European Epidemiological Patterns of Cannabis- and Substance-Related Congenital Neurological Anomalies: Geospatiotemporal and Causal Inferential Study
Abstract
1. Introduction
2. Methods
2.1. Data
2.2. National Assignment
2.3. Derived Data
2.4. Data Imputation
2.5. Statistics
2.6. Covariate Selection
2.7. Panel and Geospatial Analysis
2.8. Causal Inference
2.9. Data Availability
2.10. Ethics
3. Results
3.1. Input Data
3.2. Bivariate Relationships
3.3. Comparing High and Low Cannabis Use Countries
3.4. Multivariable Analysis
3.4.1. Multivariable Relationships—Panel Regression
3.4.2. Multivariable Relationships—Geotemporospatial Regression
3.5. Causal Inference—E-Values
4. Discussion
4.1. Main Results
4.2. Choice of Anomalies
4.3. Qualitative Causal Inference
4.4. Quantitative Causal Inference
4.5. Mechanisms
4.5.1. Morphogen Gradients
Cannabinoid Inhibition of Morphogens
4.5.2. Epigenomic Controls of Brain formation and Neurological Development
4.5.3. Downs Syndrome Cell Adhesion Molecule
4.5.4. Discs-Large Associated Protein 2
4.5.5. Retinoic Acid in Forebrain Development—Direct and Epigenomic Effects
4.5.6. Cerebellins
4.5.7. Slit-Robo
4.6. Exponential Genotoxic Effects
4.7. Generalizability
4.8. Strengths and Limitations
4.9. Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reece, A.S.; Hulse, G.K. Cannabinoid- and Substance- Relationships of European Congenital Anomaly Patterns: A Space-Time Panel Regression and Causal Inferential Study. Environ. Epigenet. 2022, 8, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Congenital Anomaly Epidemiological Correlates of Δ8THC across USA 2003–2016: Panel Regression and Causal Inferential Study. Environ. Epigenet. 2022; in press. [Google Scholar]
- Graham, J.D.P. (Ed.) Cannabis and Health. In Cannabis and Health, 1st ed.; Academic Press: London, UK; New York, NY, USA; San Francisco, CA, USA, 1976; Volume 1, pp. 271–320. [Google Scholar]
- Geber, W.F.; Schramm, L.C. Teratogenicity of marihuana extract as influenced by plant origin and seasonal variation. Arch. Int. Pharmacodyn. Ther. 1969, 177, 224–230. [Google Scholar] [PubMed]
- Geber, W.F.; Schramm, L.C. Effect of marihuana extract on fetal hamsters and rabbits. Toxicol. Appl. Pharmacol. 1969, 14, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Forrester, M.B.; Merz, R.D. Risk of selected birth defects with prenatal illicit drug use, Hawaii, 1986–2002. J. Toxicol. Environ. Health 2007, 70, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Van Gelder, M.M.H.J.; Donders, A.R.T.; Devine, O.; Roeleveld, N.; Reefhuis, J. Using bayesian models to assess the effects of under-reporting of cannabis use on the association with birth defects, national birth defects prevention study, 1997–2005. Paediatr. Perinat. Epidemiol. 2014, 28, 424–433. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Cannabis Consumption Patterns Explain the East-West Gradient in Canadian Neural Tube Defect Incidence: An Ecological Study. Glob. Pediatr. Health 2019, 6, 1–12. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. Broad Spectrum epidemiological contribution of cannabis and other substances to the teratological profile of northern New South Wales: Geospatial and causal inference analysis. BMC Pharmacol. Toxicol. 2020, 21, 75–103. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Geotemporospatial and causal inference epidemiological analysis of US survey and overview of cannabis, cannabidiol and cannabinoid genotoxicity in relation to congenital anomalies 2001–2015. BMC Pediatr. 2022, 22, 47–124. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. Cannabis in Pregnancy–Rejoinder, Exposition and Cautionary Tales. Psychiatr. Times 2020, 37. Available online: https://www.psychiatrictimes.com/view/cannabis-pregnancy-rejoinder-exposition-cautionary-tales (accessed on 15 January 2022).
