Degradation Characteristics and Remediation Ability of Contaminated Soils by Using β-HCH Degrading Bacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Enrichment, Screening, and Isolation of Degrading Bacteria
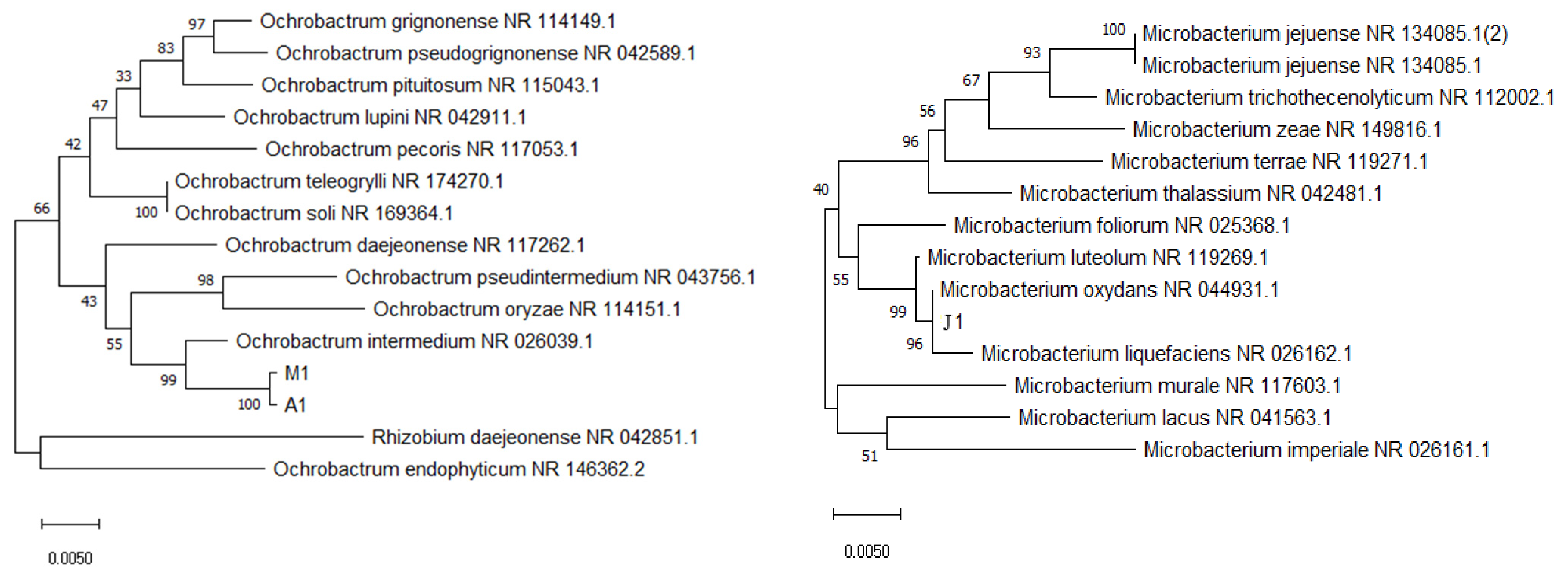
2.3. Strain Identification
2.4. Bacterial Growth Curve and Preparation of Bacterial Suspension
2.5. Degradability and Degradation Characteristics of the Strains
2.5.1. Degradability of Strains
2.5.2. Degradation Characteristics of the Strains
2.6. Construction of Complex Bacteria
2.7. Simulation of Contaminated Soil Remediation Trials
2.8. Sample Pre-Treatment and Detection
3. Results and Discussion
3.1. Bacterial Isolation, Screening and Identification
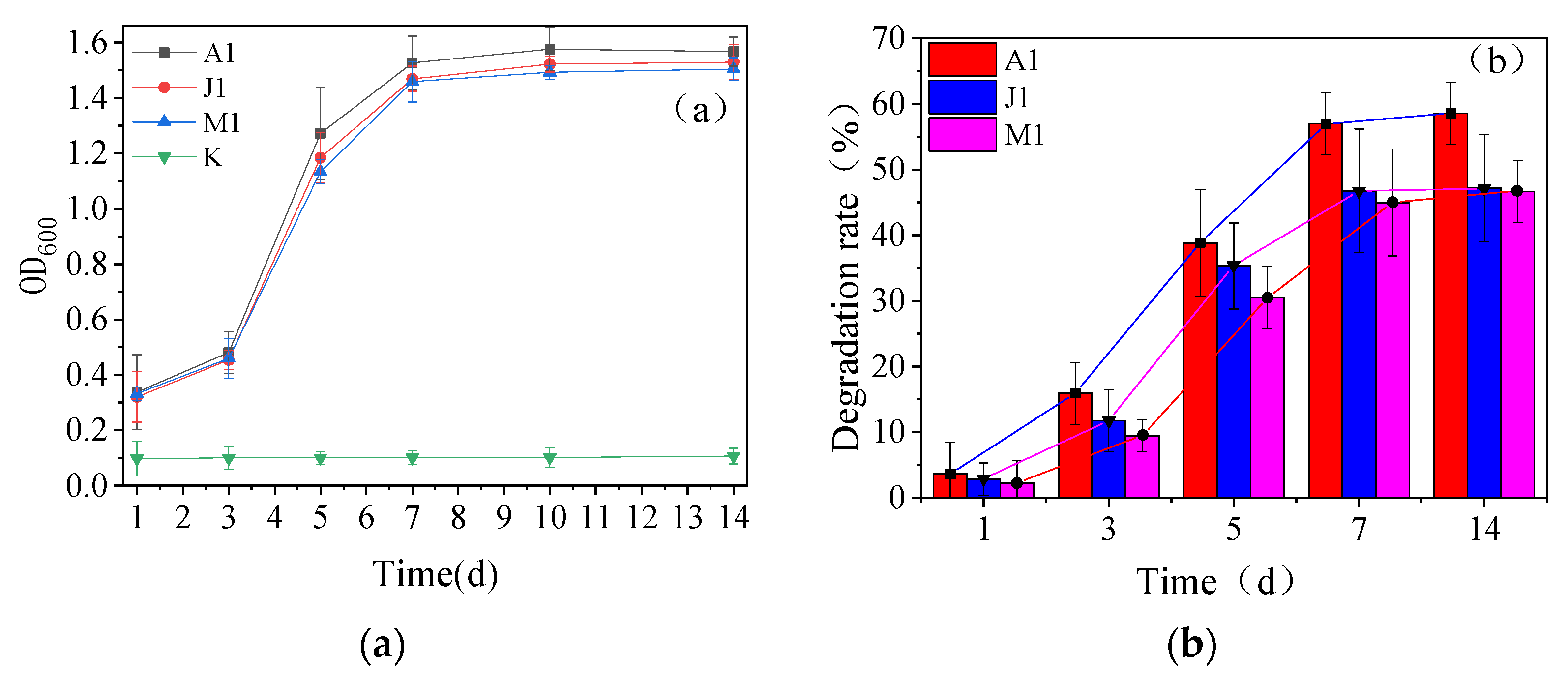
3.2. Growth Curve and Degradation Ability of Strains
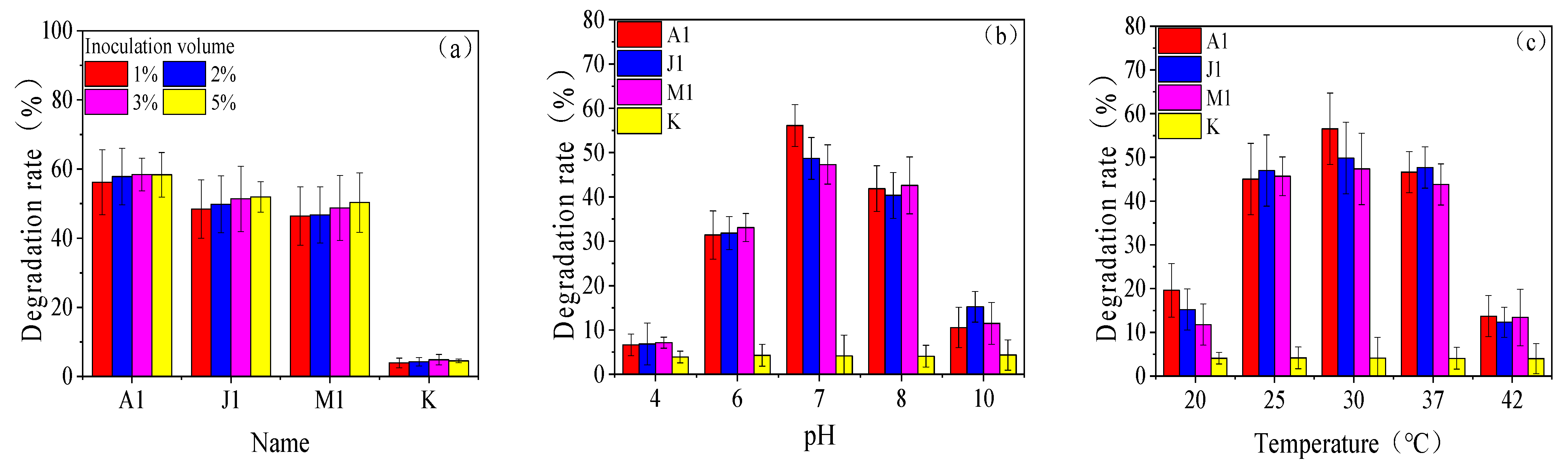
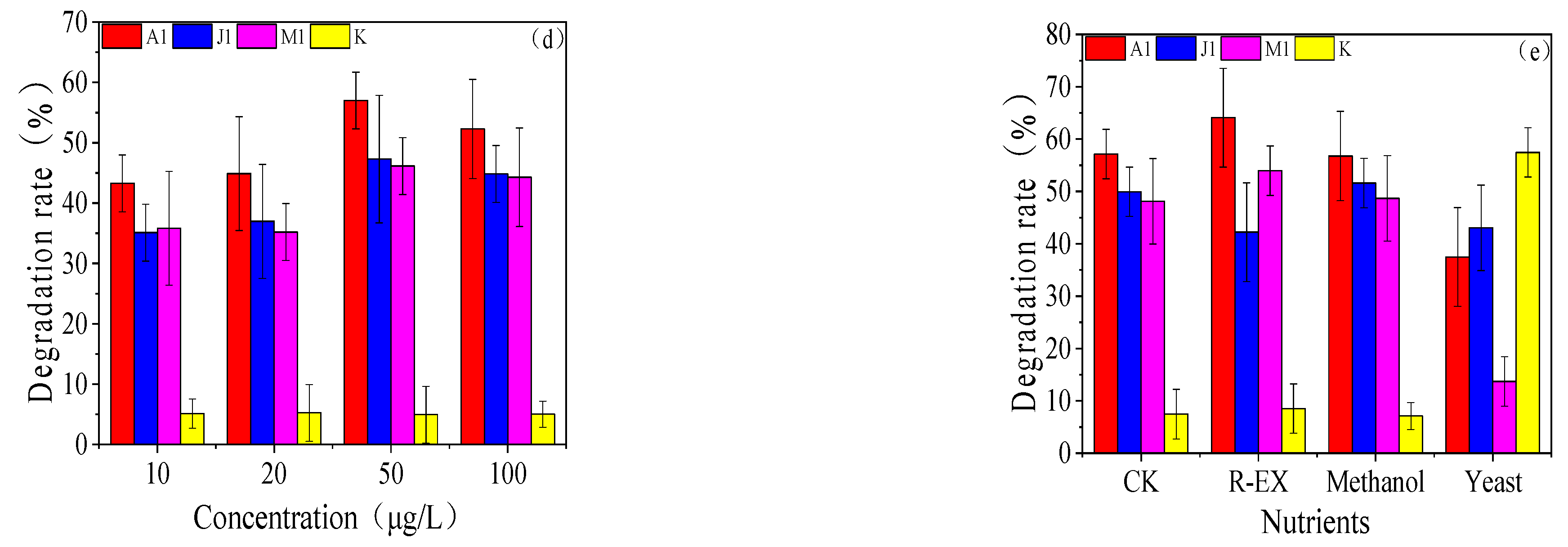
3.3. Degradation Characteristics of the Strains
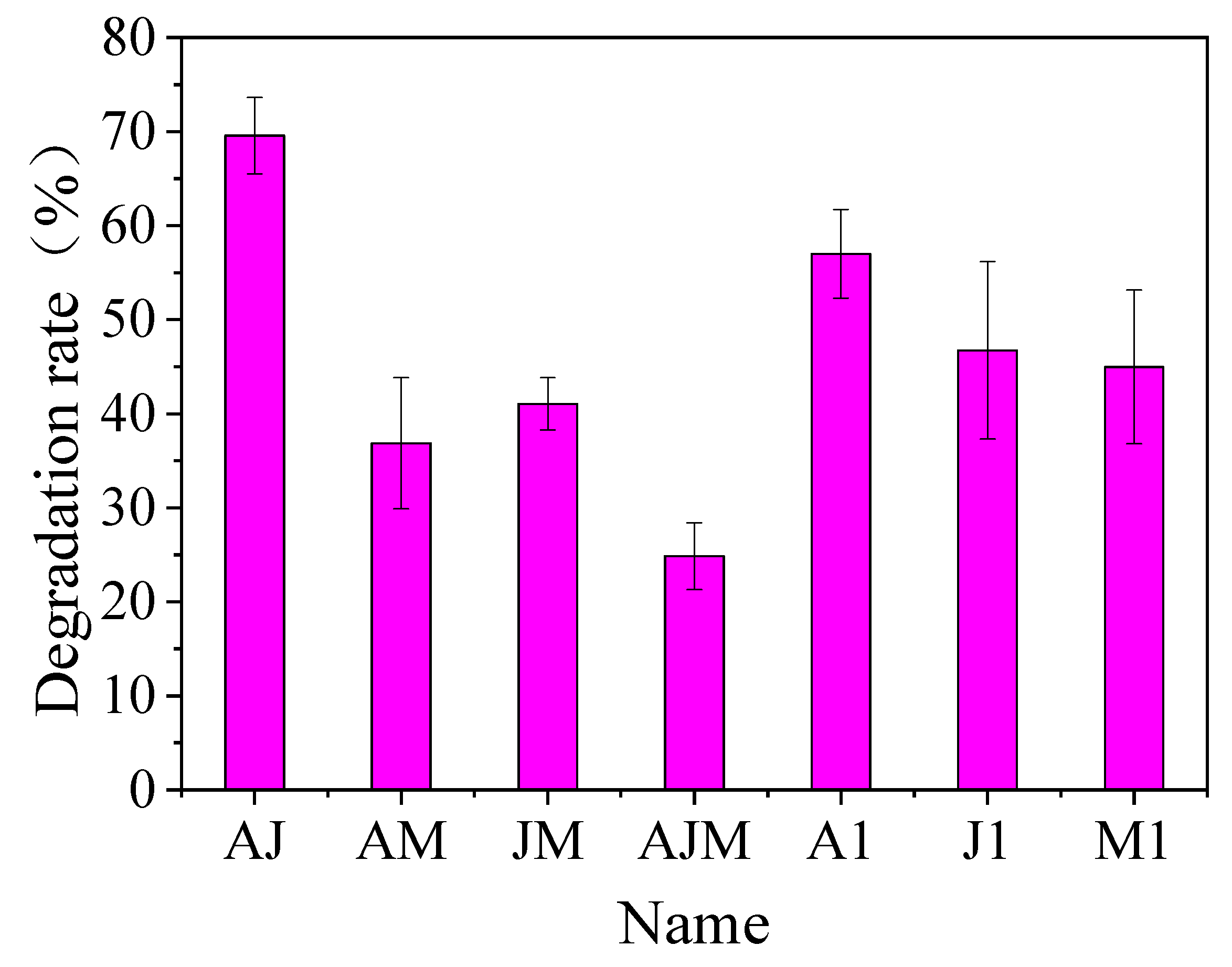
3.4. Study of Optimal Mix of Degradation Bacteria
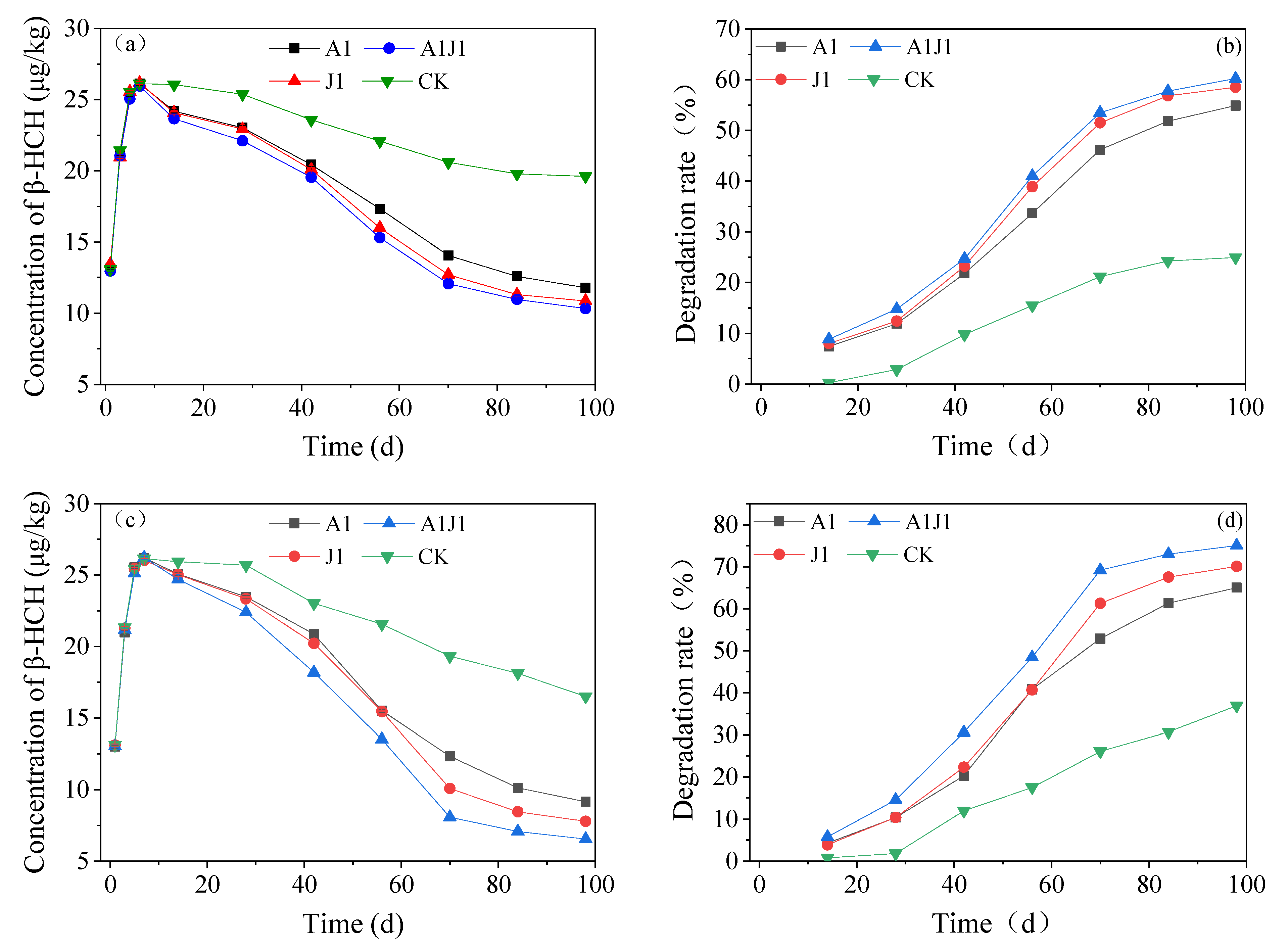
3.5. Remediation Study of Simulated β-HCH Contaminated Soil
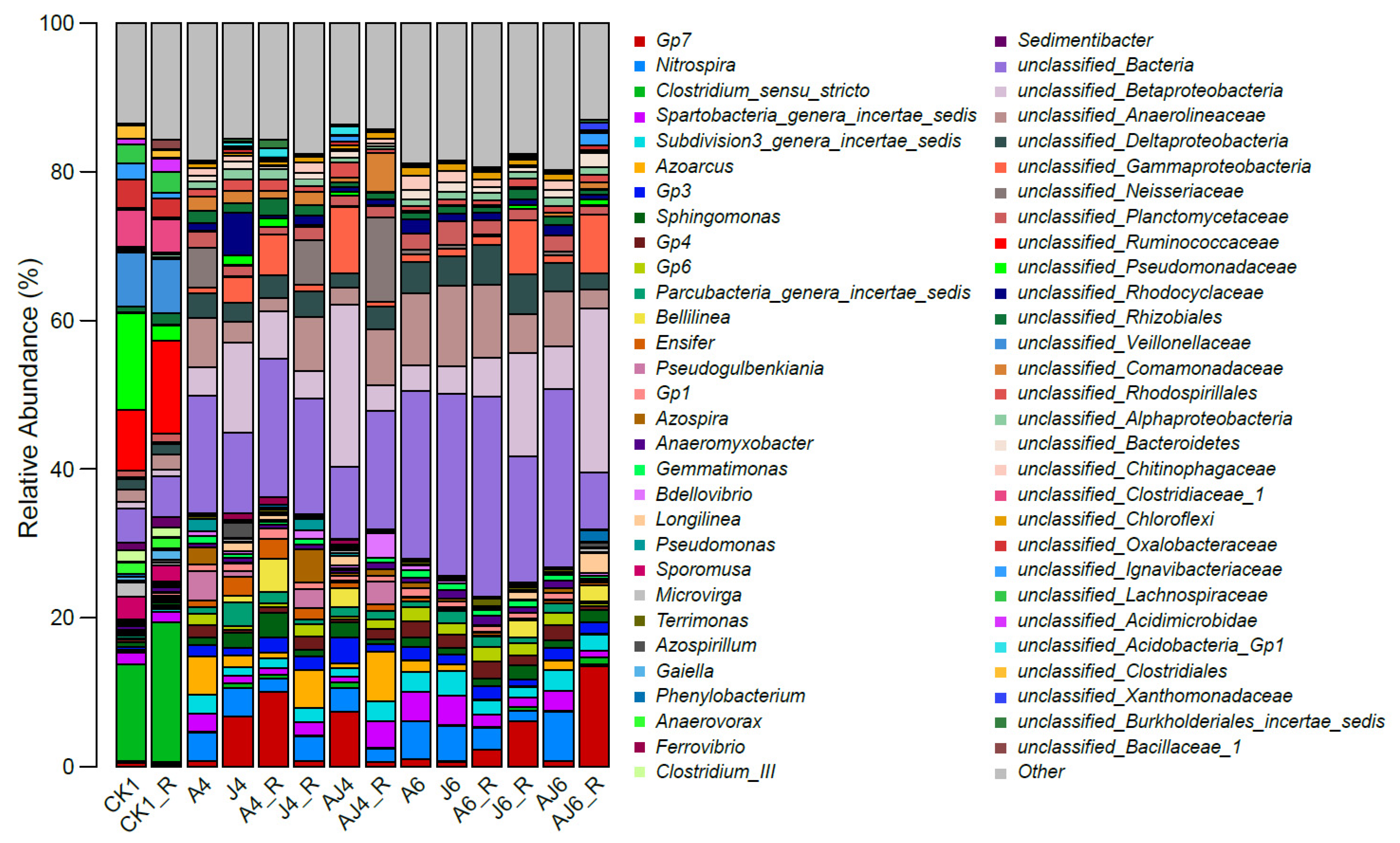
3.6. Characteristics of Microbial Community Response
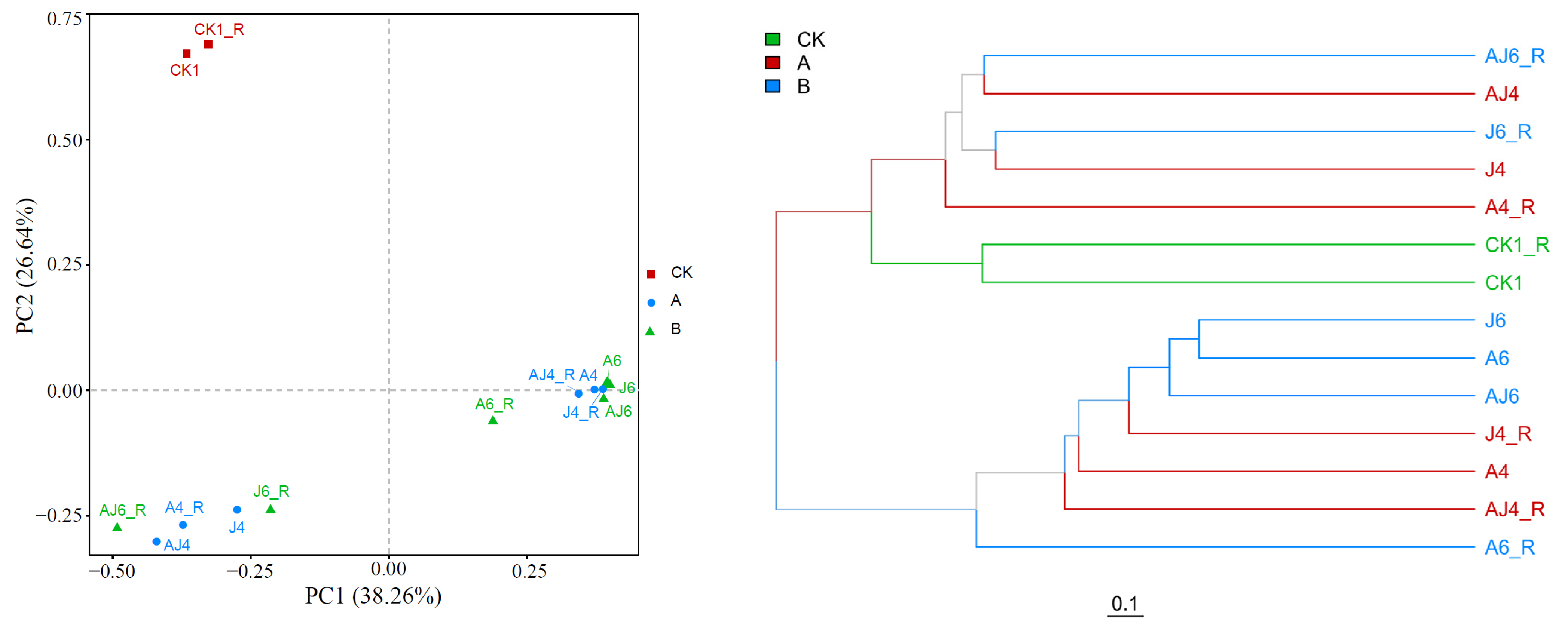
3.6.1. Alterations in Abundance and Diversity of Microbial Community
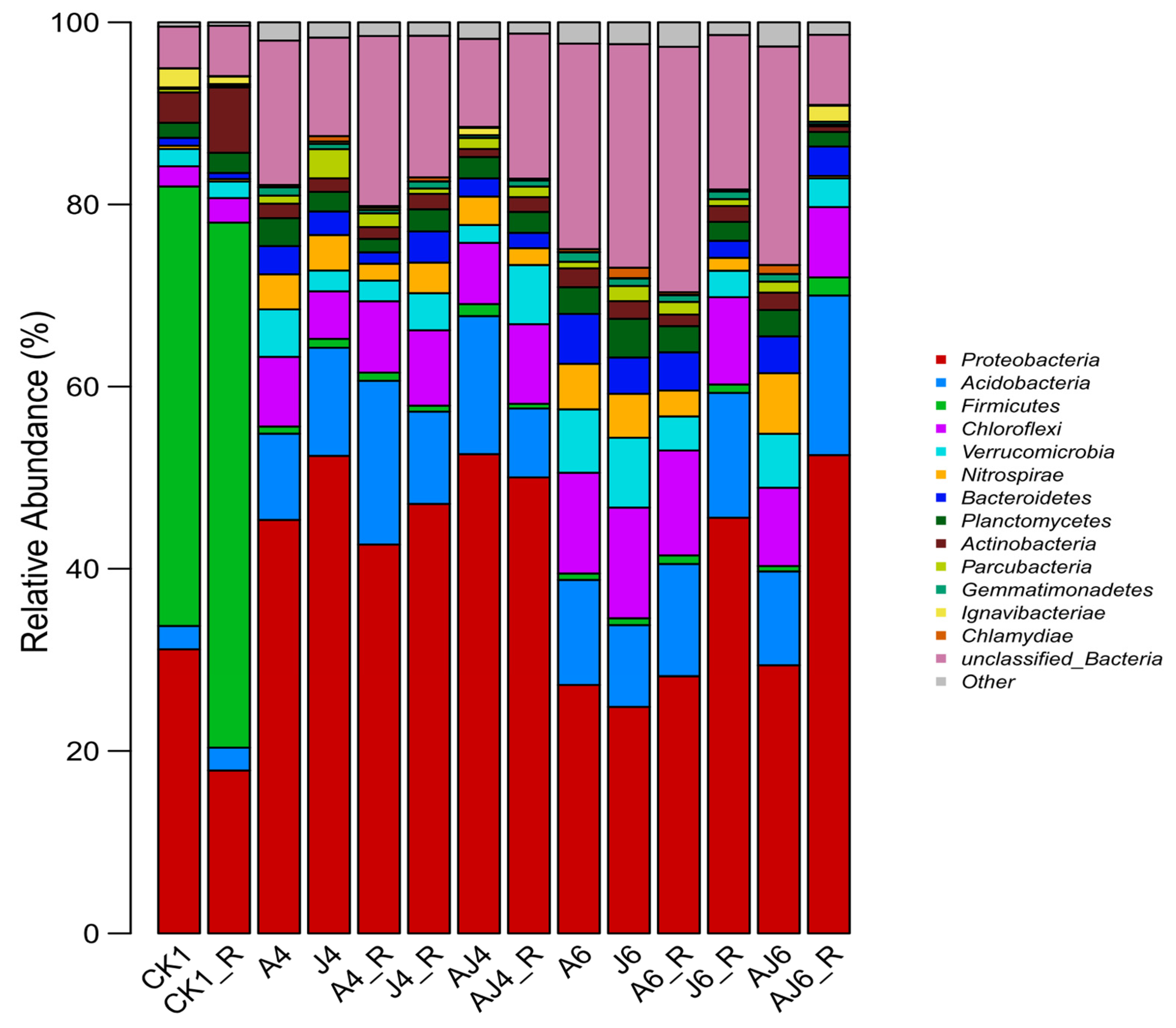
3.6.2. Phylum-Level Characteristics of Microbial Community Structure
3.6.3. Genus-Level Characteristics of Microbial Community Structure
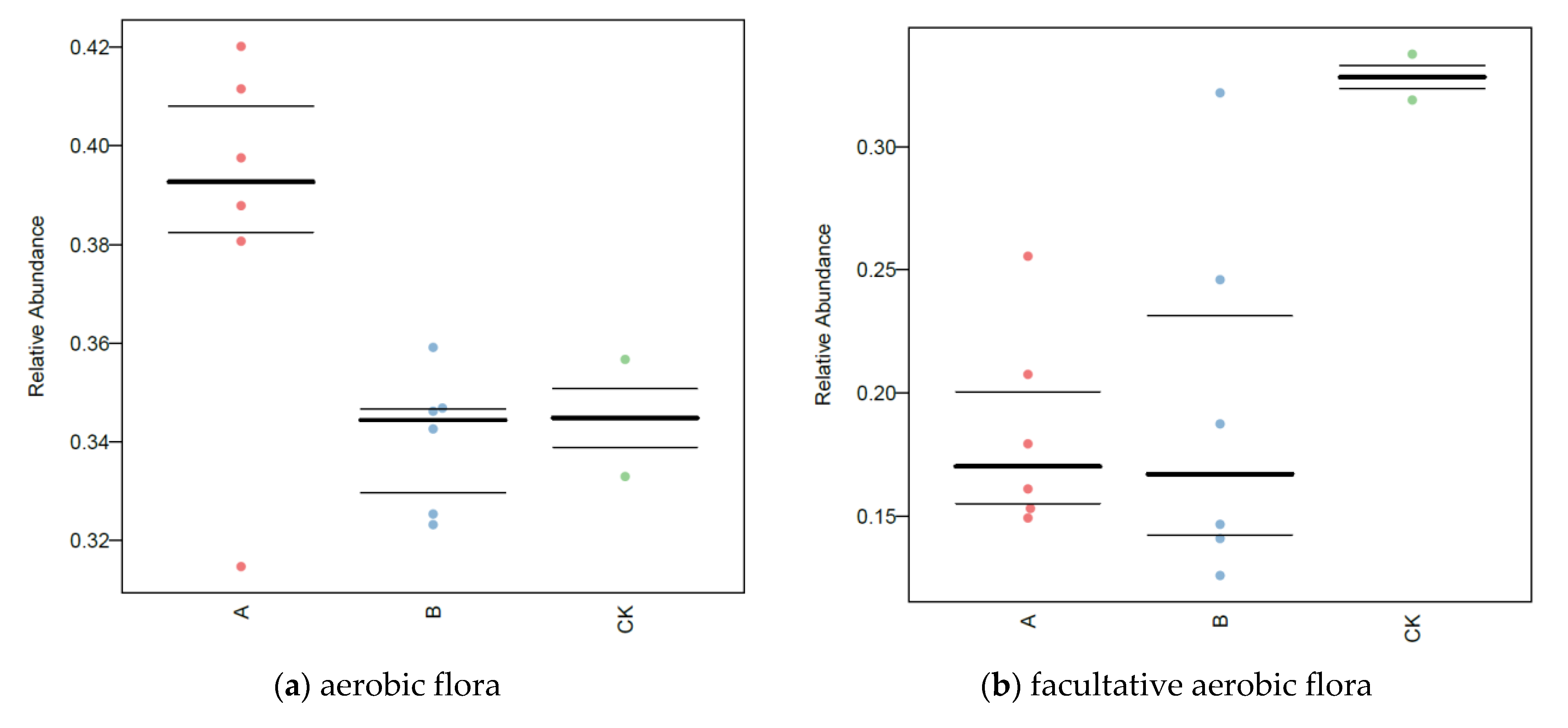
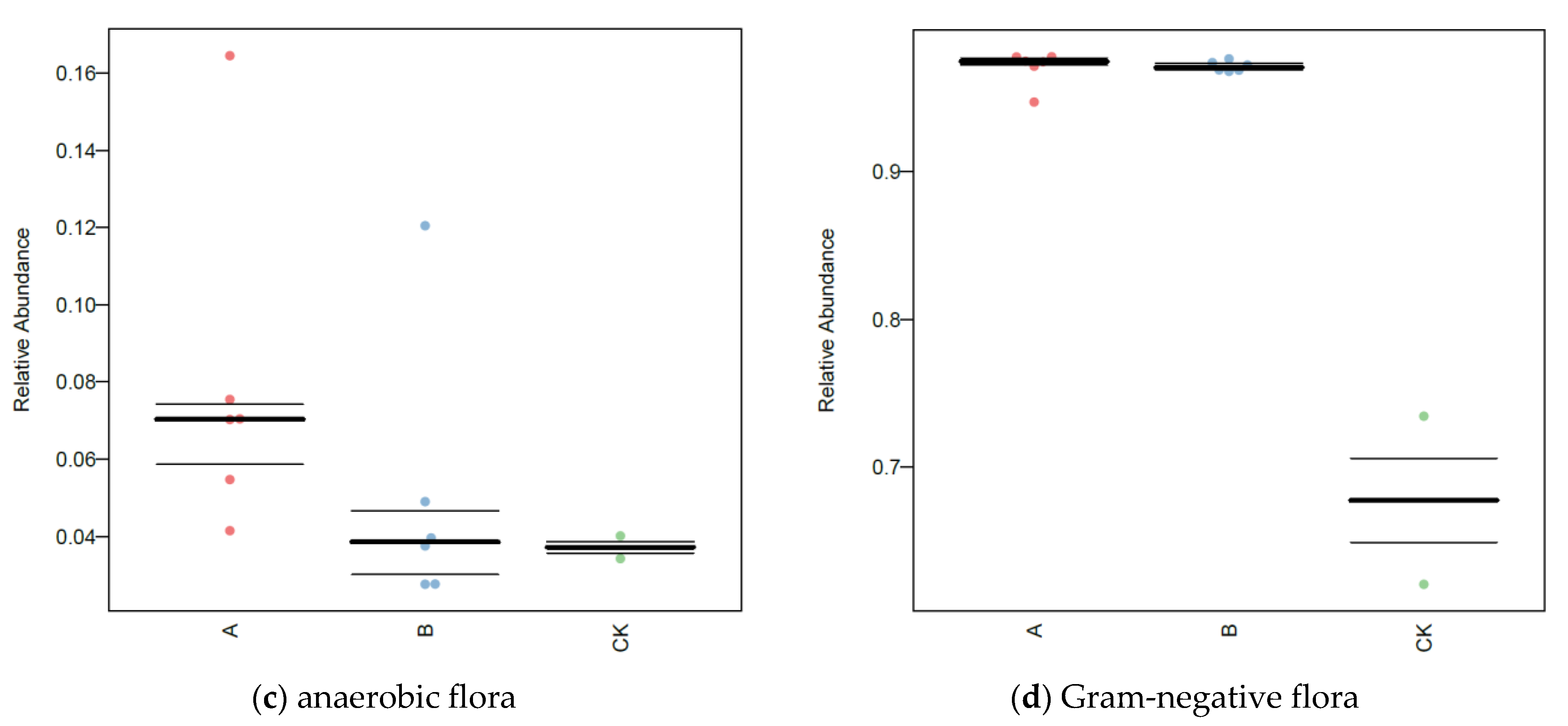
3.7. BugBase Analysis of Soil Microorganisms
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phillips, T.M.; Seech, A.G.; Lee, H.; Trevors, J.T. Biodegradation of hexachlorocyclohexane (HCH) by microorganisms. Biodegradation 2005, 16, 363–392. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yun, X.T.; Ruan, Z.Y.; Lu, C.J.; Shi, Y.; Qin, Q.; Men, Z.M.; Zou, D.Z.; Du, X.M.; Xing, B.S.; et al. Review of hexachlorocyclohexane (HCH) and dichlorodiphenyltrichloroethane (DDT) contamination in Chinese soils. Sci. Total Environ. 