Assessment of Diagnosis, Prognosis and Immune Infiltration Response to the Expression of the Ferroptosis-Related Molecule HAMP in Clear Cell Renal Cell Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
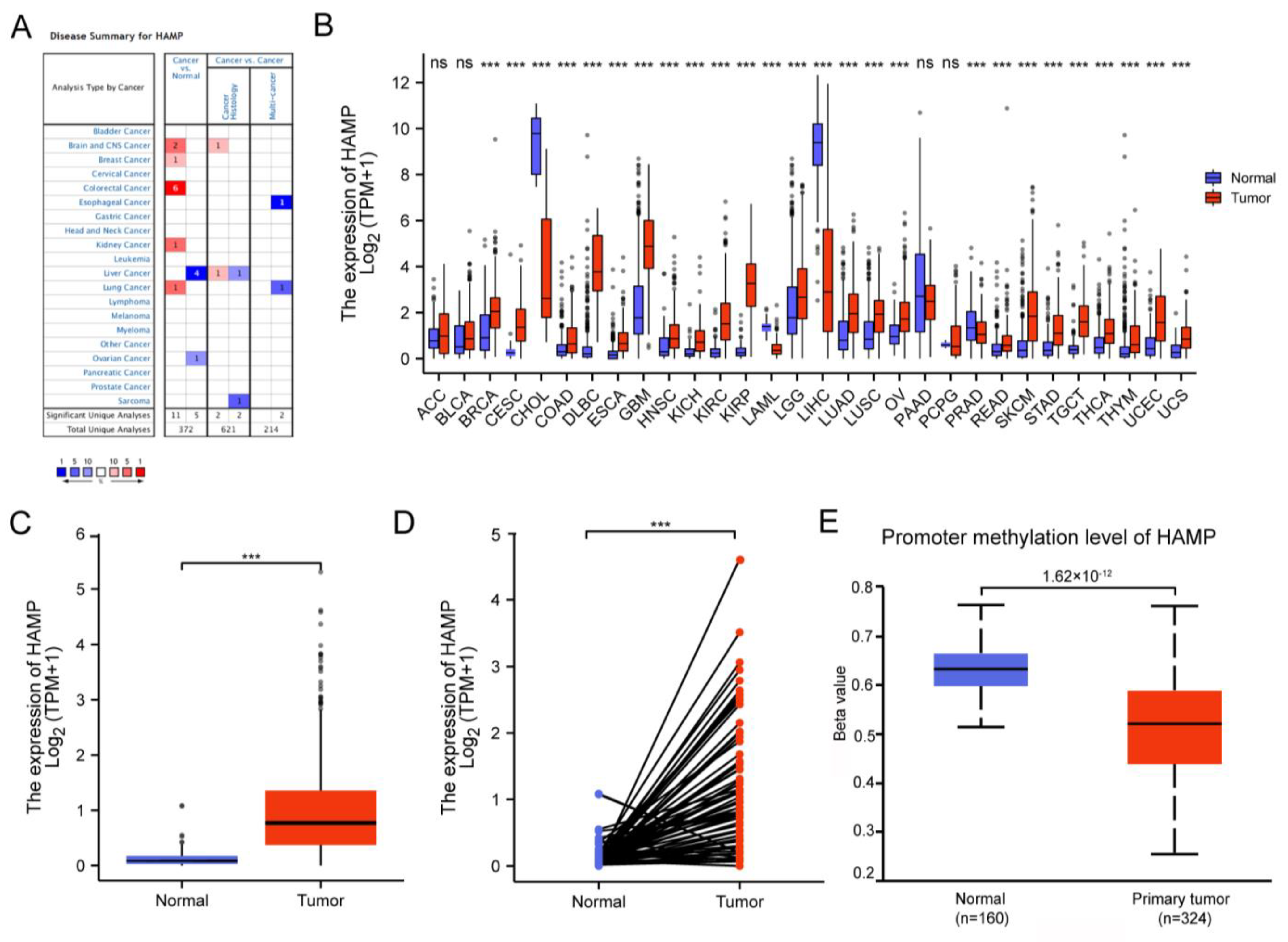
2.2. Oncomine Database Analysis
2.3. Timer Database Analysis
2.4. Ualcan
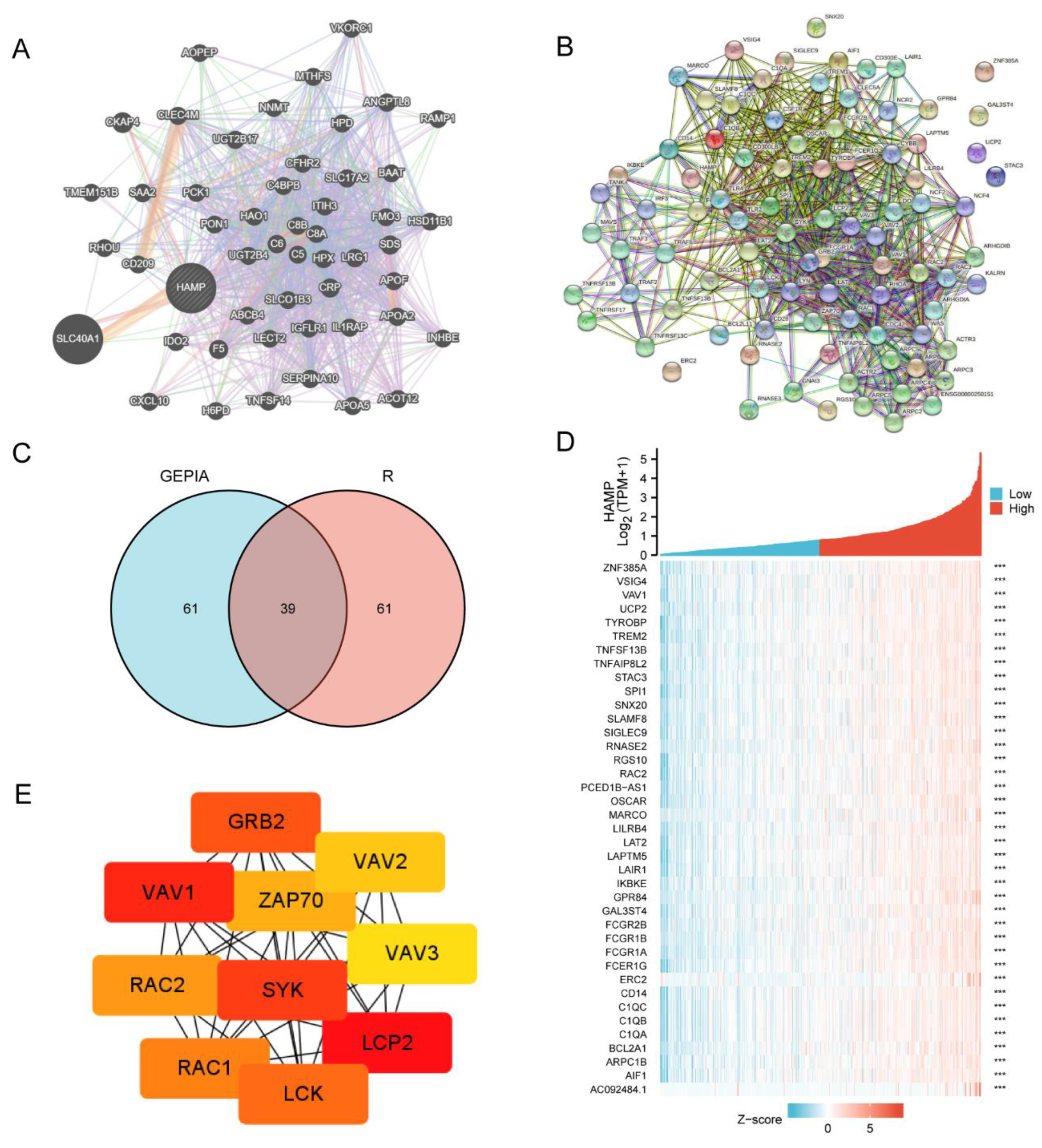
2.5. GeneMANIA Database Analysis
2.6. STRING Databases Analysis
2.7. GEPIA Database Analysis
2.8. Cytoscape Software
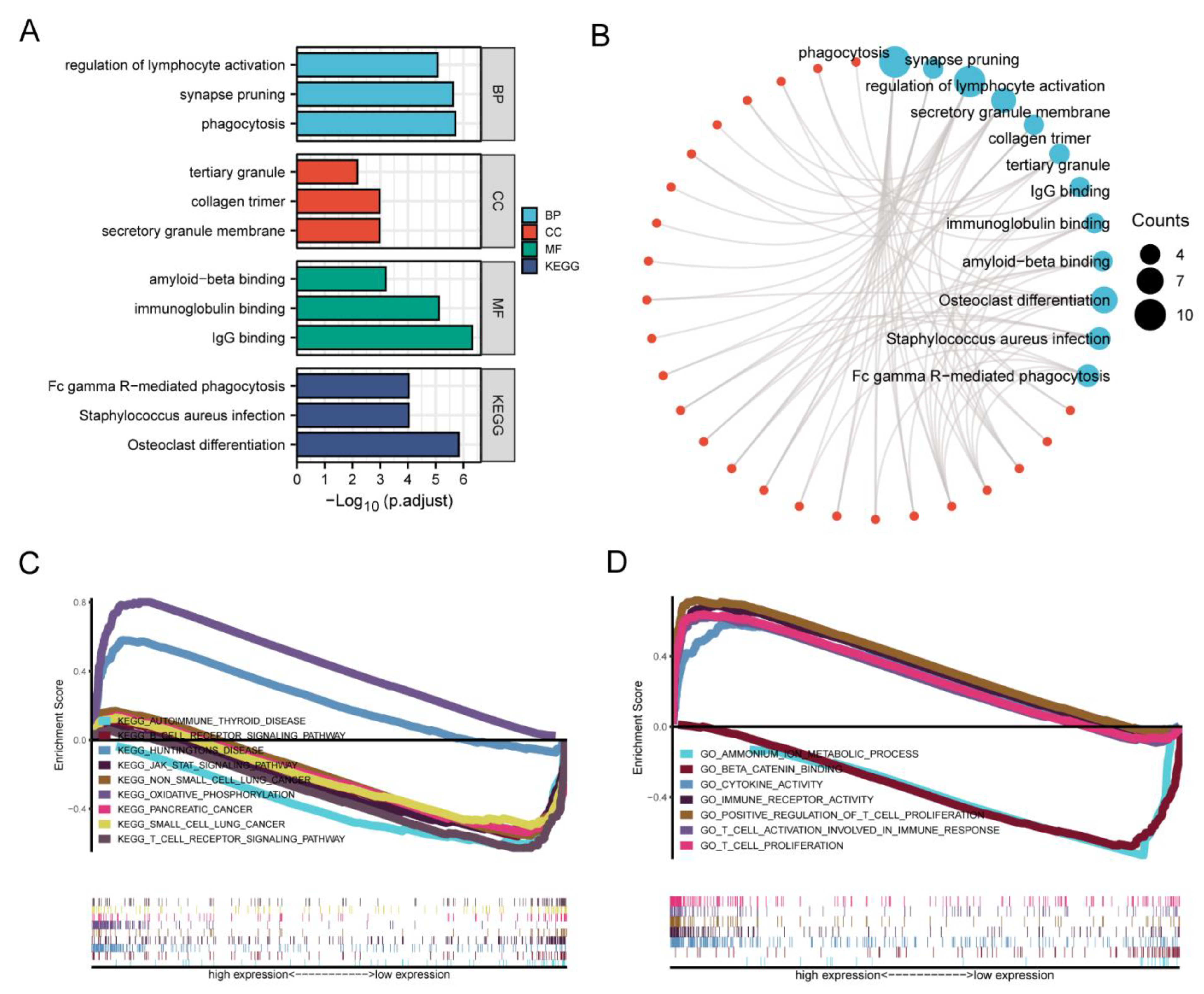
2.9. Gene Set Enrichment Analyis
2.10. Statistical Analysis
3. Results
3.1. Differential Expression of HAMP in KIRC
3.2. The Clinical Correlation and Prognostic Value of HAMP in KIRC
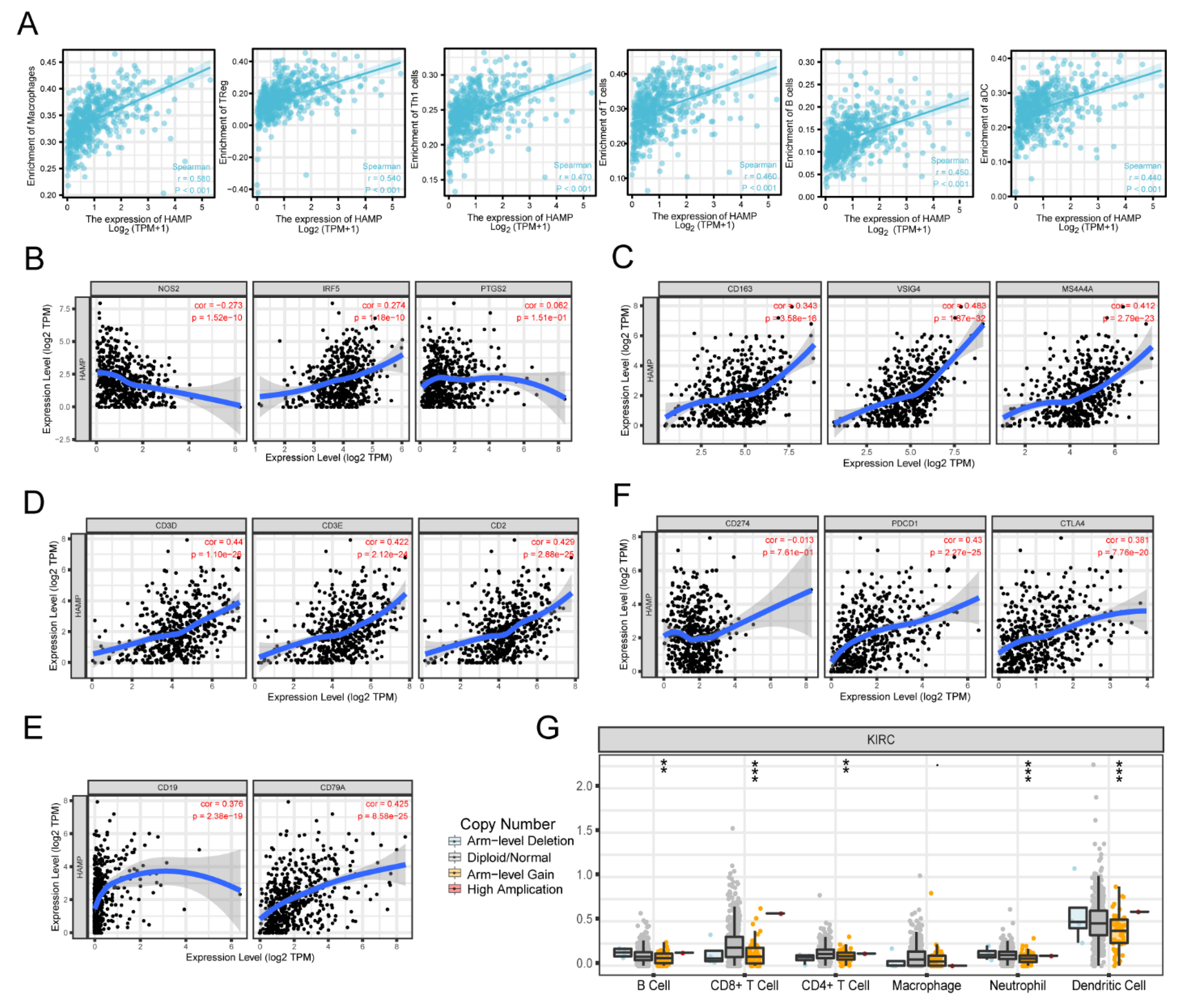
3.3. Correlation between Immune Cells and HAMP Expression Levels in KIRC
3.4. The Network of HAMP Expression in KIRC
3.5. Enrichment Analysis of HAMP in KIRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
References
- Klümper, N.; Ralser, D.J.; Bawden, E.G.; Landsberg, J.; Zarbl, R.; Kristiansen, G.; Toma, M.; Ritter, M.; Hölzel, M.; Ellinger, J.; et al. LAG3 (LAG-3, CD223) DNA methylation correlates with LAG3 expression by tumor and immune cells, immune cell infiltration, and overall survival in clear cell renal cell carcinoma. J. Immunother. Cancer 2020, 8, e000552. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, C.; Xiao, M.; Han, Y.; Zhang, S.; Xu, B. Bioinformatics Analysis of the Prognostic and Biological Significance of ZDHHC-Protein Acyltransferases in Kidney Renal Clear Cell Carcinoma. Front. Oncol. 2020, 10, 565414. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, H.; Yang, C.; Liu, L.; Deng, S.; Li, M. Comprehensive Analysis of ATP6V1s Family Members in Renal Clear Cell Carcinoma with Prognostic Values. Front. Oncol. 2020, 10, 567970. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zheng, L.; Lu, Y.; Xia, Q.; Zhou, P.; Liu, Z. Comprehensive analysis on the expression levels and prognostic values of LOX family genes in kidney renal clear cell carcinoma. Cancer Med. 2020, 9, 8624–8638. [Google Scholar] [CrossRef] [PubMed]
