Synanthropic and Wild Animals as Sentinels of Zoonotic Agents: A Study of Leptospira Genotypes Circulating in Northeastern Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.1.1. Myocastor Coypus Eradication Action Policy: Sampling Period 2009
2.1.2. Synanthropic and Wild Animals Public Veterinary Survey: Sampling from 2015 to 2022
2.2. Molecular Analysis
2.3. Genotyping
2.4. Data Anaylisis
Statistical Analysis
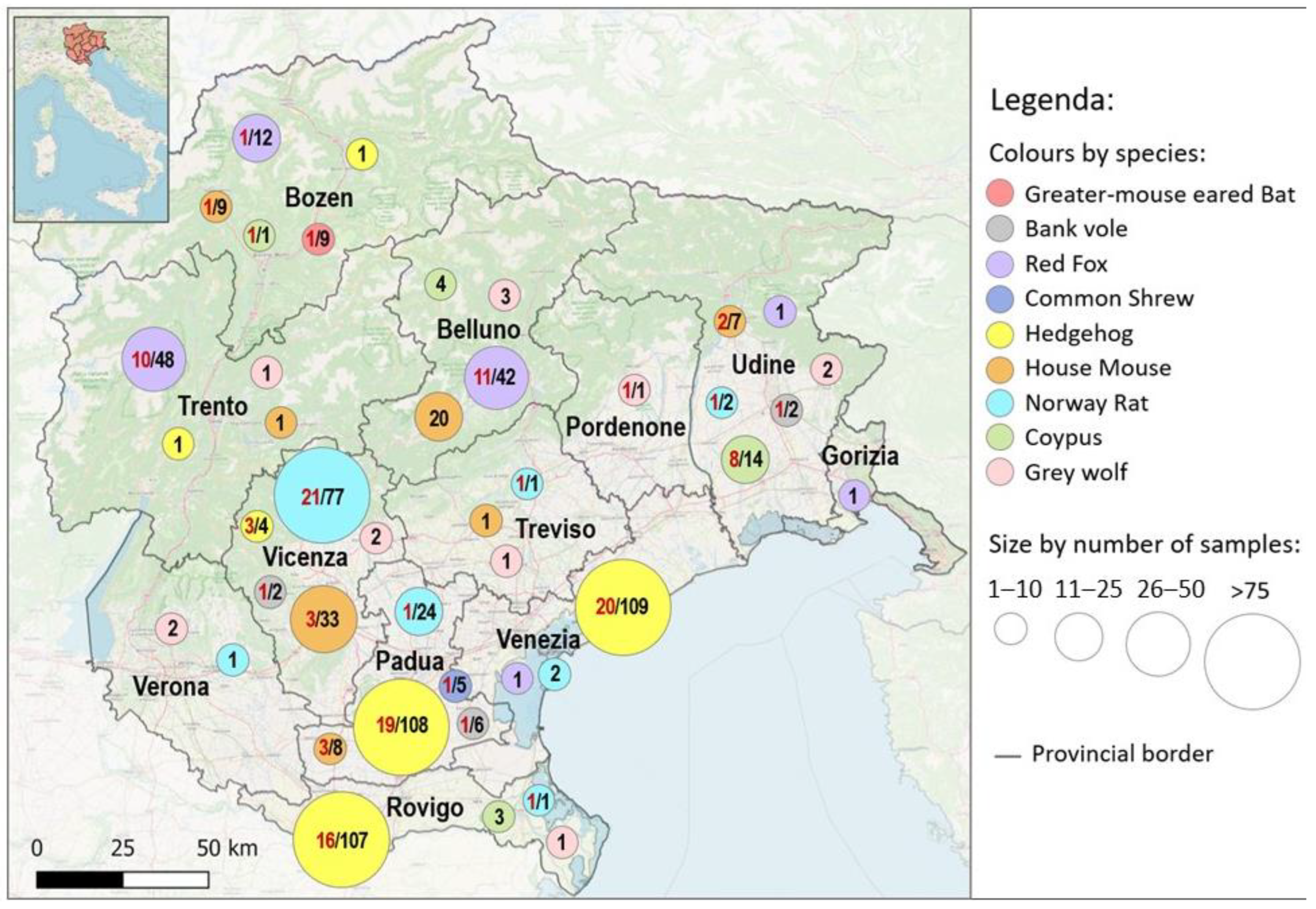
3. Results
3.1. Sampling and Molecular Analysis
3.1.1. Myocastor Coypus Eradication Action Policy: Sampling Period 2009
3.1.2. Synanthropic and Wild Animals: Sampling from 2015 to 2022
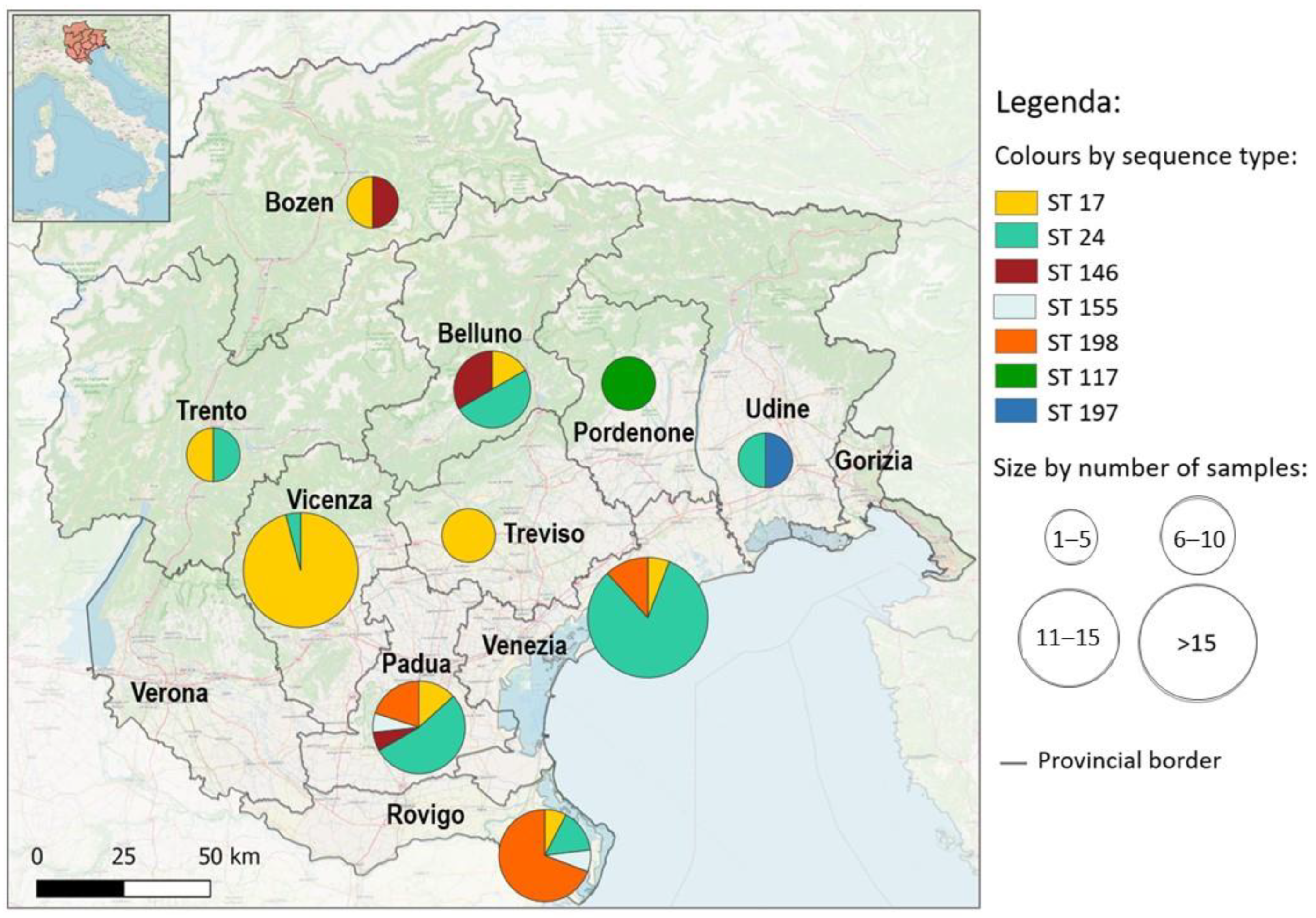
3.2. Genotyping
3.3. Evaluation of Seasonality as an Effect on Leptospira Positivity
4. Discussion
4.1. Myocastor Coypus
4.2. Synanthropic and Wild Animals
4.3. Leptospira spp. and Seasonality
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Haake, D.A.; Levett, P.N. Leptospirosis in Humans. In Leptospira and Leptospirosis. Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2015; Volume 387. [Google Scholar] [CrossRef]
- Levett, P.N. Systematics of Leptospiraceae. In Leptospira and Leptospirosis. Current Topics in Microbiology and Immunology; Adler, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 387. [Google Scholar] [CrossRef]
- WHO. World Organization for Animal Health LEPTOSPIROSIS. Man. Diagnostic Tests Vaccines Terr. Anim. 2018, 2018, 503–516. [Google Scholar]
- Thibeaux, R.; Girault, D.; Bierque, E.; Soupé-Gilbert, M.E.; Rettinger, A.; Douyère, A.; Meyer, M.; Iraola, G.; Picardeau, M.; Goarant, C. Biodiversity of environmental Leptospira: Improving identification and revisiting the diagnosis. Front. Microbiol. 2018, 9, 816. [Google Scholar] [CrossRef] [PubMed]
- Shieh, W.J.; Edwards, C.; Spiegel, R. Leptospirosis. In Ropical Infectious Diseases: Principles, Pathogens, and Practice; Guerrant, R.L., Walker, D.H., Weller, P.F., Eds.; Churchill Livingstone: Philadelphia, PA, USA, 1999; pp. 547–555. [Google Scholar]
- Verma, V.; Goyal, M.; Kala, D.; Gupta, S.; Kumar, D.; Kaushal, A. Recent advances in the diagnosis of leptospirosis. Front. Biosci.-Landmark 2020, 25, 1655–1681. [Google Scholar] [CrossRef]
