S.; Hu, L.X.; He, L.Y.; Chen, Z.F.; Chen, F.R.; Yang, Y.Q. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Effect of flow configuration and plant species. Sci. Total Environ. 2016, 571, 974–982. [Google Scholar] [CrossRef]
- Du, L.; Zhao, Y.; Wang, C.; Zhang, H.; Chen, Q.; Zhang, X.; Zhang, L.; Wu, J.; Wu, Z.; Zhou, Q. Removal performance of antibiotics and antibiotic resistance genes in swine wastewater by in-tegrated vertical-flow constructed wetlands with zeolite substrate. Sci. Total Environ. 2020, 721, 137765. [Google Scholar] [CrossRef]
- Chen, J.; Wei, X.D.; Liu, Y.S.; Ying, G.G.; Liu, S.S.; He, L.Y.; Su, H.C.; Hu, L.X.; Chen, F.R.; Yang, Y.Q. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Optimization of wetland substrates and hydraulic loading. Sci. Total Environ. 2016, 565, 240–248. [Google Scholar] [CrossRef]
- Huang, X.; Liu, C.; Li, K.; Liu, F.; Liao, D.; Liu, L.; Zhu, J.; Liao, J. Occurrence and distribution of veterinary antibiotics and tetracycline resistance genes in farmland soils around swine feedlots in Fujian Province, China. Environ. Sci. Pollut. Res. 2013, 20, 9066–9074. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.P.; Mao, D.Q.; Luo, Y.; Wang, L.; Xu, B.; Xu, L. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aqua-culture environment. Water Res. 2012, 46, 2355–2364. [Google Scholar] [CrossRef]
- Liu, L.; Liu, Y.; Wang, Z.; Liu, C.X.; Huang, X.; Zhu, G.F. Behavior of tetracycline and sulfamethazine with corresponding resistance genes from swine wastewater in pilot-scale constructed wetlands. J. Hazard. Mater. 2014, 278, 304–310. [Google Scholar] [CrossRef]
- Ma, J.; Cui, Y.; Li, A.; Zou, X.; Ma, C.; Chen, Z. Antibiotics and antibiotic resistance genes from wastewater treated in constructed wetlands. Eco-Log. Eng. 2022, 177, 106548. [Google Scholar] [CrossRef]
- Liu, L.; Liu, C.; Zheng, J.; Huang, X.; Wang, Z.; Liu, Y.; Zhu, G. Elimination of veterinary antibiotics and antibiotic resistance genes from swine wastewater in the vertical flow constructed wetlands. Chemosphere 2013, 91, 1088–1093. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xin, Y.; Huang, X.; Liu, X. Response of antibiotic resistance genes in constructed wetlands during treatment of livestock wastewater with different exogenous inducers: Antibiotic and antibiotic-resistant bacteria. Bioresour. Technol. 2020, 314, 123779. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Wu, H.; Zhang, J.; Brix, H. Simultaneous elimination of antibiotics resistance genes and dissolved organic matter in treatment wetlands: Characteristics and associated relationship. Chem. Eng. J. 2021, 415, 128966. [Google Scholar] [CrossRef]
- Ma, J.; Cui, Y.; Li, A.; Zhang, W.; Liang, J.; Wang, S.; Zhang, L. Evaluation of the fate of nutrients, antibiotics, and antibiotic resistance genes in sludge treatment wetlands. Sci. Total Environ. 2020, 712, 136370. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, Y.S.; Su, H.C.; Ying, G.G.; Liu, F.; Liu, S.S.; He, L.Y.; Chen, Z.F.; Yang, Y.Q.; Chen, F.R. Removal of antibiotics and antibiotic resistance genes in rural wastewater by an integrated constructed wetland. Environ. Sci. Pollution Res. 2015, 22, 1794–1803. [Google Scholar] [CrossRef]
- Chen, C.; Xia, K. Fate of land applied emerging organic contaminants in waste materials. Curr. Pollut. Rep. 2017, 3, 38–54. [Google Scholar] [CrossRef]
- Duan, M.; Li, H.; Gu, J.; Tuo, X.; Sun, W.; Qian, X.; Wang, X. Effects of biochar on reducing the abundance of oxytetracycline, antibiotic resistance genes, and human pathogenic bacteria in soil and lettuce. Environ. Pollut. 2017, 224, 787–795. [Google Scholar] [CrossRef]
- Zhu, B.; Chen, Q.; Chen, S.; Zhu, Y.G. Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? Environ. Int. 2017, 98, 152–159. [Google Scholar] [CrossRef]
