Adaptation of Rhizosphere Microbial Communities to Continuous Exposure to Multiple Residual Antibiotics in Vegetable Farms
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Standards
2.2. Site Selection and Rhizosphere Sample Collection
2.3. DNA Extraction, PCR Amplification, and Illumina High-Throughput Sequencing
2.4. Soil Antibiotics Extraction and Quantification
2.5. Determination of Soil Physiochemical Properties
2.6. Statistical Analysis
3. Results
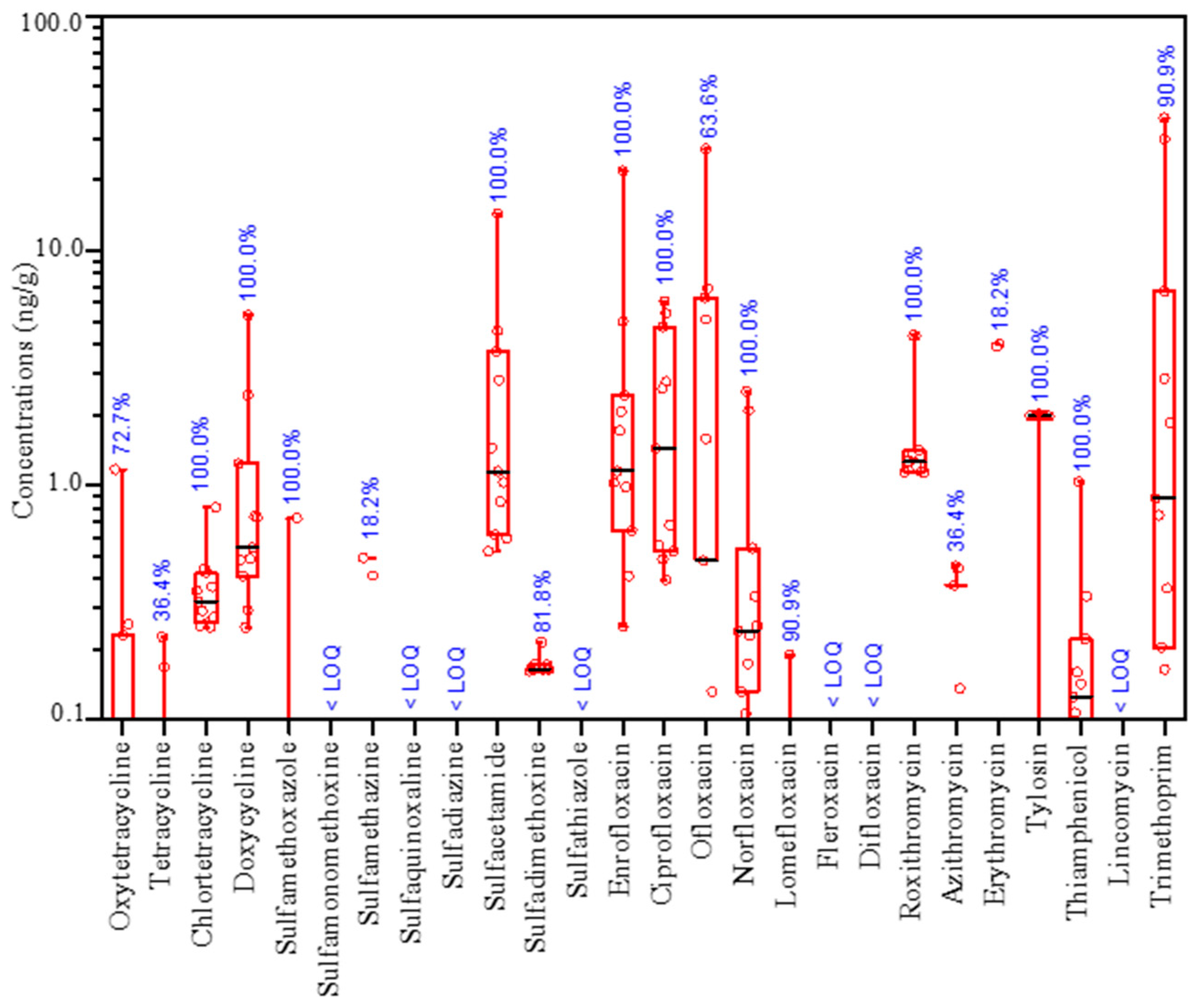
3.1. Occurrence of Antibiotics in Rhizosphere Soil
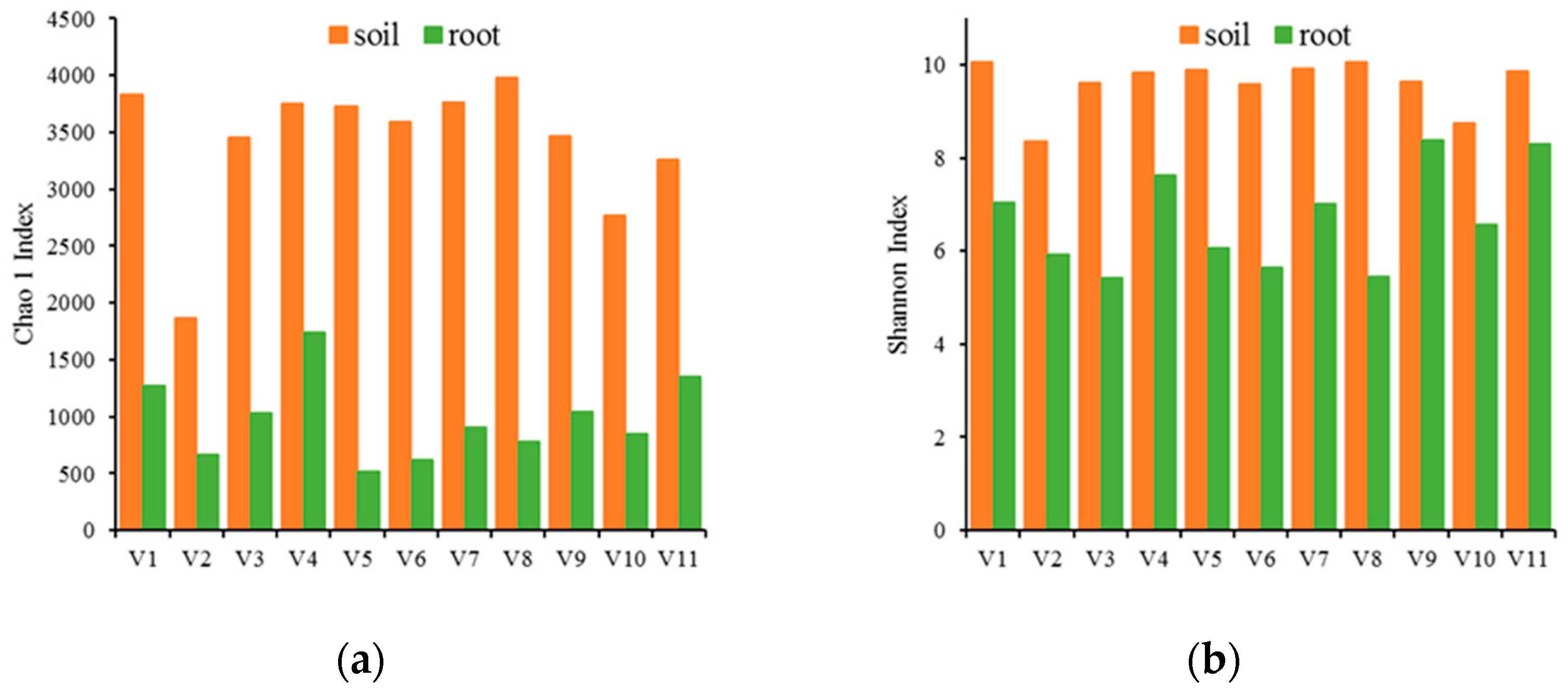
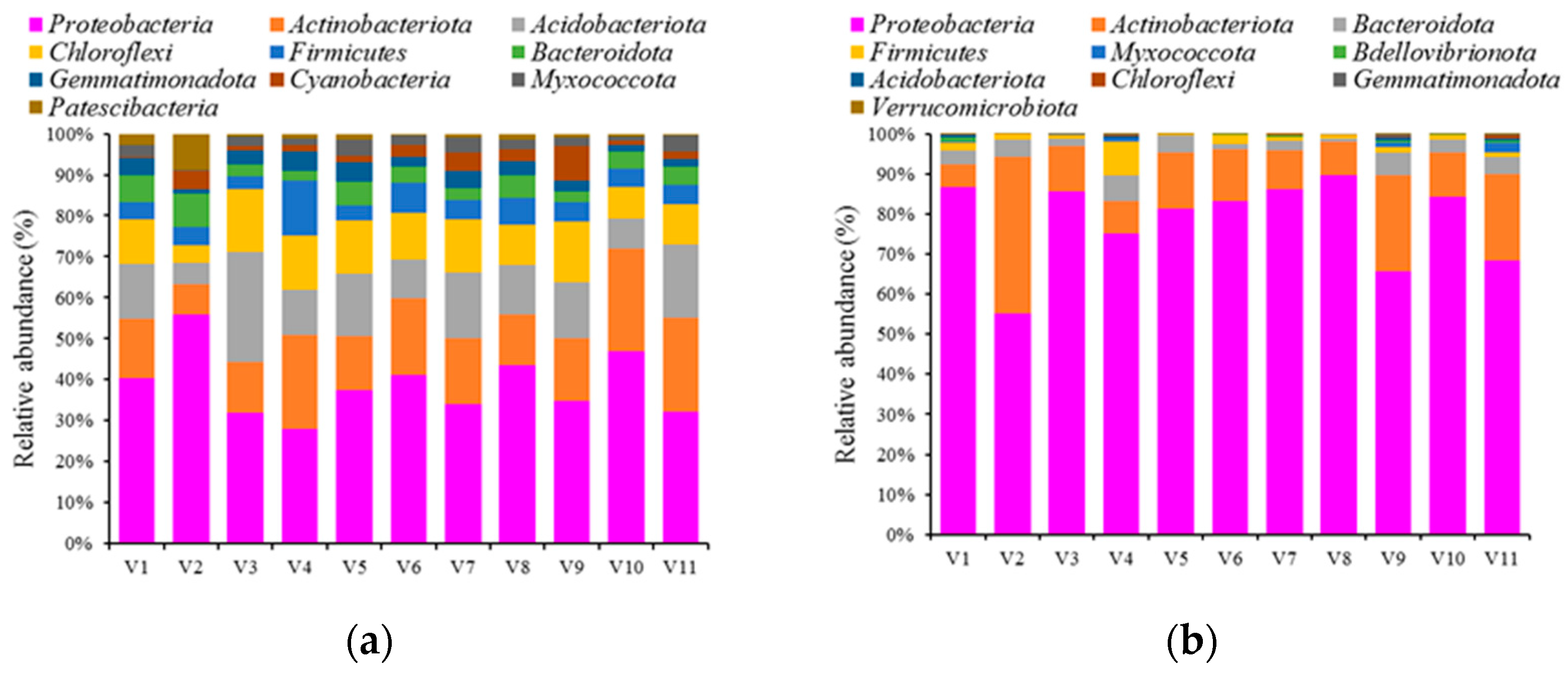
3.2. Bacterial Community Structure in the Rhizosphere
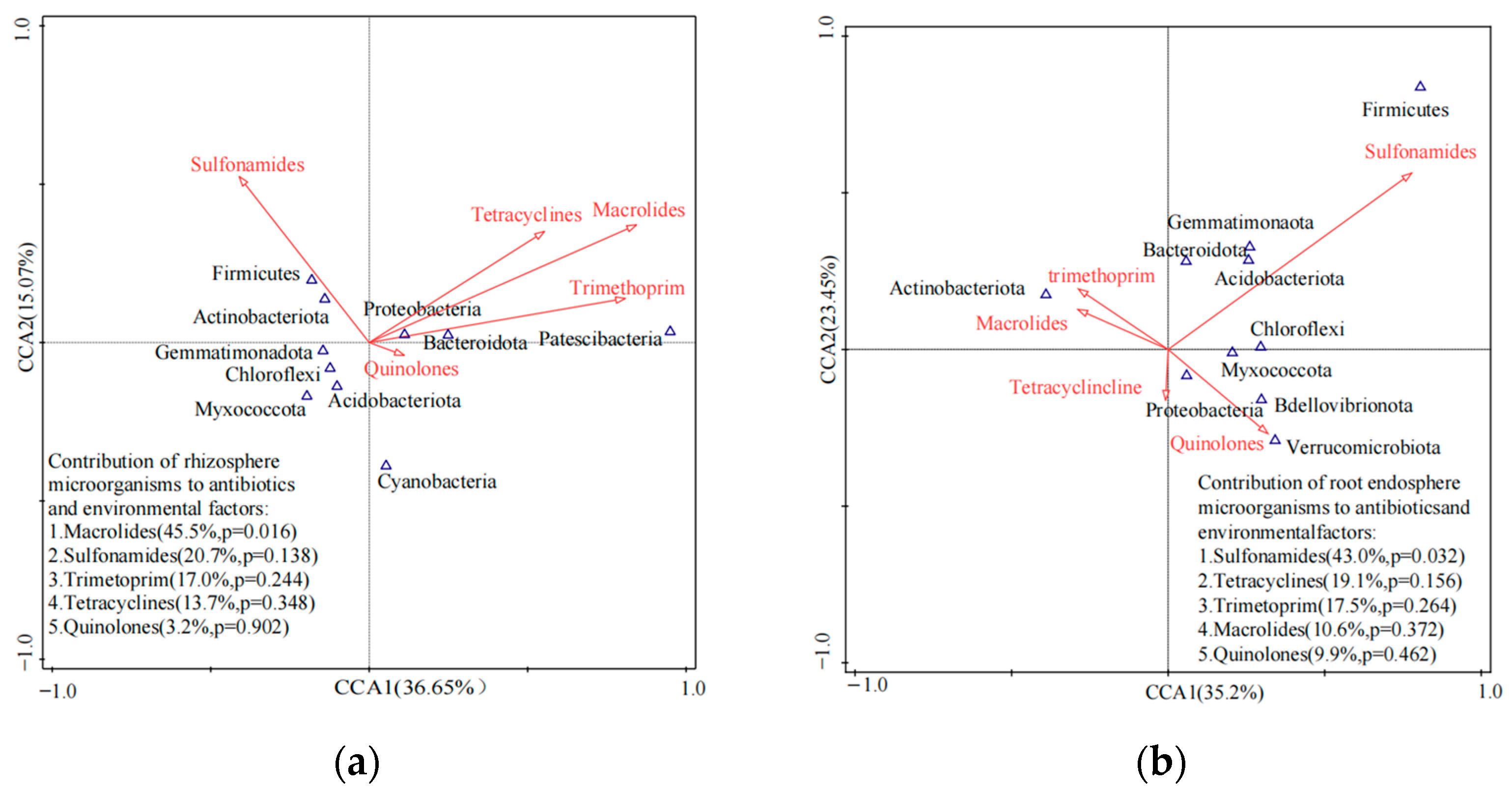
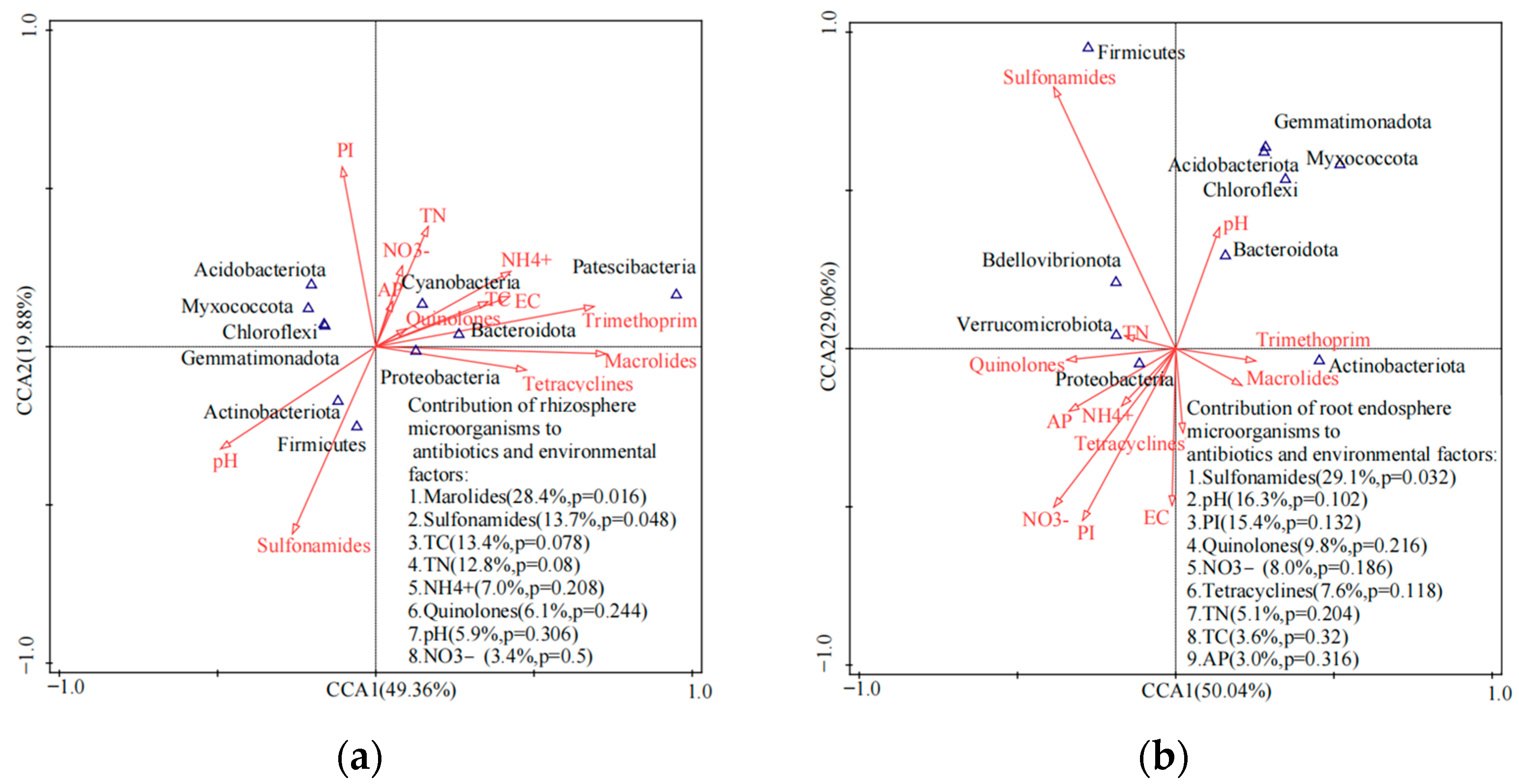
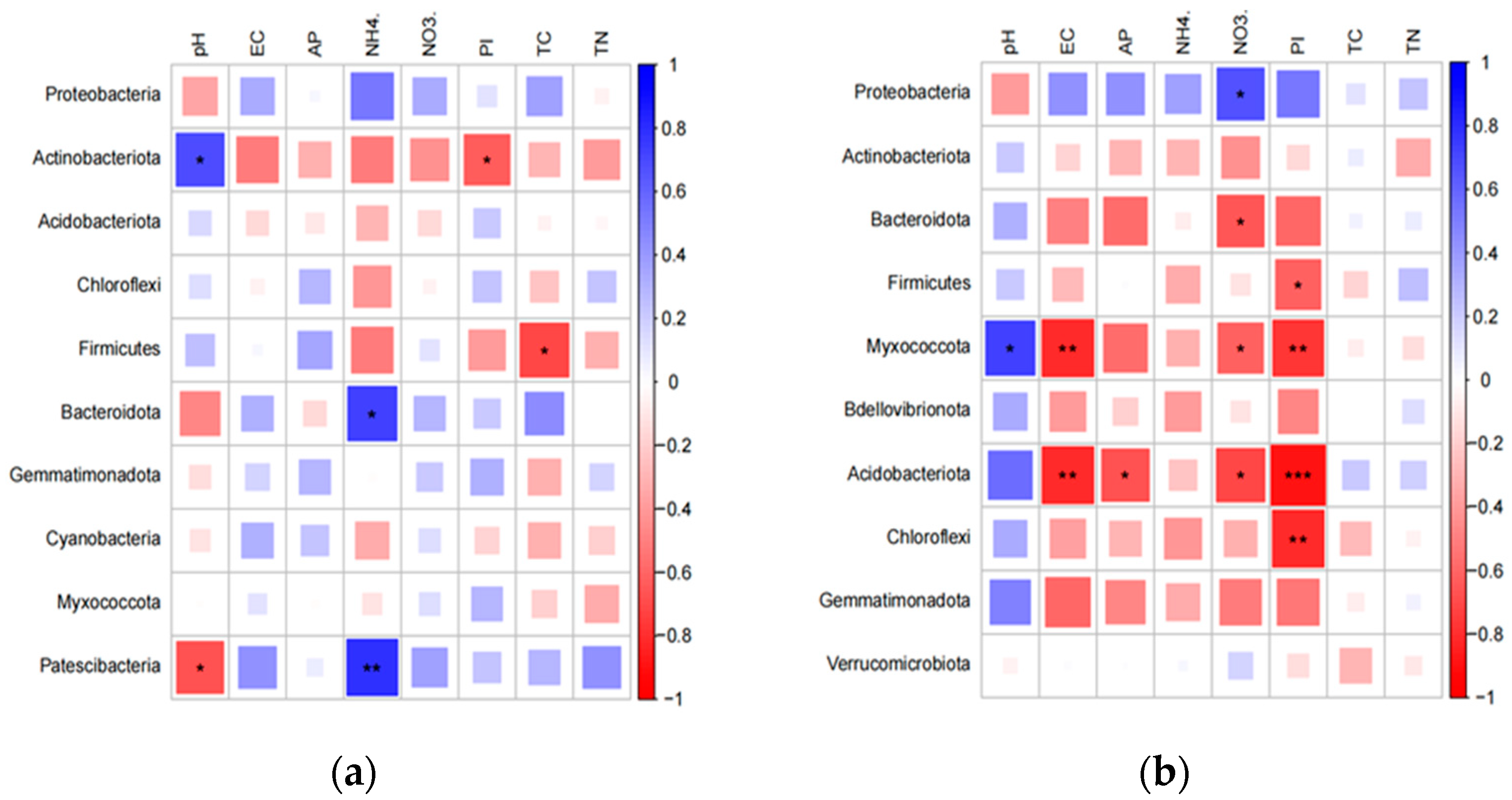
3.3. Responses of Microbial Communities in the Rhizosphere to Antibiotics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.S.; Zhang, H.B.; Luo, Y.M.; Song, J. Occurrence and assessment of veterinary antibiotics in swine manures: A case study in East China. Chin. Sci. Bull. 2012, 57, 606–614. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, J.; Lu, C.; Liao, Q.; Gudda, F.O.; Ling, W. Antibiotics in animal manure and manure-based fertilizers: Occurrence and ecological risk assessment. Chemosphere 2020, 255, 127006. [Google Scholar] [CrossRef]
- Gao, F.Z.; He, L.Y.; He, L.X.; Zou, H.Y.; Zhang, M.; Wu, D.L.; Liu, Y.S.; Shi, Y.J.; Bai, H.; Ying, G.G. Untreated swine wastes changed antibiotic resistance and microbial community in the soils and impacted abundances of antibiotic resistance genes in the vegetables. Sci. Total Environ. 2020, 741, 140482. [Google Scholar] [CrossRef]
- Ghirardini, A.; Grillini, V.; Verlicchi, P. A review of the occurrence of selected micropollutants and microorganisms in different raw and treated manure-Environmental risk due to antibiotics after application to soil. Sci. Total Environ. 2020, 707, 136118. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Johnson, T.A.; Su, J.Q.; Qiao, M.; Guo, G.X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef]
- Chen, C.; Pankow, C.A.; Oh, M.; Heath, L.S.; Zhang, L.; Du, P.; Xia, K.; Pruden, A. Effect of antibiotic use and composting on antibiotic resistance gene abundance and resistome risks of soils receiving manure-derived amendments. Environ. Int. 2019, 128, 233–243. [Google Scholar] [CrossRef]
- Zhang, N.; Juneau, P.; Huang, R.; He, Z.; Sun, B.; Zhou, J.; Liang, Y. Coexistence between antibiotic resistance genes and metal resistance genes in manure-fertilized soils. Geoderma 2021, 382, 114760. [Google Scholar] [CrossRef]