- Meng, T.; Huang, R.; Zeng, Z.; Huang, Z.; Yin, H.; Jiao, C.; Yan, P.; Hu, P.; Zhu, X.; Li, Z.; et al. Identification of Prognostic and Metastatic Alternative Splicing Signatures in Kidney Renal Clear Cell Carcinoma. Front. Bioeng. Biotechnol. 2019, 7, 270. [Google Scholar] [CrossRef]
- Song, J.; Liu, Y.D.; Su, J.; Yuan, D.; Sun, F.; Zhu, J. Systematic analysis of alternative splicing signature unveils prognostic predictor for kidney renal clear cell carcinoma. J. Cell. Physiol. 2019, 234, 22753–22764. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Angeli, J.P.F.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Zou, Y.; Palte, M.J.; Deik, A.A.; Li, H.; Eaton, J.K.; Wang, W.; Tseng, Y.Y.; Deasy, R.; Kost-Alimova, M.; Dančík, V.; et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat. Commun. 2019, 10, 1617. [Google Scholar] [CrossRef]
- Gao, M.; Fan, K.; Chen, Y.; Zhang, G.; Chen, J.; Zhang, Y. Understanding the mechanistic regulation of ferroptosis in cancer: The gene matters. J. Genet. Genom. 2022, 49, 913–926. [Google Scholar] [CrossRef]
- Bogdan, A.R.; Miyazawa, M.; Hashimoto, K.; Tsuji, Y. Regulators of Iron Homeostasis: New Players in Metabolism, Cell Death, and Disease. Trends Biochem. Sci. 2016, 41, 274–286. [Google Scholar] [CrossRef]
- Nicolas, G.; Bennoun, M.; Devaux, I.; Beaumont, C.; Grandchamp, B.; Kahn, A.; Vaulont, S. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc. Natl. Acad. Sci. USA 2001, 98, 8780–8785. [Google Scholar] [CrossRef]
- Kemna, E.H.; Tjalsma, H.; Willems, H.L.; Swinkels, D.W. Hepcidin: From discovery to differential diagnosis. Haematologica 2008, 93, 90–97. [Google Scholar] [CrossRef]
- Wang, J.; Liu, W.; Li, J.C.; Li, M.; Li, B.; Zhu, R. Hepcidin Downregulation Correlates with Disease Aggressiveness and Immune Infiltration in Liver Cancers. Front. Oncol. 2021, 11, 714756. [Google Scholar] [CrossRef]