- Zhi, S.; Ding, G.; Li, A.; Guo, H.; Shang, Z.; Ding, Y.; Zhang, K. Fate of antibiotic resistance genes during high solid anaerobic digestion with pig manure: Focused on different starting modes. Bioresour. Technol. 2021, 328, 124849. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Wang, P.; Yuan, Q.; Zhou, W. Study on photocatalytic degradation and reaction kinetics of tetracycline antibiotics in biogas slurry. Trans. Chin. Soc. Agric. Eng. 2018, 34, 193–198. [Google Scholar]
- Berg, J.; Thorsen, M.K.; Holm, P.E.; Jensen, J.; Nybroe, O.; Brandt, K.K. Cu exposure under field conditions coselects for antibiotic resistance as determined by a novel cultivation-independent bacterial community tolerance assay. Environ. Sci. Technol. 2010, 44, 8724–8728. [Google Scholar] [CrossRef] [PubMed]
- Truu, M.; Juhanson, J.; Truu, J. Microbial biomass, activity and community com-position in constructed wetlands. Sci. Total Environ. 2009, 407, 3958–3971. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Ren, S.; Niu, T.; Guo, Y.; Qi, S.; Han, X.; Liu, D.; Pan, F. Distribution of antibiotic-resistant bacteria in chicken manure and manure-fertilized vegetables. Environ. Sci. Pollut. Res. 2014, 21, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Sun, M.; Feng, Y.; Wan, J.; Xie, S.; Tian, D.; Zhao, Y.; Wu, J.; Hu, F.; Li, H.; et al. Effect of biochar amendment on the control of soil sulfonamides, antibiotic-resistant bacteria, and gene enrichment in lettuce tissues. J. Hazard. Mater. 2016, 309, 219–227. [Google Scholar] [CrossRef]
- Guo, X.P.; Li, J.; Yang, F.; Yang, J.; Yin, D. Prevalence of sulfonamide and tetracycline resistance genes in drinking water treatment plants in the Yangtze River Delta, China. Sci. Total Environ. 2014, 493, 626–631. [Google Scholar] [CrossRef]
- Yang, Y.; Li, B.; Zou, S.H.; Fang, H.H.; Zhang, T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014, 62, 97–106. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Philipp Rauf, P.; Huettel, B.; Reinhardt, R.; Elmon Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- He, S.; Wang, Y.M.; Li, C.S.; Li, Y.; Zhou, J. The nitrogen removal performance and microbial communities in a two-stage deep sequencing constructed wetland for advanced treatment of secondary effluent. Bioresour. Technol. 2018, 248, 82–88. [Google Scholar] [CrossRef]
- He, T.; Wei, G.; Luan, Z.Y.; Xie, S.G. Spatiotemporal variation of bacterial and archaeal communities in a pilot-scale constructed wetland for surface water treatment. Appl. Microbiol. Biotechnol. 2015, 100, 1479–1488. [Google Scholar] [CrossRef]
- Chen, Q.L.; An, X.L.; Zhu, Y.G.; Xie, S. Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere. Environ. Sci. Technol. 2017, 51, 8149–8157. [Google Scholar] [CrossRef] [PubMed]
- Duran, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 2018, 175, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Hu, H.W.; Chen, Q.L.; Singh, B.K.; Yan, H.; Chen, D.; He, J.Z. Transfer of antibiotic resistance from manure-amended soils to vegetable microbiomes. Environ. Int. 2019, 130, 104912. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.; Saldierna, G.J.; Shay, J. Transmission of bacterial endophyte. Microorganisms 2017, 5, 70. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]


| Wastewater Types | Botany Types | Variable Factors | Target ARGs | Removal Effects | References |
|---|---|---|---|---|---|
| Domestic wastewater | Cyperus alternifolius L. | Artificial aeration and mixing design | sul1, sul2, tetG, tetO, ermB, qnrS, qnrD, cmlA and floR | 87.8~99.1% | [30] |
| Domestic wastewater | Thalia dealbata Fraser. and Iris tectorum Maxim. | Flow patterns and plant types | sul1, sul2, sul3, tetG, tetM, tetO, tetX, ermB, ermC, cmlA and floR | 63.9~84.0% | [36] |
| Domestic wastewater | Cyperus alternifolius L. | Substrate and hydraulic load | sul1, sul2, sul3, tetG, tetM, tetO, tetX, ermB, ermC, qnrB, qnrD, qnrS, cmlA, fexA, fexB, floR, intl1 and intl2 | 50.0~85.8% | [38] |
| Pig farm wastewater | P. australis | Vertical Flow Artificial Wetland | sul1, sul2 and sul3 | 89%, 88% and 84% | [42] |
| Pig farm wastewater | Hybrid pennisetum | Filler type | tetM, tetO and tetW | 50% | [43] |