- Tong, C.; Xiao, D.; Xie, L.; Yang, J.; Zhao, R.; Hao, J.; Huo, Z.; Zeng, Z.; Xiong, W. Swine manure facilitates the spread of antibiotic resistome including tigecycline-resistant tet(X) variants to farm workers and receiving environment. Sci. Total Environ. 2022, 808, 152157. [Google Scholar] [CrossRef]
- Xi, X.P.; Wang, M.; Chen, Y.S.; Yu, S.; Hong, Y.W.; Ma, J.; Wu, Q.; Lin, Q.Y.; Xu, X.R. Adaption of the microbial community to continuous exposuresof multiple residual antibiotics in sediments from a salt-water aquacultural farm. J. Hazard. Mater. 2015, 290, 96–105. [Google Scholar] [CrossRef]
- Piotrowska-Długosz, A. The Effects of Antibiotics on the Structure, Diversity, and Function of a Soil Microbial Community. In Antibiotics and Antibiotics Resistance Genes in Soils; Soil Biology; Hashmi, M., Strezov, V., Varma, A., Eds.; Springer: Cham, Switzerland, 2017; Volume 51. [Google Scholar] [CrossRef]
- Zhao, Y.; Cocerva, T.; Cox, S.; Tardif, S.; Su, J.Q.; Zhu, Y.G.; Brandt, K.K. Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci. Total Environ. 2019, 656, 512–520. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, C.; Lian, K.; Liu, C. Effects of chronic exposure of antibiotics on microbial community structure and functions in hyporheic zone sediments. J. Hazard. Mater. 2021, 416, 126141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sui, Q.; Tong, J.; Zhong, H.; Wang, Y.; Chen, M.; Wei, Y. Soil types influence the fate of antibiotic-resistant bacteria and antibiotic resistance genes following the land application of sludge composts. Environ. Int. 2018, 118, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; You, G.; Zhang, M.; Peng, D.; Jiang, Z.; Qi, S.; Yang, S.; Hou, J. Antibiotic resistance genes alternation in soils modified with neutral and alkaline salts: Interplay of salinity stress and response strategies of microbes. Sci. Total Environ. 2022, 809, 152246. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Hu, J.C.; Chen, Y.S.; Xu, J.H.; Yang, B.F.; Jiang, J.P. Ecological effects of antibiotics on aquaculture ecosystems based on microbial community in sediments. Ocean. Coast. Manag. 2022, 224, 106173. [Google Scholar] [CrossRef]
- Qiu, D.; Xu, N.; Zhang, Q.; Zhou, W.; Wang, Y.; Zhang, Z.; Yu, Y.; Lu, T.; Sun, L.; Zhou, N.Y.; et al. Negative effects of abamectin on soil microbial communities in the short term. Front. Microbiol. 2022, 13, 1053153. [Google Scholar] [CrossRef]