- Fang, Z.; Zhu, Z.; Zhang, H.; Peng, Y.; Liu, J.; Lu, H.; Li, J.; Liang, L.; Xia, S.; Wang, Q.; et al. GDF11 contributes to hepatic hepcidin (HAMP) inhibition through SMURF1-mediated BMP-SMAD signalling suppression. Br. J. Haematol. 2020, 188, 321–331. [Google Scholar] [CrossRef]
- Vela, D.; Vela-Gaxha, Z. Differential regulation of hepcidin in cancer and non-cancer tissues and its clinical implications. Exp. Mol. Med. 2018, 50, e436. [Google Scholar] [CrossRef]
- Shen, Y.; Li, X.; Su, Y.; Badshah, S.A.; Zhang, B.; Xue, Y.; Shang, P. HAMP Downregulation Contributes to Aggressive Hepatocellular Carcinoma via Mechanism Mediated by Cyclin4-Dependent Kinase-1/STAT3 Pathway. Diagnostics 2019, 9, 48. [Google Scholar] [CrossRef]
- Wei, F.Z.; Mei, S.W.; Wang, Z.J.; Chen, J.N.; Zhao, F.Q.; Li, J.; Xiao, T.X.; Zhao, W.; Ma, Y.B.; Yuan, W.; et al. HAMP as a Prognostic Biomarker for Colorectal Cancer Based on Tumor Microenvironment Analysis. Front. Oncol. 2022, 12, 884474. [Google Scholar] [CrossRef]
- Ping, S.; Wang, S.; Zhao, Y.; He, J.; Li, G.; Li, D.; Wei, Z.; Chen, J. Identification and validation of a ferroptosis-related gene signature for predicting survival in skin cutaneous melanoma. Cancer Med. 2022, 11, 3529–3541. [Google Scholar] [CrossRef]
- Armitage, A.E.; Eddowes, L.A.; Gileadi, U.; Cole, S.; Spottiswoode, N.; Selvakumar, T.A.; Ho, L.P.; Townsend, A.R.; Drakesmith, H. Hepcidin regulation by innate immune and infectious stimuli. Blood 2011, 118, 4129–4139. [Google Scholar] [CrossRef]
- Bessman, N.J.; Mathieu, J.R.R.; Renassia, C.; Zhou, L.; Fung, T.C.; Fernandez, K.C.; Austin, C.; Moeller, J.B.; Zumerle, S.; Louis, S.; et al. Dendritic cell-derived hepcidin sequesters iron from the microbiota to promote mucosal healing. Science 2020, 368, 186–189. [Google Scholar] [CrossRef]
- Vyoral, D.; Petrák, J. Hepcidin: A direct link between iron metabolism and immunity. Int. J. Biochem. Cell Biol. 2005, 37, 1768–1773. [Google Scholar] [CrossRef] [PubMed]