2020, 749, 141212. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, J.; de Borst, B.; Weber, R.; Stobiecki, T.; Forter, M. HCH and lindane contaminated sites: European and global need for a permanent solution for a long-time neglected issue. Environ. Pollut. 2019, 248, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Akbor, M.A.; Nahar, A.; Hasan, M.; Islam, A.R.M.T.; Siddique, M.A.B. Level of pesticides contamination in the major river systems: A review on South Asian countries perspective. Heliyon 2021, 7, e07270. [Google Scholar] [CrossRef]
- Zhao, L.; Teng, Y.; Lou, Y.M. Status of organochlorine pesticide contaminated sitis in China and progress in site remediation. Soil 2018, 50, 435–445. (In Chinese) [Google Scholar]
- Aamir, M.; Khan, S.; Nawab, J.; Qamar, Z.; Khan, A. Tissue distribution of HCH and DDT congeners and human health risk associated with consumption of fish collected from Kabul River, Pakistan. Ecotoxicol. Environ. Saf. 2016, 125, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Shah, H.K.; Sharma, T.; Banerjee, B.D. Organochlorine pesticides induce inflammation, ROS production, and DNA damage in human epithelial ovary cells: An in vitro study. Chemosphere 2020, 246, 125691. [Google Scholar] [CrossRef]
- Srivastava, V.; Srivastava, T.; Kumar, M.S. Fate of the persistent organic pollutant (POP) Hexachlorocyclohexane (HCH) and remediation challenges. Int. Biodeterior. Biodegrad. 2019, 140, 43–56. [Google Scholar] [CrossRef]
- Yang, P.F.; Macdonald, R.W.; Hung, H.; Muir, D.C.G.; Kallenborn, R.; Nikolaev, A.N.; Ma, W.L.; Liu, L.Y.; Li, Y.F. Modeling historical budget for β-Hexachlorocyclohexane (HCH) in the Arctic Ocean: A contrast to α-HCH. Environ. Sci. Ecotechnol. 2023, 14, 100229. [Google Scholar] [CrossRef]
- Checa-Fernández, A.; Santos, A.; Romero, A.; Domínguez, C.M. Remediation of real soil polluted with hexachlorocyclohexanes (α-HCH and β-HCH) using combined thermal and alkaline activation of persulfate: Optimization of the operating conditions. Sep. Purif. Technol. 2021, 270, 118795. [Google Scholar] [CrossRef]