| Pig farm wastewater | Arundo donax | Filler type | sulI, sulII, sulIII, tetM, tetO and tetW | 67.5%, 85.6%, 95.6%, 87.9%, 97.9% and 98.5% | [37] |
| Synthetic pig farm wastewater | P. australis | Water flow method | sulI, sulII, tetM, tetW and tetO | 99.9% (Sulfonamides); 99.9% (Tetracycline) | [41] |
| Livestock wastewater | P. australis | Exogenous antibiotics and resistant bacteria | 73 ARGs | >60% | [44] |
| Pig farm wastewater after digestion | Iris pseudacorus | With or without aeration | tetA, tetM, tetO and tetW | 87.88% | [45] |
| Urban wastewater | P. australis | Operating conditions | intI1, qnrS, sul1, sul2, blaTEM and ermB | −7.67~92.9% | [32] |
| Wetlands wastewater | P. australis | With or without aeration | sul1, sul2, tetA, tetC, ermB and intl1 | 12.3~39.2% | [46] |
| Pig farm wastewater | Pontederia cordata and M. verticillatum L. | Water flow method | sul3, intI1, sul2, sul1, tetO, ermB, intI2, tetB/P, ermC, tetM and tetX | 87~99% | [47] |
| Wastewater Types | Botany Types | Analysis Method | Target ARGs | Influencing Factors and Conclusions | References |
|---|---|---|---|---|---|
| Domestic wastewater | Cyperus alternifolius L. | Correlation factor method | sul1, sul2, tetG, tetO, ermB, qnrS, qnrD, cmlA and floR | Dissolved oxygen, antibiotic levels significantly affect microorganisms and thus ARGs | [30] |
| Domestic wastewater | Thalia dealbata Fraser. and Iris tectorum Maxim. | Comparison test | sul1, sul2, sul3, tetG, tetM, tetO, tetX, ermB, ermC, cmlA and floR | Plant type had a significant effect | [36] |
| Domestic wastewater | Cyperus alternifolius L. | Analyzing Data | sul1, sul2, sul3, tetG, tetM, tetO, tetX, ermB, ermC, qnrB, qnrD, qnrS, cmlA, fexA, fexB, floR, intl1 and intl2 | Microbial activity is significantly correlated with pollutant removal | [38] |
| Pig farm wastewater | Arundo donax | Correlation coefficient | sulI, sulII, sulIII, tetM, tetO and tetW | The removal rate of ARGs was significantly and negatively correlated with the absolute abundance of 16S and ARGs but not with the relative abundance of ARGs | [37] |
| Synthetic pig farm wastewater | P. australis | Comparison test | sulI, sulII, tetM, tetO and tetW | pH 7-8 is optimal, added oxygen content does not contribute to the abatement of ARGs, and the effect of antibiotics is not significant | [41,55] |
| Livestock wastewater | P. australis | Comparison test | 73 target ARGs | Abundance of ARGs promoted by oxytetracycline and exogenous drug-resistant bacteria | [44] |
| Pig farm wastewater after digestion | Iris pseudacorus | Correlation coefficient method | tetA, tetM, tetO and tetW | Soluble organic matter composition and content, COD were significantly correlated with tetA, tetM, tetO and not with tetW; oxygen content | [45] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Du, D.; Ding, Y.; Zhang, K.; Zhi, S. Removal of Antibiotic Resistance Genes from Animal Wastewater by Ecological Treatment Technology Based on Plant Absorption. Int. J. Environ. Res. Public Health 2023, 20, 4357. https://doi.org/10.3390/ijerph20054357
Wang H, Du D, Ding Y, Zhang K, Zhi S. Removal of Antibiotic Resistance Genes from Animal Wastewater by Ecological Treatment Technology Based on Plant Absorption. International Journal of Environmental Research and Public Health. 2023; 20(5):4357. https://doi.org/10.3390/ijerph20054357
Chicago/Turabian StyleWang, Han, Delin Du, Yongzhen Ding, Keqiang Zhang, and Suli Zhi. 2023. "Removal of Antibiotic Resistance Genes from Animal Wastewater by Ecological Treatment Technology Based on Plant Absorption" International Journal of Environmental Research and Public Health 20, no. 5: 4357. https://doi.org/10.3390/ijerph20054357
APA StyleWang, H., Du, D., Ding, Y., Zhang, K., & Zhi, S. (2023). Removal of Antibiotic Resistance Genes from Animal Wastewater by Ecological Treatment Technology Based on Plant Absorption. International Journal of Environmental Research and Public Health, 20(5), 4357. https://doi.org/10.3390/ijerph20054357