- Mazzotta, E.; Lucchese, L.; Salata, C.; Furlanello, T.; Baroni, E.; Zotti, A.; Venturi, G.; Fincato, A.; Marchione, S.; Capello, K.; et al. Are Small Animal Practitioners Occupationally Exposed to Leptospirosis? Results of a Serological Survey. Int. J. Environ. Res. Public Health 2022, 19, 1797. [Google Scholar] [CrossRef]
- Le Turnier, P.; Epelboin, L. Update on leptospirosis. Rev. Med. Interne 2019, 40, 306–312. [Google Scholar] [CrossRef]
- Karpagam, K.B.; Ganesh, B. Leptospirosis: A neglected tropical zoonotic infection of public health importance—An updated review. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 835–846. [Google Scholar] [CrossRef]
- Lau, C.L.; Smythe, L.D.; Craig, S.B.; Weinstein, P. Climate change, flooding, urbanisation and leptospirosis: Fuelling the fire? Trans. R. Soc. Trop. Med. Hyg. 2010, 104, 631–638. [Google Scholar] [CrossRef]
- Vitale, M.; Agnello, S.; Chetta, M.; Amato, B.; Vitale, G.; Bella, C.D.; Vicari, D.; Presti, V.D.M.L. Human leptospirosis cases in Palermo Italy. The role of rodents and climate. J. Infect. Public Health 2018, 11, 209–214. [Google Scholar] [CrossRef]
- Adler, B. History of Leptospirosis and Leptospira. In Leptospira and Leptospirosis. Current Topics in Microbiology and Immunology; Adler, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 387. [Google Scholar] [CrossRef]
- Birtles, R. Leptospira Infections. In Infectious Diseases of Wild Mammals and Birds in Europe; Gavier-Widén, D., Duff, J.P., Meredith, A., Eds.; Wiley: Hoboken, NJ, USA, 2012; pp. 402–408. ISBN 9781405199056. [Google Scholar]
- Moinet, M.; Fournier-Chambrillon, C.; André-Fontaine, G.; Aulagnier, S.; Mesplède, A.; Blanchard, B.; Descarsin, V.; Dumas, P.; Dumas, Y.; Coïc, C.; et al. Leptospirosis in free-ranging endangered European Mink (Mustela lutreola) and other small carnivores (mustelidae, viverridae) from Southwestern France. J. Wildl. Dis. 2010, 46, 1141–1151. [Google Scholar] [CrossRef]
- Bregoli, M.; Pesaro, S.; Ustulin, M.; Vio, D.; Beraldo, P.; Galeotti, M.; Cocchi, M.; Lucchese, L.; Bertasio, C.; Boniotti, M.B.; et al. Environmental Exposure of Wild Carnivores to Zoonotic Pathogens: Leptospira Infection in the First Free Living Wolf (Canis lupus Linnaeus, 1758) Found Dead in the Friuli Venezia Giulia Region. Public Health 2021, 18, 2512. [Google Scholar] [CrossRef]