- Barra Caracciolo, A.; Visca, A.; Rauseo, J.; Spataro, F.; Garbini, G.L.; Grenni, P.; Mariani, L.; Mazzurco Miritana, V.; Massini, G.; Patrolecco, L. Bioaccumulation of antibiotics and resistance genes in lettuce following cattle manure and digestate fertilization and their effects on soil and phyllosphere microbial communities. Environ. Pollut. 2022, 315, 120413. [Google Scholar] [CrossRef]
- Martin-Laurent, F.; Topp, E.; Billet, L.; Batisson, I.; Malandain, C.; Besse-Hoggan, P.; Morin, S.; Artigas, J.; Bonnineau, C.; Kergoat, L.; et al. Environmental risk assessment of antibiotics in agroecosystems: Ecotoxicological effects on aquatic microbial communities and dissemination of antimicrobial resistances and antibiotic biodegradation potential along the soil-water continuum. Environ. Sci. Pollut. Res. Int. 2019, 26, 18930–18937. [Google Scholar] [CrossRef]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef]
- Liu, Z.; Klümper, U.; Liu, Y.; Yang, Y.; Wei, Q.; Lin, J.G.; Gu, J.D.; Li, M. Metagenomic and metatranscriptomic analyses reveal activity and hosts of antibiotic resistance genes in activated sludge. Environ. Int. 2019, 129, 208–220. [Google Scholar] [CrossRef]
- Bernier, S.P.; Surette, M.G. Concentration-dependent activity of antibiotics in natural environments. Front. Microbiol. 2013, 4, 20. [Google Scholar] [CrossRef]
- Seyoum, M.M.; Obayomi, O.; Bernstein, N.; Williams, C.F.; Gillor, O. Occurrence and distribution of antibiotics and corre-sponding antibiotic resistance genes in different soil types irrigated with treated wastewater. Sci. Total Environ. 2021, 782, 146835. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Mao, D.Q.; Ghirardini, H.H.; Zheng, L.Y.; Chen, Z.Y.; Gao, Y.T.; Duan, Y.T.; Guo, J.H.; Luo, Y.; Ren, H.Q. Colonization of gut microbiota by plasmid-carrying bacteria is facilitated by evolutionary adaptation to antibiotic treatment. ISME J. 2021, 16, 1284–1293. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.E.; Barea, J.M.; Mcneill, A.M.; Combaret, C.P. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Hu, H.W.; Chen, Q.L.; Singh, B.K.; Yan, H.; Chen, D.; He, J.Z. Transfer of antibiotic resistance from ma-nure-amended soils to vegetable microbiomes. Environ. Int. 2019, 130, 104912. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qiu, T.; Gao, M.; Sun, Y.; Cheng, S.; Gao, H.; Wang, X. Diversity and abundance of antibiotic resistance genes in rhizo-sphere soil and endophytes of leafy vegetables: Focusing on the effect of the vegetable species. J. Hazard. Mater. 2021, 415, 125595. [Google Scholar] [CrossRef]