- Lim, D.; Kim, K.S.; Jeong, J.H.; Marques, O.; Kim, H.J.; Song, M.; Lee, T.H.; Kim, J.I.; Choi, H.S.; Min, J.J.; et al. The hepcidin-ferroportin axis controls the iron content of Salmonella-containing vacuoles in macrophages. Nat. Commun. 2018, 9, 2091. [Google Scholar] [CrossRef] [PubMed]
- Sacco, A.; Battaglia, A.M.; Botta, C.; Aversa, I.; Mancuso, S.; Costanzo, F.; Biamonte, F. Iron Metabolism in the Tumor Microenvironment-Implications for Anti-Cancer Immune Response. Cells 2021, 10, 303. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Li, Q.; Du, Y.; Yun, J.; Xie, Y.; DeBerardinis, R.J.; Xiao, G. Genomic regression analysis of coordinated expression. Nat. Commun. 2017, 8, 2187. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Cai, C.; Du, J.; Liu, B.; Cui, L.; Fan, X.; Wu, Q.; Fang, J.; Xie, L. TCMIO: A Comprehensive Database of Traditional Chinese Medicine on Immuno-Oncology. Front. Pharmacol. 2020, 11, 439. [Google Scholar] [CrossRef]
- Shi, J.; Jiang, D.; Yang, S.; Zhang, X.; Wang, J.; Liu, Y.; Sun, Y.; Lu, Y.; Yang, K. LPAR1, Correlated with Immune Infiltrates, Is a Potential Prognostic Biomarker in Prostate Cancer. Front. Oncol. 2020, 10, 846. [Google Scholar] [CrossRef]
- Chen, F.; Chandrashekar, D.S.; Varambally, S.; Creighton, C.J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 2019, 10, 5679. [Google Scholar] [CrossRef]
- Fernald, G.H.; Capriotti, E.; Daneshjou, R.; Karczewski, K.J.; Altman, R.B. Bioinformatics challenges for personalized medicine. Bioinformatics 2011, 27, 1741–1748. [Google Scholar] [CrossRef]
- Altman, M.C.; Gill, M.A.; Whalen, E.; Babineau, D.C.; Shao, B.; Liu, A.H.; Jepson, B.; Gruchalla, R.S.; O’Connor, G.T.; Pongracic, J.A.; et al. Transcriptome networks identify mechanisms of viral and nonviral asthma exacerbations in children. Nat. Immunol. 2019, 20, 637–651. [Google Scholar] [CrossRef]