- Dietrich, M.; Mühldorfer, K.; Tortosa, P.; Markotter, W. Leptospira and Bats: Story of an Emerging Friendship. PLoS Pathog 2015, 11, 1005176. [Google Scholar] [CrossRef] [PubMed]
- Schertler, A.; Rabitsch, W.; Moser, D.; Wessely, J.; Essl, F. The potential current distribution of the coypu (Myocastor coypus) in Europe and climate change induced shifts in the near future. NeoBiota 2020, 58, 129–160. [Google Scholar] [CrossRef]
- Žele-Vengušt, D.; Lindtner-Knific, R.; Mlakar-Hrženjak, N.; Jerina, K.; Vengušt, G. Exposure of Free-Ranging Wild Animals to Zoonotic Leptospira interrogans Sensu Stricto in Slovenia. Animals 2021, 11, 2722. [Google Scholar] [CrossRef]
- Zanzani, S.A.; Di Cerbo, A.; Gazzonis, A.L.; Epis, S.; Invernizzi, A.; Tagliabue, S.; Manfredi, M.T. Parasitic and bacterial infections of myocastor coypus in a metropolitan area of northwestern Italy. J. Wildl. Dis. 2016, 52, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Ayral, F.; Kodjo, A.; Guédon, G.; Boué, F.; Richomme, C. Muskrats are greater carriers of pathogenic Leptospira than coypus in ecosystems with temperate climates. PLoS ONE 2020, 15, e0228577. [Google Scholar] [CrossRef] [PubMed]
- Millán, J.; Cevidanes, A.; Chirife, A.D.; Candela, M.G.; León-Vizcaíno, L. Risk factors of Leptospira infection in Mediterranean periurban micromammals. Zoonoses Public Health 2018, 65, e79–e85. [Google Scholar] [CrossRef]
- Schuller, S.; Francey, T.; Hartmann, K.; Hugonnard, M.; Kohn, B.; Nally, J.E.; Sykes, J. European consensus statement on leptospirosis in dogs and cats. J. Small Anim. Pract. 2015, 56, 159–179. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.; Obiegala, A.; Imholt, C.; Drewes, S.; Saathoff, M.; Freise, J.; Runge, M.; Jacob, J.; Mayer-Scholl, A.; Ulrich, R.G.; et al. Influence of Season, Population and Individual Characteristics on the Prevalence of Leptospira spp. in Bank Voles in North-West Germany. Biology 2021, 10, 933. [Google Scholar] [CrossRef]
- Bertasio, C.; Papetti, A.; Scaltriti, E.; Tagliabue, S.; D’Incau, M.; Boniotti, M.B. Serological Survey and Molecular Typing Reveal New Leptospira Serogroup Pomona Strains among Pigs of Northern Italy. Pathogens 2020, 9, 332. [Google Scholar] [CrossRef]