- Chen, J.; Cai, Y.; Deng, W.; Xing, S.; Liao, X. Transmission of tetracycline resistance genes and microbiomes from manure-borne black soldier fly larvae frass to rhizosphere soil and pakchoi endophytes. Front. Microbiol. 2022, 13, 1014910. [Google Scholar] [CrossRef]
- Obermeier, M.M.; Wicaksono, W.A.; Taffner, J.; Bergna, A.; Poehlein, A.; Cernava, T.; Lindstaedt, S.; Lovric, M.; Bogotá, C.A.M.; Berg, G. Plant resistome profiling in evolutionary old bog vegetation provides new clues to understand emergence of mul-ti-resistance. ISME J. 2021, 15, 921–937. [Google Scholar] [CrossRef]
- Sun, Y.; Qiu, T.; Gao, M.; Shi, M.; Zhang, H.; Wang, X. Inorganic and organic fertilizers application enhanced antibiotic resistome in greenhouse soils growing vegetables. Ecotoxicol. Environ. Saf. 2019, 179, 24–30. [Google Scholar] [CrossRef]
- Gremion, F.; Chatzinotas, A.; Harms, H. Comparative 16S rDNA and 16S rRNA sequence analysis indicates that Actinobacteria might be a dominant part of the metabolically active bacteria in heavy metal-contaminated bulk and rhizosphere soil. Environ. Microbiol. 2003, 5, 896–907. [Google Scholar] [CrossRef]
- Huang, Y.J.; Cheng, M.M.; Li, W.H.; Wu, L.H.; Chen, Y.S.; Luo, Y.M.; Christie, P.; Zhang, H.B. Simultaneous extraction of four classes of antibiotics in soil, manure and sewage sludge and analysis by liquid chromatography-tandem mass spectrometry with the isotope-labelled internal standard method. Anal. Methods 2013, 5, 3721. [Google Scholar]
- Chen, Y.S.; Xu, J.H.; Lv, Z.Y.; Huang, L.M.; Jiang, J.P. Impacts of biochar and oyster shells waste on the immobilization of arsenic in highly contaminated soils. J. Environ. Manag. 2018, 217, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, J.B.; Mazurek, R.; Gąsiorek, M.; Zaleski, T. Pollution indices as useful tools for the comprehensive evaluation of the degree of soil contamination—A review. Environ. Geochem. Health 2018, 40, 2395–2420. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, X.; Chen, J.; Li, Y.; Liu, X.; Feng, Y.; Sun, Y. Source, occurrence and risks of twenty antibiotics in vegetables and soils from facility agriculture through fixed-point monitoring and numerical simulation. J. Environ. Manag. 2022, 319, 115652. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, J.; Gbadegesin, L.A.; He, Y. Modelling approaches for linking the residual concentrations of antibiotics in soil with antibiotic properties and land-use types in the largest urban agglomerations in China: A review. Sci. Total Environ. 2022, 838 Pt 2, 156141. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.F.; Li, X.W.; Wang, J.F.; Christakos, G.; Hu, M.G.; An, L.H.; Li, F.S. Spatial estimation of antibiotic residues in surface soils in a typical intensive vegetable cultivation area in China. Sci. Total Environ. 2012, 430, 126–131. [Google Scholar] [CrossRef]