- Dominguez, C.M.; Checa-Fernandez, A.; Romero, A.; Santos, A. Degradation of HCHs by thermally activated persulfate in soil system: Effect of temperature and oxidant concentration. J. Environ. Chem. Eng. 2021, 9, 105668. [Google Scholar] [CrossRef]
- Balazs, H.E.; Schmid, C.A.O.; Feher, I.; Podar, D.; Szatmari, P.M.; Marincaş, O.; Balázs, Z.R.; Schröder, P. HCH phytoremediation potential of native plant species from a contaminated urban site in Turda, Romania. J. Environ. Manag. 2018, 223, 286–296. [Google Scholar] [CrossRef] [PubMed]
- Kaur, I.; Gaur, V.K.; Regar, R.K.; Roy, A.; Srivastava, P.K.; Gaur, R.; Manickam, N.; Barik, S.K. Plants exert beneficial influence on soil microbiome in a HCH contaminated soil revealing advantage of microbe-assisted plant-based HCH remediation of a dumpsite. Chemosphere 2021, 280, 130690. [Google Scholar] [CrossRef] [PubMed]
- Lal, R.; Dadhwal, M.; Kumari, K.; Sharma, P.; Singh, A.; Kumari, H.; Jit, S.; Gupta, S.K.; Nigam, A.; Lal, D.; et al. Pseudomonas sp. to Sphingobium indicum: A journey of microbial degradation and bioremediation of Hexachlorocyclohexane. Indian J. Microbiol. 2008, 48, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Manickam, N.; Bajaj, A.; Saini, H.S.; Shanker, R. Surfactant mediated enhanced biodegradation of hexachlorocyclohexane (HCH) isomers by Sphingomonas sp. NM05. Biodegradation 2012, 23, 673–682. [Google Scholar] [CrossRef]
- Ma, A.Z.; Wu, J.; Wang, T.; Zhang, G.S.; Li, S.P. Isolation and characterization of a HCH degradation Sphingomonas sp. stain BHC-A. Acta Microbiol. Sin. 2005, 45, 728–732. [Google Scholar]
- Sineli, P.E.; Tortella, G.; Dávila Costa, J.S.; Benimeli, C.S.; Cuozzo, S.A. Evidence of α-, β- and γ-HCH mixture aerobic degradation by the native actinobacteria Streptomyces sp. M7. World J. Microb. Biot. 2016, 32, 81. [Google Scholar] [CrossRef]