- Levett, P.N.; Morey, R.E.; Galloway, R.L.; Turner, D.E.; Steigerwalt, A.G.; Mayer, L.W. Detection of pathogenic leptospires by real-time quantitative PCR. J. Med. Microbiol. 2005, 54, 45–49. [Google Scholar] [CrossRef]
- Smythe, L.D.; Smith, I.L.; Smith, G.A.; Dohnt, M.F.; Symonds, M.L.; Barnett, L.J.; McKay, D.B. A quantitative PCR (TaqMan) assay for pathogenic Leptospira spp. BMC Infect. Dis. 2002, 2, 13. [Google Scholar] [CrossRef] [PubMed]
- Boonsilp, S.; Thaipadungpanit, J.; Amornchai, P.; Wuthiekanun, V.; Bailey, M.S.; Holden, M.T.G.; Zhang, C.; Jiang, X.; Koizumi, N.; Taylor, K.; et al. A Single Multilocus Sequence Typing (MLST) Scheme for Seven Pathogenic Leptospira Species. PLoS Negl. Trop. Dis. 2013, 7, e1954. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Maiden, M.C.J. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010, 11, 595. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Rabe-Hesketh, S.; Dorie, V.; Gelman, A.; Liu, J. A Nondegenerate Penalized Likelihood Estimator for Variance Parameters in Multilevel Models. Psychometrika 2013, 78, 685–709. [Google Scholar] [CrossRef]
- Fischer, S.; Mayer-Scholl, A.; Imholt, C.; Spierling, N.G.; Heuser, E.; Schmidt, S.; Reil, D.; Rosenfeld, U.M.; Jacob, J.; Nöckler, K.; et al. Leptospira Genomospecies and Sequence Type Prevalence in Small Mammal Populations in Germany. Vector-Borne Zoonotic Dis. 2018, 18, 188–199. [Google Scholar] [CrossRef]
- Obiegala, A.; Woll, D.; Karnath, C.; Silaghi, C.; Schex, S.; Eßbauer, S.; Pfeffer, M. Prevalence and Genotype Allocation of Pathogenic Leptospira Species in Small Mammals from Various Habitat Types in Germany. PLoS Negl. Trop. Dis. 2016, 10, e0004501. [Google Scholar] [CrossRef]
- Yusof, M.A.; Mohd-Taib, F.S.; Ishak, S.N.; Md-Nor, S.; Md-Sah, S.A.; Mohamed, N.Z.; Azhari, N.N.; Neela, V.; Sekawi, Z. Microhabitat Factors Influenced the Prevalence of Pathogenic Leptospira spp. in Small Mammal Host. Ecohealth 2019, 16, 260–274. [Google Scholar] [CrossRef]
- Monahan, A.M.; Callanan, J.J.; Nally, J.E. Review paper: Host-pathogen interactions in the kidney during chronic leptospirosis. Vet. Pathol. 2009, 46, 792–799. [Google Scholar] [CrossRef]