- Vega, I.A.-d.; Paz-Cabrera, M.C.; Rother, M.B.; Wiegant, W.W.; Checa-Rodríguez, C.; Hernández-Fernaud, J.R.; Huertas, P.; Freire, R.; van Attikum, H.; Smits, V.A.J. PHF2 regulates homology-directed DNA repair by controlling the resection of DNA double strand breaks. Nucleic Acids Res. 2020, 48, 4915–4927. [Google Scholar]
- Liu, Y.; Huan, W.; Wu, J.; Zou, S.; Qu, L. IGFBP6 Is Downregulated in Unstable Carotid Atherosclerotic Plaques According to an Integrated Bioinformatics Analysis and Experimental Verification. J. Atheroscler. Thromb. 2020, 27, 1068–1085. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Oguro, S.; Ino, Y.; Shimada, K.; Hatanaka, Y.; Matsuno, Y.; Esaki, M.; Nara, S.; Kishi, Y.; Kosuge, T.; Hiraoka, N. Clinical significance of tumor-infiltrating immune cells focusing on BTLA and Cbl-b in patients with gallbladder cancer. Cancer Sci. 2015, 106, 1750–1760. [Google Scholar] [CrossRef]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef]
- Fan, J.; Salathia, N.; Liu, R.; Kaeser, G.E.; Yung, Y.C.; Herman, J.L.; Kaper, F.; Fan, J.B.; Zhang, K.; Chun, J.; et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods 2016, 13, 241–244. [Google Scholar] [CrossRef]
- Giglio, M.; Tauber, R.; Nadendla, S.; Munro, J.; Olley, D.; Ball, S.; Mitraka, E.; Schriml, L.M.; Gaudet, P.; Hobbs, E.T.; et al. ECO, the Evidence & Conclusion Ontology: Community standard for evidence information. Nucleic Acids Res. 2019, 47, D1186–D1194. [Google Scholar]
- D’Angelo, S.P.; Shoushtari, A.N.; Keohan, M.L.; Dickson, M.A.; Gounder, M.M.; Chi, P.; Loo, J.K.; Gaffney, L.; Schneider, L.; Patel, Z.; et al. Combined KIT and CTLA-4 Blockade in Patients with Refractory GIST and Other Advanced Sarcomas: A Phase Ib Study of Dasatinib plus Ipilimumab. Clin. Cancer Res. 2017, 23, 2972–2980. [Google Scholar] [CrossRef]
- Hu, Z.; Zhou, J.; Jiang, J.; Yuan, J.; Zhang, Y.; Wei, X.; Loo, N.; Wang, Y.; Pan, Y.; Zhang, T.; et al. Genomic characterization of genes encoding histone acetylation modulator proteins identifies therapeutic targets for cancer treatment. Nat. Commun. 2019, 10, 733. [Google Scholar] [CrossRef]