- Luo, S.; Liang, Y.P.; Zeng, H.H.; Qin, L.T. Screening and Identification of Bacteria β-HCH Rhizosphere Degradation in Constructed Wetland Plants. J. Henan Univ. Sci. Technol. (Nat. Sci.) 2021, 42, 78–87. (In Chinese) [Google Scholar]
- Verma, H.; Kumar, R.; Oldach, P.; Sangwan, N.; Khurana, J.P.; Gilbert, J.A.; Lal, R. Comparative genomic analysis of nine Sphingobium strains: Insights into their evolution and hexachlorocyclohexane (HCH) degradation pathways. BMC Genom. 2014, 15, 1014. [Google Scholar] [CrossRef]
- Peng, G.S. Study on the characteristics of bacterial community and environmental factors in the removal of β-HCH in constructed wetlands. Master’s Thesis, Guilin University of Technology, Guilin, China, 2020. (In Chinese). [Google Scholar]
- Yin, T.R.; Te, S.H.; Reinhard, M.; Yang, Y.; Chen, H.T.; He, Y.L.; Gin, K.Y.H. Biotransformation of Sulfluramid (N-ethyl perfluorooctane sulfonamide) and dynamics of associated rhizospheric microbial community in microcosms of wetland plants. Chemosphere 2018, 211, 378–389. [Google Scholar] [CrossRef]
- Aparicio, J.D.; Raimondo, E.E.; Saez, J.M.; Costa-Gutierrez, S.B.; Álvarez, A.; Benimeli, C.S.; Polti, M.A. The current approach to soil remediation: A review of physicochemical and biological technologies, and the potential of their strategic combination. J. Environ. Chem. Eng. 2022, 10, 107141. [Google Scholar] [CrossRef]
- Wang, J.L.; Chen, X.Y.; Yan, W.D.; Ning, C.; Gsell, T. Both artificial root exudates and natural Koelreuteria paniculata exudates modify bacterial community structure and enhance phenanthrene biodegradation in contaminated soils. Chemosphere 2021, 263, 128041. [Google Scholar] [CrossRef]
- Chen, Q.; Zeng, H.H.; Liang, Y.P.; Qin, L.T.; Peng, G.S.; Huang, L.L.; Song, X.H. Purification Effects on β-HCH Removal and Bacterial Community Differences of Vertical-Flow Constructed Wetlands with Different Vegetation Plantations. Sustainability 2021, 13, 13244. [Google Scholar] [CrossRef]
- Suharti, S.; Oktafia, H.; Sudarman, A.; Baik, M.; Wiryawan, K.G. Effect of cyanide-degrading bacteria inoculation on performance, rumen fermentation characteristics of sheep fed bitter cassava (Manihot esculenta Crantz) leaf meal. Ann. Agric. Sci.-CAIRO 2021, 66, 131–136. [Google Scholar] [CrossRef]