- Boey, K.; Shiokawa, K.; Rajeev, S. Leptospira infection in rats: A literature review of global prevalence and distribution. PLoS Negl. Trop. Dis. 2019, 13, e0007499. [Google Scholar] [CrossRef]
- Mohd-Taib, F.S.; Ishak, S.N.; Yusof, M.A.; Azhari, N.N.; Md-Lasim, A.; Md. Nor, S.; Mohd-Sah, S.A.; Neela, V.K. Leptospirosis: An insight into community structure of small mammal’s host in urban environment. Trop. Biomed. 2020, 37, 142–154. [Google Scholar]
- Aviat, F.; Blanchard, B.; Michel, V.; Blanchet, B.; Branger, C.; Hars, J.; Mansotte, F.; Brasme, L.; De Champs, C.; Bolut, P.; et al. Leptospira exposure in the human environment in France: A survey in feral rodents and in fresh water. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Michel, V.; Ruvoen-Clouet, N.; Menard, A.; Sonrier, C.; Fillonneau, C.; Rakotovao, F.; Ganière, J.P.; André-Fontaine, G. Role of the coypu (Myocastor coypus) in the epidemiology of leptospirosis in domestic animals and humans in France. Eur. J. Epidemiol. 2001, 17, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Ashford, R.W.W. When Is a Reservoir Not a Reservoir? Emerg. Infect. Dis. 2003, 9, 1495–1496. [Google Scholar] [CrossRef] [PubMed]
- Ayral, F.; Djelouadji, Z.; Raton, V.; Zilber, A.-L.; Gasqui, P.; Faure, E.; Baurier, F.; Vourc’h, G.; Kodjo, A.; Combes, B. Hedgehogs and Mustelid Species: Major Carriers of Pathogenic Leptospira, a Survey in 28 Animal Species in France (20122015). PLoS ONE 2016, 11, e0162549. [Google Scholar] [CrossRef]
- Żmudzki, J.; Arent, Z.; Jabłoński, A.; Nowak, A.; Zębek, S.; Stolarek, A.; Bocian, Ł.; Brzana, A.; Pejsak, Z. Seroprevalence of 12 serovars of pathogenic Leptospira in red foxes (Vulpes vulpes) in Poland. Acta Vet. Scand. 2018, 60, 34. [Google Scholar] [CrossRef]
- Slavica, A.; Cvetnić, Ž.; Milas, Z.; Janicki, Z.; Turk, N.; Konjević, D.; Severin, K.; Tončić, J.; Lipej, Z. Incidence of leptospiral antibodies in different game species over a 10-year period (1996–2005) in Croatia. Eur. J. Wildl. Res. 2008, 54, 305–311. [Google Scholar] [CrossRef]
- Millán, J.; Candela, M.G.; López-Bao, J.V.; Pereira, M.; Jiménez, M.Á.; León-Vizcaíno, L. Leptospirosis in wild and domestic carnivores in natural areas in Andalusia, Spain. Vector-Borne Zoonotic Dis. 2009, 9, 549–554. [Google Scholar] [CrossRef]