- Tasho, R.P.; Cho, J.Y. Veterinary antibiotics in animal waste, its distribution in soil and uptake by plants: A review. Sci. Total Environ. 2016, 563–564, 366–376. [Google Scholar] [CrossRef]
- Bueno, I.; Rodríguez, A.; Beaudoin, A.; Arnold, W.A.; Wammer, K.H.; de la Torre, A.; Singer, R.S. Identifying the spatiotemporal vulnerability of soils to antimicrobial contamination through land application of animal manure in Minnesota, United States. Sci. Total Environ. 2022, 832, 155050. [Google Scholar] [CrossRef]
- Ben, Y.; Fu, C.; Hu, M.; Liu, L.; Wong, M.H.; Zheng, C. Human health risk assessment of antibiotic resistance associated with anti-biotic residues in the environment: A review. Environ. Res. 2019, 169, 483–493. [Google Scholar] [CrossRef]
- Li, C.; Chen, J.; Wang, J.; Ma, Z.; Han, P.; Luan, Y.; Lu, A. Occurrence of antibiotics in soils and manures from greenhouse vegetable production bases of Beijing.; China and an associated risk assessment. Sci. Total Environ. 2015, 521–522, 101–107. [Google Scholar] [CrossRef]
- Xiang, L.; Wu, X.L.; Jiang, Y.N.; Yan, Q.Y.; Li, Y.W.; Huang, X.P.; Cai, Q.Y.; Mo, C.H. Occurrence and risk assessment of tetracycline anti-biotics in soil from organic vegetable farms in a subtropical city, south China. Environ. Sci. Pollut. Res. Int. 2016, 23, 13984–13995. [Google Scholar] [CrossRef]
- Li, J.; Xin, Z.; Zhang, Y.; Chen, J.; Yan, J.; Li, H. Long-term manure application increased the levels of an-tibiotics and antibiotic resistance genes in a greenhouse soil. Appl. Soil Ecol. 2017, 121, 193–200. [Google Scholar] [CrossRef]
- Wei, R.; Ge, F.; Zhang, L.; Hou, X.; Cao, Y.; Gong, L.; Chen, M.; Wang, R.; Bao, E. Occurrence of 13 veterinary drugs in animal ma-nure-amended soils in Eastern China. Chemosphere 2016, 144, 2377–2383. [Google Scholar] [CrossRef]
- Zhang, H.; Luo, Y.; Wu, L.; Huang, Y.; Christie, P. Residues and potential ecological risks of veterinary antibiotics in manures and composts associated with protected vegetable farming. Environ. Sci. Pollut. Res. Int. 2015, 22, 5908–5918. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cheng, D.; Xie, J.; Zhang, Y.; Wan, Y.; Zhang, Y.; Shi, X. Impacts of farmland application of antibiotic-contaminated ma-nures on the occurrence of antibiotic residues and antibiotic resistance genes in soil: A meta-analysis study. Chemosphere 2022, 300, 134529. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.B.; Zakaria, M.P.; Latif, P.A.; Saari, N. Occurrence of veterinary antibiotics and progesterone in broiler manure and agri-cultural soil in Malaysia. Sci. Total Environ. 2014, 488–489, 261–267. [Google Scholar] [CrossRef]