- Zheng, J.Y.; Wang, L.; Zhao, J.S.; Niu, Y.H.; Xiao, H.B.; Wang, Z.; Yu, S.X.; Shi, Z.H. Forty-year-old orchards promote carbon storage by changing aggregate-associated enzyme activities and microbial communities. Catena 2022, 213, 106195. [Google Scholar] [CrossRef]
- Gilmour, K.A.; Davie, C.T.; Gray, N. Survival and activity of an indigenous iron-reducing microbial community from MX80 bentonite in high temperature / low water environments with relevance to a proposed method of nuclear waste disposal. Sci. Total Environ. 2022, 814, 152660. [Google Scholar] [CrossRef] [PubMed]
- Vijgen, J.; Abhilash, P.C.; Yi, F.L.; Lal, R.; Forter, M.; Torres, J.; Singh, N.; Yunus, M.; Tian, C.G.; Schäffer, A.; et al. Hexachlorocyclohexane (HCH) as new Stockholm Convention POPs—A global perspective on the management of Lindane and its waste isomers. Environ. Sci. Pollut. Res. 2010, 18, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.K.; Zhu, Y.Y.; Zhu, L.S.; Zhang, J.; Li, B.; Wang, J.H.; Wang, J.; Zhang, C.; Cheng, C. Effects of the herbicide mesotrione on soil enzyme activity and microbial communities. Ecotoxicol. Environ. Saf. 2018, 164, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Papale, M.; Giannarelli, S.; Francesconi, S.; Marco, G.D.; Mikkonen, A.; Conte, A.; Rizzo, C.; Domenico, E.D.; Michaud, L.; Giudice, A.L. Enrichment, isolation and biodegradation potential of psychrotolerant polychlorinated-biphenyl degrading bacteria from the Kongsfjorden (Svalbard Islands, High Arctic Norway). Mar. Pollut. Bull. 2016, 114, 849–859. [Google Scholar] [CrossRef]
- Rabani, M.S.; Sharma, R.; Singh, R.; Gupta, M.K. Characterization and identification of naphthalene degrading bacteria isolated from petroleum contaminated sites and their possible use in bioremediation. Polycycl. Aromat. Compd. 2022, 42, 978–989. [Google Scholar] [CrossRef]