- Åkerstedt, J.; Lillehaug, A.; Larsen, I.L.; Eide, N.E.; Arnemo, J.M.; Handeland, K. Serosurvey for canine distemper virus, canine adenovirus, leptospira interrogans, andtoxoplasma gondii in free-ranging canids in Scandinavia And Svalbard. J. Wildl. Dis. 2010, 46, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Cilia, G.; Bertelloni, F.; Angelini, M.; Cerri, D.; Fratini, F. Leptospira Survey in Wild Boar (Sus scrofa) Hunted in Tuscany, Central Italy. Pathogens 2020, 9, 377. [Google Scholar] [CrossRef]
- Vengust, G.; Lindtner-Knific, R.; Zele, D.; Bidovec, A. Leptospira antibodies in wild boars (Sus scrofa) in Slovenia. Eur. J. Wildl. Res. 2008, 54, 749–752. [Google Scholar] [CrossRef]
- Cano-Manuel, F.J.; López-Olvera, J.; Fandos, P.; Soriguer, R.C.; Pérez, J.M.; Granados, J.E. Long-term monitoring of 10 selected pathogens in wild boar (Sus scrofa) in Sierra Nevada National Park, southern Spain. Vet. Microbiol. 2014, 174, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Stritof Majetic, Z.; Galloway, R.; Ruzic Sabljic, E.; Milas, Z.; Mojcec Perko, V.; Habus, J.; Margaletic, J.; Pernar, R.; Turk, N. Epizootiological survey of small mammals as Leptospira spp. reservoirs in Eastern Croatia. Acta Trop. 2014, 131, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.C.T.R.B.; Colocho, R.A.B.; Pereira, C.R.; Lage, A.P.; Heinemann, M.B.; Dorneles, E.M.S. Canine leptospirosis in stray and sheltered dogs: A systematic review. Anim. Health Res. Rev. 2022, 23, 39–58. [Google Scholar] [CrossRef] [PubMed]
- Salaün, L.; Mérien, F.; Gurianova, S.; Baranton, G.; Picardeau, M. Application of multilocus variable-number tandem-repeat analysis for molecular typing of the agent of leptospirosis. J. Clin. Microbiol. 2006, 44, 3954–3962. [Google Scholar] [CrossRef] [PubMed]
- Bertasio, C.; Boniotti, M.B.; Lucchese, L.; Ceglie, L.; Bellinati, L.; Mazzucato, M.; Furlanello, T.; D’Incau, M.; Natale, A. Detection of New Leptospira Genotypes Infecting Symptomatic Dogs: Is a New Vaccine Formulation Needed? Pathogens 2020, 9, 484. [Google Scholar] [CrossRef] [PubMed]
| Province | Species | Serogroups | Serovar | N | Total | MAT Range | Prevalence (%) | CI |
|---|---|---|---|---|---|---|---|---|
| TN | L interrogans | Australis | Bratislava | 2 | 30 | 1:200–1:1600 | 6.7 | 0.8–22.1 |