- Reece, A.S.; Hulse, G.K. Quadruple convergence–rising cannabis prevalence, intensity, concentration and use disorder treatment. Lancet Reg. Health-Eur. 2021, 10, 100245–100246. [Google Scholar] [CrossRef] [PubMed]
- Manthey, J.; Freeman, T.P.; Kilian, C.; Lopez-Pelayo, H.; Rehm, J. Public health monitoring of cannabis use in Europe: Prevalence of use, cannabis potency, and treatment rates. Lancet Reg. Health-Eur. 2021, 10, 100227–200237. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.S.; Hulse, G.K. Epidemiological Associations of Various Substances and Multiple Cannabinoids with Autism in USA. Clin. Pediatr. Open Access 2019, 4, 1–20. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. Effect of Cannabis Legalization on US Autism Incidence and Medium Term Projections. Clin. Pediatr. Open Access 2019, 4, 1–17. [Google Scholar] [CrossRef]
- Reece, A.S.; Hulse, G.K. Gastroschisis and Autism-Dual Canaries in the Californian Coalmine. JAMA Surg. 2019, 154, 366–367. [Google Scholar] [CrossRef]
- Volkow, N.D.; Han, B.; Compton, W.M.; Blanco, C. Marijuana Use During Stages of Pregnancy in the United States. Ann. Intern. Med. 2017, 166, 763–764. [Google Scholar] [CrossRef]
- Volkow, N.D.; Compton, W.M.; Wargo, E.M. The Risks of Marijuana Use During Pregnancy. JAMA 2017, 317, 129–130. [Google Scholar] [CrossRef]
- Volkow, N.D.; Han, B.; Compton, W.M.; McCance-Katz, E.F. Self-reported Medical and Nonmedical Cannabis Use among Pregnant Women in the United States. JAMA 2019, 322, 167–169. [Google Scholar] [CrossRef]
- Hölzel, B.N.; Pfannkuche, K.; Allner, B.; Allner, H.T.; Hescheler, J.; Derichsweiler, D.; Hollert, H.; Schiwy, A.; Brendt, J.; Schaffeld, M.; et al. Following the adverse outcome pathway from micronucleus to cancer using H2B-eGFP transgenic healthy stem cells. Arch. Toxicol. 2020, 94, 3265–3280. [Google Scholar] [CrossRef] [PubMed]
- Russo, C.; Ferk, F.; Misik, M.; Ropek, N.; Nersesyan, A.; Mejri, D.; Holzmann, K.; Lavorgna, M.; Isidori, M.; Knasmuller, S. Low doses of widely consumed cannabinoids (cannabidiol and cannabidivarin) cause DNA damage and chromosomal aberrations in human-derived cells. Arch. Toxicol. 2018, 93, 179–188. [Google Scholar] [CrossRef]
- Tahir, S.K.; Trogadis, J.E.; Stevens, J.K.; Zimmerman, A.M. Cytoskeletal organization following cannabinoid treatment in undifferentiated and differentiated PC12 cells. Biochem. Cell Biol. 1992, 70, 1159–1173. [Google Scholar] [CrossRef] [PubMed]
- Vela, G.; Martin, S.; Garcia-Gil, L.; Crespo, J.A.; Ruiz-Gayo, M.; Fernandez-Ruiz, J.J.; Garcia-Lecumberri, C.; Pelaprat, D.; Fuentes, J.A.; Ramos, J.A.; et al. Maternal exposure to delta9-tetrahydrocannabinol facilitates morphine self-administration behavior and changes regional binding to central mu opioid receptors in adult offspring female rats. Brain Res. 1998, 807, 101–109. [Google Scholar] [CrossRef]
- Busch, F.W.; Seid, D.A.; Wei, E.T. Mutagenic activity of marihuana smoke condensates. Cancer Lett. 1979, 6, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Koller, V.J.; Ferk, F.; Al-Serori, H.; Misik, M.; Nersesyan, A.; Auwarter, V.; Grummt, T.; Knasmuller, S. Genotoxic properties of representatives of alkylindazoles and aminoalkyl-indoles which are consumed as synthetic cannabinoids. Food Chem. Toxicol. 2015, 80, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.M.; Raj, A.Y. Influence of cannabinoids on somatic cells in vivo. Pharmacology 1980, 21, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Shoyama, Y.; Sugawa, C.; Tanaka, H.; Morimoto, S. Cannabinoids act as necrosis-inducing factors in Cannabis sativa. Plant Signal Behav. 2008, 3, 1111–1112. [Google Scholar] [CrossRef]
- Price, P.J.; Suk, W.A.; Spahn, G.J.; Freeman, A.E. Transformation of Fischer rat embryo cells by the combined action of murine leukemia virus and (-)-trans-9-tetrahydrocannabinol. Proc. Soc. Exp. Biol. Med. 1972, 140, 454–456. [Google Scholar] [CrossRef]
- Fish, E.W.; Murdaugh, L.B.; Zhang, C.; Boschen, K.E.; Boa-Amponsem, O.; Mendoza-Romero, H.N.; Tarpley, M.; Chdid, L.; Mukhopadhyay, S.; Cole, G.J.; et al. Cannabinoids Exacerbate Alcohol Teratogenesis by a CB1-Hedgehog Interaction. Sci. Rep. 2019, 9, 16057. [Google Scholar] [CrossRef]
- Koller, V.J.; Auwarter, V.; Grummt, T.; Moosmann, B.; Misik, M.; Knasmuller, S. Investigation of the in vitro toxicological properties of the synthetic cannabimimetic drug CP-47,497-C8. Toxicol. Appl. Pharmacol. 2014, 277, 164–171. [Google Scholar] [CrossRef]