- Lee, S.T.; Wiemels, J.L. Genome-wide CpG island methylation and intergenic demethylation propensities vary among different tumor sites. Nucleic Acids Res. 2016, 44, 1105–1117. [Google Scholar] [CrossRef]
- Schmutte, C.; Yang, A.S.; Nguyen, T.T.; Beart, R.W.; Jones, P.A. Mechanisms for the involvement of DNA methylation in colon carcinogenesis. Cancer Res. 1996, 56, 2375–2381. [Google Scholar]
- Sohn, B.H.; Hwang, J.E.; Jang, H.J.; Lee, H.S.; Oh, S.C.; Shim, J.J.; Lee, K.W.; Kim, E.H.; Yim, S.Y.; Lee, S.H.; et al. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin. Cancer Res. 2017, 23, 4441–4449. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Jing, C.; Xiao, C.; Li, T. Long Non-Coding RNA Profile Study Identifies an Immune-Related lncRNA Prognostic Signature for Kidney Renal Clear Cell Carcinoma. Front. Oncol. 2020, 10, 1430. [Google Scholar] [CrossRef] [PubMed]
- Maj, T.; Wang, W.; Crespo, J.; Zhang, H.; Wang, W.; Wei, S.; Zhao, L.; Vatan, L.; Shao, I.; Szeliga, W.; et al. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat. Immunol. 2017, 18, 1332–1341. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, A.; Swanson, K.D.; Csizmadia, E.; Solanki, A.; Landon-Brace, N.; Gehring, M.P.; Helenius, K.; Olson, B.M.; Pyzer, A.R.; Wang, L.C.; et al. Cabozantinib Eradicates Advanced Murine Prostate Cancer by Activating Antitumor Innate Immunity. Cancer Discov. 2017, 7, 750–765. [Google Scholar] [CrossRef]
- Tanaka, A.; Sakaguchi, S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017, 27, 109–118. [Google Scholar] [CrossRef]
- Chang, W.H.; Lai, A.G. An immunoevasive strategy through clinically-relevant pan-cancer genomic and transcriptomic alterations of JAK-STAT signaling components. Mol. Med. 2019, 25, 46. [Google Scholar] [CrossRef]


| Cell Type | Gene Markers | KIRC | |||
|---|---|---|---|---|---|
| None | Purity | ||||
| Cor | P | Cor | P | ||