- Guo, J.; Wu, X.K.; Liu, S.Y.; Zhang, Y.; Cai, Z.Q. Screening of three ammonia-oxidizing bacteria and construction of compounding agent CCZU C6 in high-efficiency ammonia-oxidizing. J. Water Process Eng. 2021, 40, 101862. [Google Scholar] [CrossRef]
- Góngora-Echeverría, V.R.; García-Escalante, R.; Rojas-Herrera, R.; Giácoman-Vallejos, G.; Ponce-Caballero, C. Pesticide bioremediation in liquid media using a microbial consortium and bacteria-pure strains isolated from a biomixture used in agricultural areas. Ecotoxicol. Environ. Saf. 2020, 200, 110734. [Google Scholar] [CrossRef] [PubMed]
- Prekrasna, L.; Pavlovska, M.; Oleinik, I.; Dykyi, E.; Slobodnik, J.; Alygizakis, N.; Solomenko, L.; Stoica, E. Bacterial communities of the Black Sea exhibit activity against persistent organic pollutants in the water column and sediments. Ecotoxicol. Environ. Saf. 2022, 234, 113367. [Google Scholar] [CrossRef] [PubMed]
- Palladini, J.; Bagnati, R.; Passoni, A.; Davoli, E.; Lanno, A.; Terzaghi, E.; Falakdin, P.; Di, G.A. Bioaccumulation of PCBs and their hydroxy and sulfonated metabolites in earthworms: Comparing lab and field results. Environ. Pollut. 2021, 293, 118507. [Google Scholar] [CrossRef]
- Nikos, C.K.; Carl, R.W. KOW: A novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 1996, 21, 425–426. [Google Scholar]
- Bocedi, A.; Lai, O.; Cattani, G.; Roncoroni, C.; Gambardella, G.; Notari, S.; Tancredi, F.; Bitonti, G.; Calabrò, S.; Ricci, G. Animal Biomonitoring for the Surveillance of Environment Affected by the Presence of Slight Contamination by β-HCH. Antioxidants 2022, 11, 527. [Google Scholar] [CrossRef]
- Manz, M.; Wenzel, K.D.; Dietze, U.; Schüürmann, G. Persistent organic pollutants in agricultural soils of central Germany. Sci. Total Environ. 2001, 277, 187–198. [Google Scholar] [CrossRef]
- Koshlaf, E.; Shahsavari, E.; Haleyur, N.; Osborn, A.M.; Ball, A.S. Effect of biostimulation on the distribution and composition of the microbial community of a polycyclic aromatic hydrocarbon-contaminated landfill soil during bioremediation. Geoderma 2019, 338, 216–225. [Google Scholar] [CrossRef]