- Sarafian, T.A.; Habib, N.; Oldham, M.; Seeram, N.; Lee, R.P.; Lin, L.; Tashkin, D.P.; Roth, M.D. Inhaled marijuana smoke disrupts mitochondrial energetics in pulmonary epithelial cells in vivo. Am. J. Physiol. 2006, 290, L1202–L1209. [Google Scholar] [CrossRef]
- Sarafian, T.A.; Kouyoumjian, S.; Khoshaghideh, F.; Tashkin, D.P.; Roth, M.D. Delta 9-tetrahydrocannabinol disrupts mitochondrial function and cell energetics. Am. J. Physiol. 2003, 284, L298–L306. [Google Scholar]
- Morimoto, S.; Tanaka, Y.; Sasaki, K.; Tanaka, H.; Fukamizu, T.; Shoyama, Y.; Shoyama, Y.; Taura, F. Identification and characterization of cannabinoids that induce cell death through mitochondrial permeability transition in Cannabis leaf cells. J. Biol. Chem. 2007, 282, 20739–20751. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Hroudova, J.; Fisar, Z. Cannabinoid-Induced Changes in the Activity of Electron Transport Chain Complexes of Brain Mitochondria. J. Mol. Neurosci. 2015, 56, 926–931. [Google Scholar] [CrossRef] [PubMed]
- Tahir, S.K.; Zimmerman, A.M. Influence of marihuana on cellular structures and biochemical activities. Pharmacol. Biochem. Behav. 1991, 40, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Fisar, Z.; Singh, N.; Hroudova, J. Cannabinoid-induced changes in respiration of brain mitochondria. Toxicol. Lett. 2014, 231, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Canto, C.; Menzies, K.J.; Auwerx, J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab. 2015, 22, 31–53. [Google Scholar] [CrossRef] [PubMed]
- Scientists Are Baffled by Spatter of Babies Born without Hands or Arms in France, as Investigation Fails to Discover a Cause. Available online: https://www.dailymail.co.uk/news/article-7242059/Scientists-baffled-babies-born-without-hands-arms-France-probe-fails-discover-cause.html (accessed on 15 January 2022).
- Agence France-Presse in Paris: France to investigate cause of upper limb defects in babies. In The Guardian; The Guardian: London, UK, 2018.
- Willsher, K. Baby arm defects prompt nationwide investigation in France. In Guardian; The Guardian: London, UK, 2018. [Google Scholar]
- Carlson, B.M. Human Embryology and Developmental Biology, 6th ed.; Elsevier: Philadelphia, PA, USA, 2019; Volume 1. [Google Scholar]
- Eurocat Data: Prevalence Charts and Tables. Available online: https://eu-rd-platform.jrc.ec.europa.eu/eurocat/eurocat-data/prevalence_en (accessed on 10 January 2022).
- Global Health Observatory. Available online: https://www.who.int/data/gho/data/indicators/indicator-details/GHO/total-(recorded-unrecorded)-alcohol-per-capita-(15-)-consumption (accessed on 10 January 2022).
- European Monitoring Centre for Drugs and Drug Addiction (EMCDDA): Statistical Bulletin 2021—Prevalence of Drug Use. Available online: https://www.emcdda.europa.eu/data/stats2021/gps_en (accessed on 10 January 2022).
- The World Bank: Crude Data: Adjusted Net National Income per Capita (Current US$). Available online: https://data.worldbank.org/indicator/NY.ADJ.NNTY.PC.CD (accessed on 10 January 2022).
- R: A Language and Environment for Statistical Computing. Available online: https://cran.r-project.org/ (accessed on 15 January 2022).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; Francios, R.; Groelmund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686–1691. [Google Scholar] [CrossRef]
- Pebesma, E. Simple Features for R: Standardized Support for Spatial Vector Data. R J. 2018, 10, 439–446. [Google Scholar] [CrossRef]
- Viridis: Default Color Maps from ‘matplotlib’. Available online: https://CRAN.R-project.org/package=viridis (accessed on 15 January 2022).
- Colorplaner: Ggplot2 Extension to Visualize Two Variables per Color Aesthetic through Colorspace Projection. Available online: https://github.com/wmurphyrd/colorplaner (accessed on 15 January 2022).
- The Nlme Package: Linear and Nonlinear Mixed Effects Models, R Version 3. Available online: https://www.researchgate.net/publication/272475067_The_Nlme_Package_Linear_and_Nonlinear_Mixed_Effects_Models_R_Version_3 (accessed on 15 January 2022).
- Broom.Mixed: Tidying Methods for Mixed Models. Available online: http://github.com/bbolker/broom.mixed (accessed on 15 January 2022).
- Broom: Convert Statistical Objects into Tidy Tibbles. Available online: https://CRAN.R-project.org/package=broom (accessed on 15 January 2022).