| PD | L interrogans | Australis | Bratislava | 21 | 41 | 1:100–1:3200 | 51.2 | 35.1–67.1 |
| L interrogans | Icterohaemorrhagiae | Icterohaemorrhagiae | 2 | 1:100–1:200 | 4.9 | 0.0–0.2 | ||
| L interrogans | Icterohaemorrhagiae | Copenhageni | 1 | 1:100 | 2.4 | 0.1–12.9 |
| Species | Positives | Negatives | N | Prevalence (%) | CI |
|---|---|---|---|---|---|
| Coypus | 9 | 13 | 22 | 40.9 | 23.2–61.2 |
| Bank Vole | 3 | 7 | 10 | 30.0 | 10.8–60.3 |
| Norway Rat | 25 | 83 | 108 | 23.1 | 15.6–32.2 |
| Red Fox | 22 | 83 | 105 | 21.0 | 13.6–30.0 |
| Common Shrew | 1 | 4 | 5 | 20.0 | 1.0–62.4 |
| Hedgehog | 58 | 272 | 330 | 17.6 | 13.6–22.1 |
| House Mouse | 9 | 70 | 79 | 11.4 | 5.3–20.5 |
| Greater-Mouse Eared Bat | 1 | 8 | 9 | 11.1 | 0.6–43.5 |
| Grey Wolf | 1 | 12 | 13 | 7.7 | 0.4–33.3 |
| Totals | 129 | 552 | 681 | 18.9 | 16.1–22.1 |
| Host | ST | Species | Serogroups | Serovar | N | Prevalence | CI 95% |
|---|---|---|---|---|---|---|---|
| Hedgehog | 24 | L interrogans | Australis | Bratislava/Jalna/Lora/Muenchen | 24 | 54.5% | 37.9–68.3 |
| 198 | Australis | 15 | 34.1% | 20.0–48.9 | |||
| 17 | L interrogans | Icterohaemorrhagiae | Icterohaemorrhagiae/Copenhageni | 3 | 6.8% | 1.4–11.8 | |
| 146 | L borgpetersenii | Javanica | Javanica/Poi/Sorexjalna | 1 | 2.3% | 0.07–11.8 | |
| 155 | L borgpetersenii | Sejroe | Polonica/Saxkoebing/Istrica | 1 | 2.3% | 0.07–11.8 | |
| House Mouse | 24 | L interrogans | Australis | Australis | 1 | 16.7% | 0.9–56.4 |
| 17 | L interrogans | Icterohaemorragiae | Icterohaemorrhagiae/Copenhageni | 3 | 50% | 18.8–81.2 | |
| 155 | L borgpetersenii | Sejroe | Polonica/Saxkoebing/Istrica | 1 | 16.7% | 0.9–56.4 | |
| 146 | L borgpetersenii | Javanica | Javanica/ Poi/ Sorexjalna | 1 | 16.7% | 0.9–56.4 | |
| Norway Rat | 17 | L interrogans | Icterohaemorrhagiae | Icterohaemorrhagiae/Copenhageni | 22 | 100% | 0.85–1 |
| Red Fox | 24 | L interrogans | Australis | Bratislava/Jalna/Lora/Muenchen | 4 | 44.4% | 18.9–73.3 |
| 17 | L interrogans | Icterohaemorragiae | Icterohaemorrhagiae/Copenhageni | 3 | 33.3% | 12.1–64.6 | |
| 146 | L borgpetersenii | Javanica | Javanica/ Poi/ Sorexjalna | 2 | 22.2% | 6.3–54.7 | |
| Bank vole | 197 | L borgpetersenii | Sejroe | 1 | 100% | 2.6–97.4 | |