- Gros, M.; Mas-Pla, J.; Boy-Roura, M.; Geli, I.; Domingo, F.; Petrović, M. Veterinary pharmaceuticals and antibiotics in manure and slurry and their fate in amended agricultural soils: Findings from an experimental field site (Baix Empordà.; NE Catalonia). Sci. Total Environ. 2019, 654, 1337–1349. [Google Scholar] [CrossRef]
- Bastos, M.C.; dos Santos, D.R.; Aubertheau, E.; Lima, J.A.M.D.; Le Guet, T.; Caner, L.; Mondamert, L.; Labanowski, J. Antibiotics and microbial resistance in Brazilian soils under manure application. Land Degrad. Dev. 2018, 29, 2472–2484. [Google Scholar] [CrossRef]
- Yang, Y.; Owino, A.A.; Gao, Y.; Yan, X.; Xu, C.; Wang, J. Occurrence, composition and risk assessment of antibiotics in soils from Kenya, Africa. Ecotoxicology 2016, 25, 1194–1201. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, L.; Yang, L.; Li, S.; Sun, L.; Yu, X. Distribution.; dynamics and determinants of antibiotics in soils in a peri-urban area of Yangtze River Delta.; Eastern China. Chemosphere 2018, 211, 261–270. [Google Scholar] [CrossRef]
- Xie, W.Y.; Shen, Q.; Zhao, F.J. Antibiotics and Antibiotic Resistance From Animal Manures to Soil: A Review. Eur. J. Soil Sci. 2018, 69, 181–195. [Google Scholar] [CrossRef]
- Le, H.T.V.; Maguire, R.O.; Xia, K. Spatial distribution and temporal change of antibiotics in soils amended with manure using two field application methods. Sci. Total Environ. 2021, 759, 143431. [Google Scholar] [CrossRef]
- Ali, A.; Ghani, M.I.; Li, Y.; Ding, H.; Meng, H.; Cheng, Z. Hiseq Base Molecular Characterization of Soil Microbial Commu-nity, Diversity Structure, and Predictive Functional Profiling in Continuous Cucumber Planted Soil Affected by Diverse Cropping Systems in an Intensive Greenhouse Region of Northern China. Int. J. Mol. Sci. 2019, 20, 2619. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.S.; Wang, W.Z.; Deng, J.B.; Xu, W.H. The residue of tetracycline antibiotics in soil and Brassica juncea var. gemmifera, and the diversity of soil bacterial community under different livestock manure treatments. Environ. Geochem. Health 2022, 45, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jewett, C.; Gilley, J.; Bartelt-Hunt, S.L.; Snow, D.D.; Hodges, L.; Li, X. Microbial communities in the rhizosphere and the root of lettuce as affected by Salmonella-contaminated irrigation water. FEMS Microbiol. Ecol. 2018, 94, fiy135. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Sun, Y.; Snow, D.; Walia, H.; Li, X. Transmission Routes of the Microbiome and Resistome from Manure to Soil and Lettuce. Environ. Sci. Technol. 2021, 55, 11102–11112. [Google Scholar] [CrossRef]
- Kovalakova, P.; Cizmas, L.; McDonald, T.J.; Marsalek, B.; Feng, M.; Sharma, V.K. Occurrence and toxicity of antibiotics in the aquatic environment: A review. Chemosphere 2020, 251, 126351. [Google Scholar] [CrossRef]
- Yang, L.Y.; Zhou, S.Y.; Lin, C.S.; Huang, X.R.; Neilson, R.; Yang, X.R. Effects of biofertilizer on soil microbial diversity and antibiotic resistance genes. Sci. Total Environ. 2022, 820, 153170. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, S.; Liu, X.; Chen, J.; Han, M.; Wan, G.Z.; Guo, W. Profiles of antibiotic resistance genes in an inland salt-lake Ebinur Lake, Xinjiang, China: The relationship with antibiotics, environmental factors, and microbial communities. Ecotoxicol. Environ. Saf. 2021, 221, 112427. [Google Scholar] [CrossRef]
- Shen, D.; Gu, X.; Zheng, Y.; Delgado-Moreno, L.; Jia, W.; Ye, Q.; Wang, W. The fate of erythromycin in soils and its effect on soil microbial community structure. Sci. Total Environ. 2022, 820, 153373. [Google Scholar] [CrossRef] [PubMed]
- Cycoń, M.; Mrozik, A.; Piotrowska-Seget, Z. Antibiotics in the Soil Environment-Degradation and Their Impact on Microbial Activity and Diversity. Front. Microbiol. 2019, 10, 338. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.; Chen, J.; Qiao, X.; Tian, R.; Zhu, M. Insight into dynamics and bioavailability of antibiotics in paddy soils by in situ soil moisture sampler. Sci. Total Environ. 2020, 703, 135562. [Google Scholar] [CrossRef]
- Sun, Y.; Guo, Y.; Shi, M.; Qiu, T.; Gao, M.; Tian, S.; Wang, X. Effect of antibiotic type and vegetable species on antibiotic accumula-tion in soil-vegetable system.; soil microbiota.; and resistance genes. Chemosphere 2021, 263, 128099. [Google Scholar] [CrossRef]
- Dickinson, A.W.; Power, A.; Hansen, M.G.; Brandt, K.K.; Piliposian, G.; Appleby, P.; O’Neill, P.A.; Jones, R.T.; Sierocinski, P.; Koskella, B.; et al. Heavy metal pollution and co-selection for antibiotic resistance: A microbial palaeontology approach. Environ. Int. 2019, 132, 105117. [Google Scholar] [CrossRef]