- Leeper, T.J. (Ed.) Margins: Marginal Effects for Model Objects. In R Package Version 0.3.26; CRAN: Houston, TX, USA, 2021; Volume 1, pp. 1–36. [Google Scholar]
- Wright, M.N.; Ziegler, A. Ranger: A Fast Implementation of Random Forests for High Dimensional Data in C++ and R. J. Stat. Softw. 2017, 77, 1–17. [Google Scholar] [CrossRef]
- Greenwell, B.M.; Boehmke, B.C. Variable Importance Plots—An Introduction to the vip Package. R J. 2021, 12, 343–366. [Google Scholar] [CrossRef]
- Package ‘plm’. Available online: https://cran.r-project.org/web/packages/plm/plm.pdf (accessed on 15 January 2022).
- The spdep Package. Available online: https://www.rdocumentation.org/packages/spdep/versions/1.2-7 (accessed on 15 January 2022).
- Millo, G.; Piras, G. Splm: Spatial Panel Data Models in R. J. Stat. Softw. 2012, 47, 1–38. [Google Scholar] [CrossRef]
- Package ‘splm’. Available online: https://cran.r-project.org/web/packages/splm/splm.pdf (accessed on 15 January 2022).
- Croissant, Y.; Millo, G. Panel Data Econometrics with R; John Wiley and Sons: Oxford, UK, 2019; Volume 1. [Google Scholar]
- Wal, W.; Geskus, R. Ipw: An R Package for Inverse Probability Weighting. J. Stat. Softw. 2011, 43, 1–23. [Google Scholar] [CrossRef]
- VanderWeele, T.J.; Ding, P. Sensitivity Analysis in Observational Research: Introducing the E-Value. Ann. Intern. Med. 2017, 167, 268–274. [Google Scholar] [CrossRef] [PubMed]
- VanderWeele, T.J.; Martin, J.N.; Mathur, M.B. E-values and incidence density sampling. Epidemiology 2020, 31, e51–e52. [Google Scholar] [CrossRef] [PubMed]
- VanderWeele, T.J.; Mathur, M.B. Commentary: Developing best-practice guidelines for the reporting of E-values. Int. J. Epidemiol. 2020, 49, 1495–1497. [Google Scholar] [CrossRef] [PubMed]
- VanderWeele, T.J.; Ding, P.; Mathur, M. Technical Considerations in the Use of the E-Value. J. Causal Inference 2019, 7, 1–11. [Google Scholar] [CrossRef]
- Pearl, J.; Mackaenzie, D. The Book of Why: The New Science of Cause and Effect; Basic Books: New York, NY, USA, 2019; Volume 1. [Google Scholar]
- Package ‘Evalue’. Available online: https://cran.r-project.org/web/packages/EValue/EValue.pdf (accessed on 15 January 2022).
- McKowen, J.; Woodward, D.; Yule, A.M.; DiSalvo, M.; Rao, V.; Greenbaum, J.; Joshi, G.; Wilens, T.E. Characterizing autistic traits in treatment-seeking young adults with substance use disorders. Am. J. Addict. 2022, 31, 108–114. [Google Scholar] [CrossRef]
- Schrott, R.; Acharya, K.; Itchon-Ramos, N.; Hawkey, A.B.; Pippen, E.; Mitchell, J.T.; Kollins, S.H.; Levin, E.D.; Murphy, S.K. Cannabis use is associated with potentially heritable widespread changes in autism candidate gene DLGAP2 DNA methylation in sperm. Epigenetics 2019, 15, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.B. The Environment and Disease: Association or Causation? Proc. R. Soc. Med. 1965, 58, 295–300. [Google Scholar] [CrossRef]
- Vallee, A.; Lecarpentier, Y.; Guillevin, R.; Vallee, J.N. Effects of cannabidiol interactions with Wnt/beta-catenin pathway and PPARgamma on oxidative stress and neuroinflammation in Alzheimer’s disease. Acta Biochim. Biophys. Sin. 2017, 49, 853–866. [Google Scholar] [CrossRef]
- Nallathambi, R.; Mazuz, M.; Namdar, D.; Shik, M.; Namintzer, D.; Vinayaka, A.C.; Ion, A.; Faigenboim, A.; Nasser, A.; Laish, I.; et al. Identification of Synergistic Interaction Between Cannabis-Derived Compounds for Cytotoxic Activity in Colorectal Cancer Cell Lines and Colon Polyps That Induces Apoptosis-Related Cell Death and Distinct Gene Expression. Cannabis Cannabinoid Res. 2018, 3, 120–135. [Google Scholar] [CrossRef] [PubMed]