| CD8+ T cell | CD8A | 0.382 | 5.72 × 10−20 | 0.353 | 5.99 × 10−15 |
| CD8B | 0.37 | 1.06 × 10−18 | 0.337 | 1.03 × 10−13 | |
| T cell (general) | CD3D | 0.44 | 1.1 × 10−26 | 0.413 | 2.27 × 10−20 |
| CD3E | 0.422 | 2.12 × 10−24 | 0.396 | 8.37 × 10−19 | |
| CD2 | 0.429 | 2.88 × 10−25 | 0.402 | 2.33 × 10−19 | |
| B cell | CD19 | 0.376 | 2.38 × 10−19 | 0.337 | 1.02 × 10−13 |
| CD79A | 0.425 | 8.58 × 10−25 | 0.396 | 1.13 × 10−12 | |
| Monocyte | CD86 | 0.546 | 0.17 × 10−42 | 0.537 | 7.34 × 10−36 |
| CD115 (CSF1R) | 0.443 | 4.87 × 10−27 | 0.419 | 4.81 × 10−21 | |
| TAM | CCL2 | 0.016 | 7.18 × 10−01 | −0.044 | 3.43 × 10−01 |
| CD68 | 0.456 | 0.01 × 10−28 | 0.477 | 1.57 × 10−27 | |
| IL10 | 0.417 | 6.93 × 10−24 | 0.375 | 7.19 × 10−17 | |
| M1 Macrophage | INOS (NOS2) | −0.273 | 1.52 × 10−10 | −0.332 | 2.43 × 10−13 |
| IRF5 | 0.274 | 1.18 × 10−10 | 0.273 | 2.64 × 10−09 | |
| COX2 (PTGS2) | 0.062 | 0.51 × 10−01 | 0.017 | 1.10 × 10−01 | |
| M2 Macrophage | CD163 | 0.343 | 3.58 × 10−16 | 0.34 | 6.06 × 10−14 |
| VSIG4 | 0.483 | 0.87 × 10−32 | 0.48 | 5.74 × 10−28 | |
| MS4A4A | 0.412 | 2.79 × 10−23 | 0.386 | 8.67 × 10−18 | |
| Neutrophils | CD66b (CEACAM8) | −0.107 | 1.34 × 10−02 | −0.103 | 2.64 × 10−02 |
| CD11b (ITGAM) | 0.434 | 7.38 × 10−26 | 0.436 | 9.54 × 10−22 | |
| CCR7 | 0.366 | 2.32 × 10−18 | 0.342 | 4.51 × 10−14 | |
| Natural killer cell | KIR2DL1 | −0.109 | 1.18 × 10−02 | −0.114 | 1.44 × 10−02 |
| KIR2DL3 | −0.074 | 8.94 × 10−02 | −0.057 | 2.25 × 10−01 | |
| KIR2DL4 | 0.177 | 3.86 × 10−05 | 0.154 | 8.88 × 10−04 | |
| KIR3DL1 | −0.135- | 1.75 × 10−03 | −0.115 | 1.34 × 10−02 | |
| KIR3DL2 | 0.01 | 8.25 × 10−01 | 0.008 | 8.57 × 10−01 | |
| KIR3DL3 | 0.026 | 5.5 × 10−01 | 0.011 | 8.16 × 10−01 | |
| KIR2DS4 | −0.096 | 2.73 × 10−02 | −0.096 | 4.03 × 10−02 | |
| Cell Type | Gene Markers | KIRC | |||
|---|---|---|---|---|---|
| Tumor | Normal | ||||
| R | P | R | P | ||
| B cell | CD19 | 0.031 | 0.48 | 0.22 | 0.06 |
| CD79A | 0.15 | 4.8 × 10−4 | 0.2 | 0.098 | |