- Yan, J.; Bassler, B.L. Surviving as a Community: Antibiotic Tolerance and Persistence in Bacterial Biofilms. Cell Host Microbe 2019, 26, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Cengiz, M.; Balcioglu, I.A.; Oruc, H.H.; Cengiz, T.G. Evaluation of the interaction between soil and antibiotics. J. Environ. Sci. Health B 2010, 45, 183–189. [Google Scholar] [CrossRef]
- Polianciuc, S.I.; Gurzău, A.E.; Kiss, B.; Ştefan, M.G.; Loghin, F. Antibiotics in the environment: Causes and consequences. Med. Pharm. Rep. 2020, 93, 231–240. [Google Scholar] [CrossRef]
- Xue, P.P.; Carrillo, Y.; Pino, V.; Minasny, B.; McBratney, A.B. Soil Properties Drive Microbial Community Structure in a Large Scale Transect in South Eastern Australia. Sci. Rep. 2018, 8, 11725. [Google Scholar] [CrossRef]
- Trivedi, P.; Delgado-Baquerizo, M.; Anderson, I.C.; Singh, B.K. Response of Soil Properties and Microbial Communities to Agri-culture: Implications for Primary Productivity and Soil Health Indicators. Front. Plant Sci. 2016, 7, 990. [Google Scholar] [CrossRef]
| pH | EC | AP | TC | TN | NH4+ | NO3− |
| 6.9 ± 0.4 | 553.4 ± 461.2 | 79.5 ± 39.6 | 16,990 ± 6210 | 3360 ± 1650 | 4.4 ± 0.6 | 80.6 ± 82.3 |
| As | Zn | Cr | Cu | Pb | Cd | Ni |
| 16.17 ± 4.0 | 330.8 ± 190.9 | 41.22 ± 13.07 | 20.3 ± 16.0 | 56.6 ± 21.5 | 0.4 ± 0.2 | 9.1 ± 3.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, J.; Chen, Y.; Feng, Y.; Li, X.; Xu, J.; Jiang, J. Adaptation of Rhizosphere Microbial Communities to Continuous Exposure to Multiple Residual Antibiotics in Vegetable Farms. Int. J. Environ. Res. Public Health 2023, 20, 3137. https://doi.org/10.3390/ijerph20043137
Qiu J, Chen Y, Feng Y, Li X, Xu J, Jiang J. Adaptation of Rhizosphere Microbial Communities to Continuous Exposure to Multiple Residual Antibiotics in Vegetable Farms. International Journal of Environmental Research and Public Health. 2023; 20(4):3137. https://doi.org/10.3390/ijerph20043137
Chicago/Turabian StyleQiu, Jincai, Yongshan Chen, Ying Feng, Xiaofeng Li, Jinghua Xu, and Jinping Jiang. 2023. "Adaptation of Rhizosphere Microbial Communities to Continuous Exposure to Multiple Residual Antibiotics in Vegetable Farms" International Journal of Environmental Research and Public Health 20, no. 4: 3137. https://doi.org/10.3390/ijerph20043137
APA StyleQiu, J., Chen, Y., Feng, Y., Li, X., Xu, J., & Jiang, J. (2023). Adaptation of Rhizosphere Microbial Communities to Continuous Exposure to Multiple Residual Antibiotics in Vegetable Farms. International Journal of Environmental Research and Public Health, 20(4), 3137. https://doi.org/10.3390/ijerph20043137