- Petko, J.; Tranchina, T.; Patel, G.; Levenson, R.; Justice-Bitner, S. Identifying novel members of the Wntless interactome through genetic and candidate gene approaches. Brain Res. Bull. 2018, 138, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Xian, X.; Tang, L.; Wu, C.; Huang, L. miR-23b-3p and miR-130a-5p affect cell growth, migration and invasion by targeting CB1R via the Wnt/beta-catenin signaling pathway in gastric carcinoma. OncoTargets Ther. 2018, 11, 7503–7512. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, M.G.; Cobbs, L.V.; Dummer, P.D.; Petros, T.J.; Halford, M.M.; Stacker, S.A.; Zou, Y.; Fishell, G.J.; Au, E. Non-canonical Wnt Signaling through Ryk Regulates the Generation of Somatostatin- and Parvalbumin-Expressing Cortical Interneurons. Neuron 2019, 103, 853–864.e854. [Google Scholar] [CrossRef] [PubMed]
- Nalli, Y.; Dar, M.S.; Bano, N.; Rasool, J.U.; Sarkar, A.R.; Banday, J.; Bhat, A.Q.; Rafia, B.; Vishwakarma, R.A.; Dar, M.J.; et al. Analyzing the role of cannabinoids as modulators of Wnt/beta-catenin signaling pathway for their use in the management of neuropathic pain. Bioorg. Med. Chem. Lett. 2019, 29, 1043–1046. [Google Scholar] [CrossRef]
- Birerdinc, A.; Jarrar, M.; Stotish, T.; Randhawa, M.; Baranova, A. Manipulating molecular switches in brown adipocytes and their precursors: A therapeutic potential. Prog. Lipid Res. 2013, 52, 51–61. [Google Scholar] [CrossRef]
- Richard, D.; Picard, F. Brown fat biology and thermogenesis. Front. Biosci. 2011, 16, 1233–1260. [Google Scholar] [CrossRef]
- Xu, T.R.; Yang, Y.; Ward, R.; Gao, L.; Liu, Y. Orexin receptors: Multi-functional therapeutic targets for sleeping disorders, eating disorders, drug addiction, cancers and other physiological disorders. Cell. Signal. 2013, 25, 2413–2423. [Google Scholar] [CrossRef]
- Fraher, D.; Ellis, M.K.; Morrison, S.; McGee, S.L.; Ward, A.C.; Walder, K.; Gibert, Y. Lipid Abundance in Zebrafish Embryos Is Regulated by Complementary Actions of the Endocannabinoid System and Retinoic Acid Pathway. Endocrinology 2015, 156, 3596–3609. [Google Scholar] [CrossRef]
- Kučukalić, S.; Ferić Bojić, E.; Babić, R.; Avdibegović, E.; Babić, D.; Agani, F.; Jakovljević, M.; Kučukalić, A.; Bravo Mehmedbašić, A.; Šabić Džananović, E.; et al. Genetic Susceptibility to Posttraumatic Stress Disorder: Analyses of the Oxytocin Receptor, Retinoic Acid Receptor-Related Orphan Receptor A and Cannabinoid Receptor 1 Genes. Psychiatr. Danub. 2019, 31, 219–226. [Google Scholar] [CrossRef]
- Lee YS, Jeong WI: Retinoic acids and hepatic stellate cells in liver disease. J. Gastroenterol. Hepatol. 2012, 27 (Suppl. S2), 75–79. [CrossRef] [PubMed]
- Aguado, T.; Romero, E.; Monory, K.; Palazuelos, J.; Sendtner, M.; Marsicano, G.; Lutz, B.; Guzmán, M.; Galve-Roperh, I. The CB1 cannabinoid receptor mediates excitotoxicity-induced neural progenitor proliferation and neurogenesis. J. Biol. Chem. 2007, 282, 23892–23898. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.J.; Walsh, F.S.; Doherty, P. The FGF receptor uses the endocannabinoid signaling system to couple to an axonal growth response. J. Cell Biol. 2003, 160, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Schrott, R.; Murphy, S.K.; Modliszewski, J.L.; King, D.E.; Hill, B.; Itchon-Ramos, N.; Raburn, D.; Price, T.; Levin, E.D.; Vandrey, R.; et al. Refraining from use diminishes cannabis-associated epigenetic changes in human sperm. Environ. Epigenet. 2021, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- GeneCards: GRIA1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GRIA1&keywords=GRIA1 (accessed on 1 June 2022).
- GeneCards: GRIK2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GRIK2&keywords=GRIK2 (accessed on 1 June 2022).
- GeneCards: GRIN. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GRIN2C&keywords=GRIN2c (accessed on 1 June 2022).
- GeneCards: GRID. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GRID2&keywords=GRID2 (accessed on 1 June 2022).
- GeneCards: GRM3. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GRM3&keywords=GRM3 (accessed on 1 June 2022).
- GeneCards: GRBR. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GABRE&keywords=GABR (accessed on 1 June 2022).
- GeneCards: GRBRA. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GABRA1&keywords=GABRA (accessed on 1 June 2022).