| T cell | CD3D | 0.24 | 2.9 × 10−08 | 0.62 | 5.2 × 10−09 |
| CD3E | 0.26 | 2.2 × 10−09 | 0.55 | 4.2 × 10−07 | |
| CD2 | 0.27 | 2.7 × 10−10 | 0.51 | 5.7 × 10−06 | |
| M1 Macrophage | INOS (NOS2) | −0.11 | 0.014 | 0.32 | 0.12 |
| IRF5 | 0.19 | 8.2 × 10−06 | −0.037 | 0.76 | |
| COX2 (PTGS2) | −0.021 | 0.64 | 0.082 | 0.49 | |
| M2 Macrophage | CD163 | 0.43 | 0 | 0.44 | 1 × 10−04 |
| VSIG4 | 0.59 | 0 | 0.49 | 1.3 × 10−05 | |
| MS4A4A | 0.37 | 0 | 0.48 | 1.9 × 10−05 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leng, J.; Xing, Z.; Li, X.; Bao, X.; Zhu, J.; Zhao, Y.; Wu, S.; Yang, J. Assessment of Diagnosis, Prognosis and Immune Infiltration Response to the Expression of the Ferroptosis-Related Molecule HAMP in Clear Cell Renal Cell Carcinoma. Int. J. Environ. Res. Public Health 2023, 20, 913. https://doi.org/10.3390/ijerph20020913
Leng J, Xing Z, Li X, Bao X, Zhu J, Zhao Y, Wu S, Yang J. Assessment of Diagnosis, Prognosis and Immune Infiltration Response to the Expression of the Ferroptosis-Related Molecule HAMP in Clear Cell Renal Cell Carcinoma. International Journal of Environmental Research and Public Health. 2023; 20(2):913. https://doi.org/10.3390/ijerph20020913
Chicago/Turabian StyleLeng, Jing, Zixuan Xing, Xiang Li, Xinyue Bao, Junzheya Zhu, Yunhan Zhao, Shaobo Wu, and Jiao Yang. 2023. "Assessment of Diagnosis, Prognosis and Immune Infiltration Response to the Expression of the Ferroptosis-Related Molecule HAMP in Clear Cell Renal Cell Carcinoma" International Journal of Environmental Research and Public Health 20, no. 2: 913. https://doi.org/10.3390/ijerph20020913
APA StyleLeng, J., Xing, Z., Li, X., Bao, X., Zhu, J., Zhao, Y., Wu, S., & Yang, J. (2023). Assessment of Diagnosis, Prognosis and Immune Infiltration Response to the Expression of the Ferroptosis-Related Molecule HAMP in Clear Cell Renal Cell Carcinoma. International Journal of Environmental Research and Public Health, 20(2), 913. https://doi.org/10.3390/ijerph20020913