| Grey Wolf | 117 | L. kirschneri | Pomona | Mozdok | 1 | 100% | 5.1–1 |
| Spring | Summer | Autumn | Winter | p | |
| Hedgehog | 14% 95%CI 5.9–27.2 | 25.3% 95%CI 16.6–35.7 | 17.6% 95%CI 11.7–24.9 | 7.7% 95%CI 2.1–18.5 | p = 0.04 |
| Norway Rat | 28.6% 95%CI 15.7–44.6 | 16.7% 95%CI 0.4–64.1 | 17.8% 95%CI 8.0–32.1 | 26.7% 95%CI 7.8–55.1 | p = 0.7 |
| House Mouse | 5.9% 95%CI 0.7–19.7 | 22.7% 95%CI 7.8–45.4 | 9.1% 95%CI 0.2–41.3 | 8.3% 95%CI 0.2–38.4 | p = 0.8 |
| Red Fox | 25% 95%CI 8.7–49.1 | 18.2% 95%CI 5.2–40.3 | 11.1% 95%CI 1.4–34.7 | 24.4% 95%CI 12.9–39.5 | p = 0.7 |
| Overall | 20% 95%CI 14.3–26.6 | 24% 95%CI 17.3–31.4 | 17% 95%CI 12.3–22.6 | 15.2% 95%CI 9.6–22.6 | p = 0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mazzotta, E.; Bellinati, L.; Bertasio, C.; Boniotti, M.B.; Lucchese, L.; Ceglie, L.; Martignago, F.; Leopardi, S.; Natale, A. Synanthropic and Wild Animals as Sentinels of Zoonotic Agents: A Study of Leptospira Genotypes Circulating in Northeastern Italy. Int. J. Environ. Res. Public Health 2023, 20, 3783. https://doi.org/10.3390/ijerph20053783
Mazzotta E, Bellinati L, Bertasio C, Boniotti MB, Lucchese L, Ceglie L, Martignago F, Leopardi S, Natale A. Synanthropic and Wild Animals as Sentinels of Zoonotic Agents: A Study of Leptospira Genotypes Circulating in Northeastern Italy. International Journal of Environmental Research and Public Health. 2023; 20(5):3783. https://doi.org/10.3390/ijerph20053783
Chicago/Turabian StyleMazzotta, Elisa, Laura Bellinati, Cristina Bertasio, Maria Beatrice Boniotti, Laura Lucchese, Letizia Ceglie, Federico Martignago, Stefania Leopardi, and Alda Natale. 2023. "Synanthropic and Wild Animals as Sentinels of Zoonotic Agents: A Study of Leptospira Genotypes Circulating in Northeastern Italy" International Journal of Environmental Research and Public Health 20, no. 5: 3783. https://doi.org/10.3390/ijerph20053783
APA StyleMazzotta, E., Bellinati, L., Bertasio, C., Boniotti, M. B., Lucchese, L., Ceglie, L., Martignago, F., Leopardi, S., & Natale, A. (2023). Synanthropic and Wild Animals as Sentinels of Zoonotic Agents: A Study of Leptospira Genotypes Circulating in Northeastern Italy. International Journal of Environmental Research and Public Health, 20(5), 3783. https://doi.org/10.3390/ijerph20053783