- GeneCards: GABRB. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GABRB1&keywords=GABRB (accessed on 1 June 2022).
- GeneCards: HTR. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=HTR2A&keywords=HTR2A (accessed on 1 June 2022).
- GeneCards: DRD. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DRD2&keywords=DRD2 (accessed on 1 June 2022).
- GeneCards: SLC6A3/DAT. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SLC6A3&keywords=SLC6A3 (accessed on 1 June 2022).
- GeneCards: ORPM1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=OPRM1&keywords=ORPM1 (accessed on 1 June 2022).
- GeneCards: OPRD1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=OPRD1&keywords=OPRD1 (accessed on 1 June 2022).
- GeneCards: OXTR. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=OXTR&keywords=OXTR (accessed on 1 June 2022).
- GeneCards: NRXN. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=NRXN1 (accessed on 1 June 2022).
- GeneCards: NLGN1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=NLGN1&keywords=NLGN1 (accessed on 1 June 2022).
- Radyushkin, K.; Hammerschmidt, K.; Boretius, S.; Varoqueaux, F.; El-Kordi, A.; Ronnenberg, A.; Winter, D.; Frahm, J.; Fischer, J.; Brose, N.; et al. Neuroligin-3-deficient mice: Model of a monogenic heritable form of autism with an olfactory deficit. Genes Brain Behav. 2009, 8, 416–425. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, L.; Chen, G. An unexpected role of neuroligin-2 in regulating KCC2 and GABA functional switch. Mol. Brain 2013, 6, 23. [Google Scholar] [CrossRef]
- Anderson, G.R.; Aoto, J.; Tabuchi, K.; Foldy, C.; Covy, J.; Yee, A.X.; Wu, D.; Lee, S.J.; Chen, L.; Malenka, R.C.; et al. Beta-Neurexins Control Neural Circuits by Regulating Synaptic Endocannabinoid Signaling. Cell 2015, 162, 593–606. [Google Scholar] [CrossRef]
- Hishimoto, A.; Liu, Q.R.; Drgon, T.; Pletnikova, O.; Walther, D.; Zhu, X.G.; Troncoso, J.C.; Uhl, G.R. Neurexin 3 polymorphisms are associated with alcohol dependence and altered expression of specific isoforms. Hum. Mol. Genet. 2007, 16, 2880–2891. [Google Scholar] [CrossRef]
- Wang, H. Endocannabinoid Mediates Excitatory Synaptic Function of β-Neurexins. Commentary: β-Neurexins Control Neural Circuits by Regulating Synaptic Endocannabinoid Signaling. Front. Neurosci. 2016, 10, 203. [Google Scholar] [CrossRef]
- Foldy, C.; Malenka, R.C.; Sudhof, T.C. Autism-associated neuroligin-3 mutations commonly disrupt tonic endocannabinoid signaling. Neuron 2013, 78, 498–509. [Google Scholar] [CrossRef]
- GeneCards: Down Syndrome Cell Adhesion Molecule. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DSCAM (accessed on 1 June 2022).
- Shibata, M.; Pattabiraman, K.; Lorente-Galdos, B.; Andrijevic, D.; Kim, S.K.; Kaur, N.; Muchnik, S.K.; Xing, X.; Santpere, G.; Sousa, A.M.M.; et al. Regulation of prefrontal patterning and connectivity by retinoic acid. Nature 2021, 598, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Pattabiraman, K.; Muchnik, S.K.; Kaur, N.; Morozov, Y.M.; Cheng, X.; Waxman, S.G.; Sestan, N. Hominini-specific regulation of CBLN2 increases prefrontal spinogenesis. Nature 2021, 598, 489–494. [Google Scholar] [CrossRef]
- Seigneur, E.; Südhof, T.C. Cerebellins are differentially expressed in selective subsets of neurons throughout the brain. J. Comp. Neurol. 2017, 525, 3286–3311. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, A.; Villalba, A.; de Juan Romero, C.; Pico, E.; Kyrousi, C.; Tzika, A.C.; Tessier-Lavigne, M.; Ma, L.; Drukker, M.; Cappello, S.; et al. Evolution of Cortical Neurogenesis in Amniotes Controlled by Robo Signaling Levels. Cell 2018, 174, 590–606.e521. [Google Scholar] [CrossRef]
- Yeh, M.L.; Gonda, Y.; Mommersteeg, M.T.; Barber, M.; Ypsilanti, A.R.; Hanashima, C.; Parnavelas, J.G.; Andrews, W.D. Robo1 modulates proliferation and neurogenesis in the developing neocortex. J. Neurosci. 2014, 34, 5717–5731. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.R.E.; Zhao, H.T.; Park, J.M.; Dipoppa, M.; Monsalve-Mercado, M.M.; Dahan, J.B.; Rodgers, C.C.; Lejeune, A.; Hillman, E.M.C.; Miller, K.D.; et al. A human-specific modifier of cortical connectivity and circuit function. Nature 2021, 599, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Alpar, A.; Tortoriello, G.; Calvigioni, D.; Niphakis, M.J.; Milenkovic, I.; Bakker, J.; Cameron, G.A.; Hanics, J.; Morris, C.V.; Fuzik, J.; et al. Endocannabinoids modulate cortical development by configuring Slit2/Robo1 signalling. Nat. Commun. 2014, 5, 4421. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Newton, C.; Perkins, I.; Friedman, H.; Klein, T.W. Cannabinoid treatment suppresses the T-helper cell-polarizing function of mouse dendritic cells stimulated with Legionella pneumophila infection. J. Pharmacol. Exp. Ther. 2006, 319, 269–276. [Google Scholar] [CrossRef]
- GeneCards: GLI3. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GLI3&keywords=GLI3 (accessed on 1 June 2022).