- Mitsuhiko, K.; Norio, N.; Fadhil, S.; Abdullah, A.R.; Mohd, S.K.; Tatsuki, T.; Takuya, M.; Kiyohiko, N. Effect of temperature on thermophilic composting of aquaculture sludge: NH3 recovery, nitrogen mass balance, and microbial community dynamics. Bioresour. Technol. 2018, 265, 207–213. [Google Scholar]
- Wang, J.; Tang, K.; Hu, X.J.; Wang, H.F.; Gudda, F.O.; Odinga, E.S.; El-Ramady, H.; Ling, W.T. Impact of hexachlorocyclohexane addition on the composition and potential functions of the bacterial community in red and purple paddy soil. Environ. Pollut. 2022, 297, 118795. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, J.F.; Zhang, L.F.; Zhang, W.Z.; Jia, J.X.; Dai, H.H.; Wang, Z.Q. Responses of nitrification performance, triclosan resistome and diversity of microbes to continuous triclosan stress in activated sludge system. J. Environ. Sci. 2020, 92, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Phandanouvong-Lozano, V.; Sun, W.; Sanders, J.M.; Hay, A.G. Biochar does not attenuate triclosan’s impact on soil bacterial communities. Chemosphere 2018, 213, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Matturro, B.; Heavner, G.L.; Richardson, R.E.; Rossetti, S. Quantitative estimation of Dehalococcoides mccartyi at laboratory and field scale: Comparative study between CARD-FISH and Real Time PCR. J. Microbiol. Meth. 2013, 93, 127–133. [Google Scholar] [CrossRef]
- Rosell, M.; Palau, J.; Mortan, S.H.; Caminal, G.; Soler, A.; Shouakar-Stash, O.; Marco-Urrea, E. Dual carbon-chlorine isotope fractionation during dichloroelimination of 1,1,2-trichloroethane by an enrichment culture containing Dehalogenimonas sp. Sci. Total Environ. 2019, 648, 422–429. [Google Scholar] [CrossRef]
- van der Zaan, B.; Hannes, F.; Hoekstra, N.; Rijnaarts, H.; de Vos, W.M.; Smidt, H.; Gerritse, J. Correlation of Dehalococcoides 16S rRNA and chloroethene-reductive dehalogenase genes with geochemical conditions in chloroethene-contaminated groundwater. Appl. Environ. Microbiol. 2010, 76, 843–850. [Google Scholar] [CrossRef]
- Saiyari, D.M.; Chuang, H.P.; Senoro, D.B.; Lin, T.F.; Whang, L.M.; Chiu, Y.T.; Chen, Y.H. A review in the current developments of genus Dehalococcoides, its consortia and kinetics for bioremediation options of contaminated groundwater. Sustain. Environ. Res. 2018, 28, 149–157. [Google Scholar] [CrossRef]
- Rivera-Araya, J.; Huynh, N.D.; Kaszuba, M.; Chávez, R.; Schlömann, M.; Levicán, G. Mechanisms of NaCl-tolerance in acidophilic iron-oxidizing bacteria and archaea: Comparative genomic predictions and insights. Hydrometallurgy 2020, 194, 105354. [Google Scholar] [CrossRef]
- Sun, F.L.; Xu, Z.T.; Fan, L.L. Response of heavy metal and antibiotic resistance genes and related microorganisms to different heavy metals in activated sludge. J. Environ. Manag. 2021, 300, 113754. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.; Shi, H.; Liang, Y.; Qin, L.; Zeng, H.; Song, X. Degradation Characteristics and Remediation Ability of Contaminated Soils by Using β-HCH Degrading Bacteria. Int. J. Environ. Res. Public Health 2023, 20, 2767. https://doi.org/10.3390/ijerph20042767
Chen Q, Shi H, Liang Y, Qin L, Zeng H, Song X. Degradation Characteristics and Remediation Ability of Contaminated Soils by Using β-HCH Degrading Bacteria. International Journal of Environmental Research and Public Health. 2023; 20(4):2767. https://doi.org/10.3390/ijerph20042767
Chicago/Turabian StyleChen, Qing, Huijun Shi, Yanpeng Liang, Litang Qin, Honghu Zeng, and Xiaohong Song. 2023. "Degradation Characteristics and Remediation Ability of Contaminated Soils by Using β-HCH Degrading Bacteria" International Journal of Environmental Research and Public Health 20, no. 4: 2767. https://doi.org/10.3390/ijerph20042767
APA StyleChen, Q., Shi, H., Liang, Y., Qin, L., Zeng, H., & Song, X. (2023). Degradation Characteristics and Remediation Ability of Contaminated Soils by Using β-HCH Degrading Bacteria. International Journal of Environmental Research and Public Health, 20(4), 2767. https://doi.org/10.3390/ijerph20042767