- Reece, A.S.; Hulse, G.K. Cannabinoid Genotoxicity and Congenital Anomalies: A Convergent Synthesis of European and USA Datasets. In Cannabis, Cannabinoids and Endocannabinoids; Preedy, V., Patel, V., Eds.; Elsevier: London, UK, 2022; Volume 1, in press. [Google Scholar]
- Reece, A.S.; Hulse, G.K. Chapter 3: Geospatiotemporal and Causal Inferential Analysis of United States Congenital Anomalies as a Function of Multiple Cannabinoid- and Substance-Exposures: Phenocopying Thalidomide and Hundred Megabase-Scale Genotoxicity. In Epidemiology of Cannabis: Genotoxicity and Neurotoxicity, Epigenomics and Aging; Elsevier: New York, NY, USA, 2023; Volume 1, in press. [Google Scholar]
- Reece, A.S.; Hulse, G.K. Effects of Cannabis on Congenital Limb Anomalies in 14 European Nations: A Geospatiotemporal and Causal Inferential Study. Environ. Epigenet. 2022, 8, 1–34. [Google Scholar] [CrossRef]


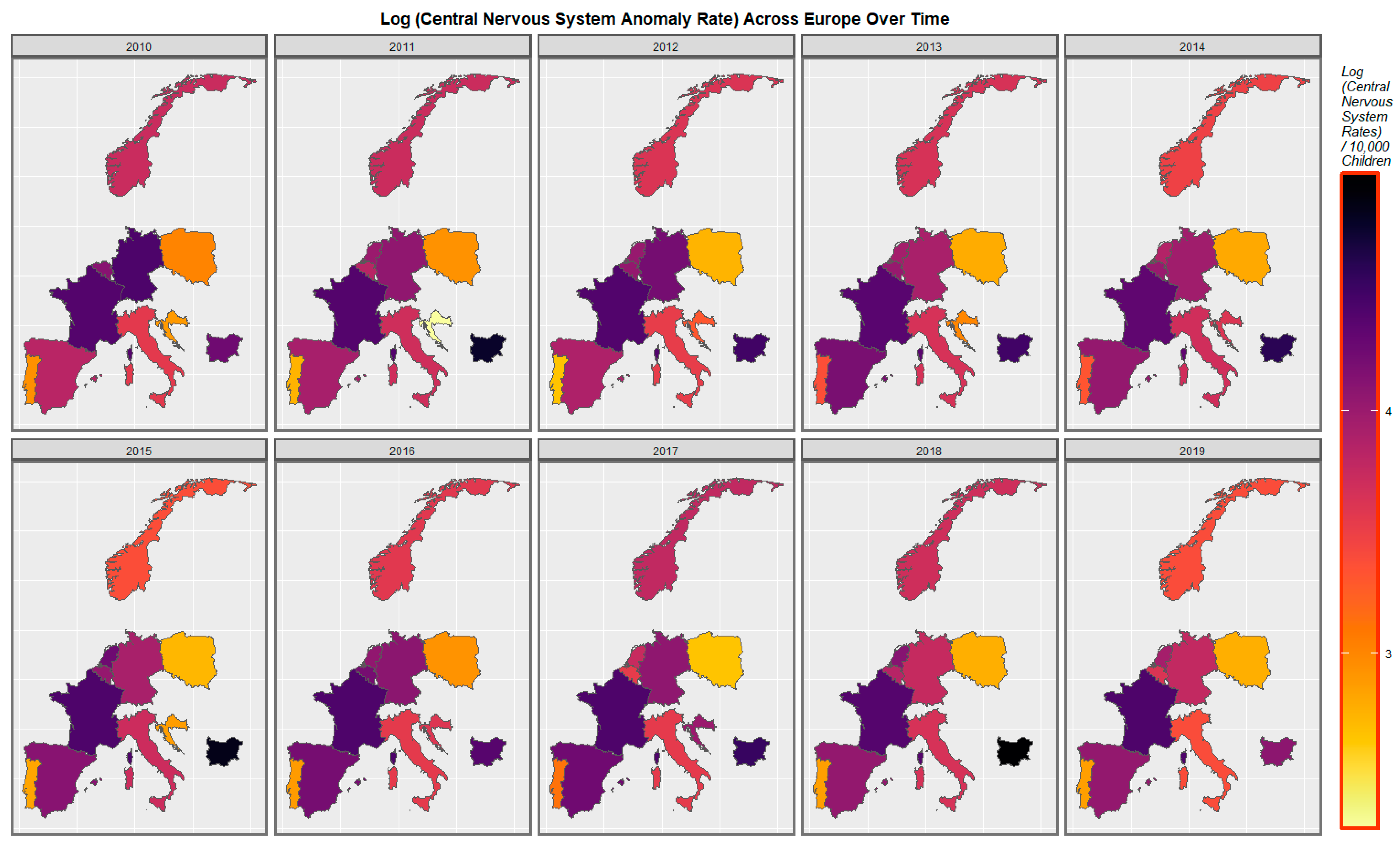
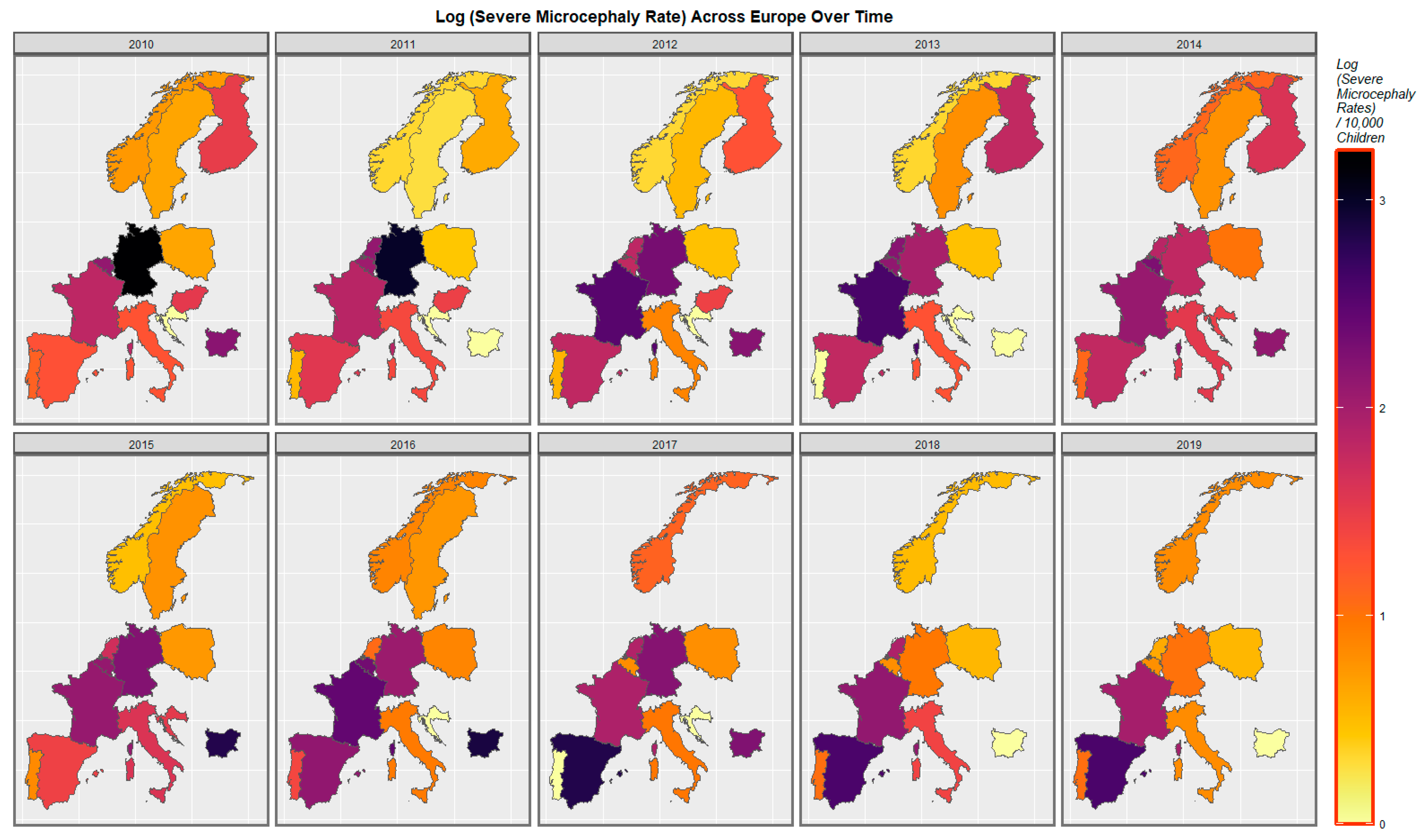

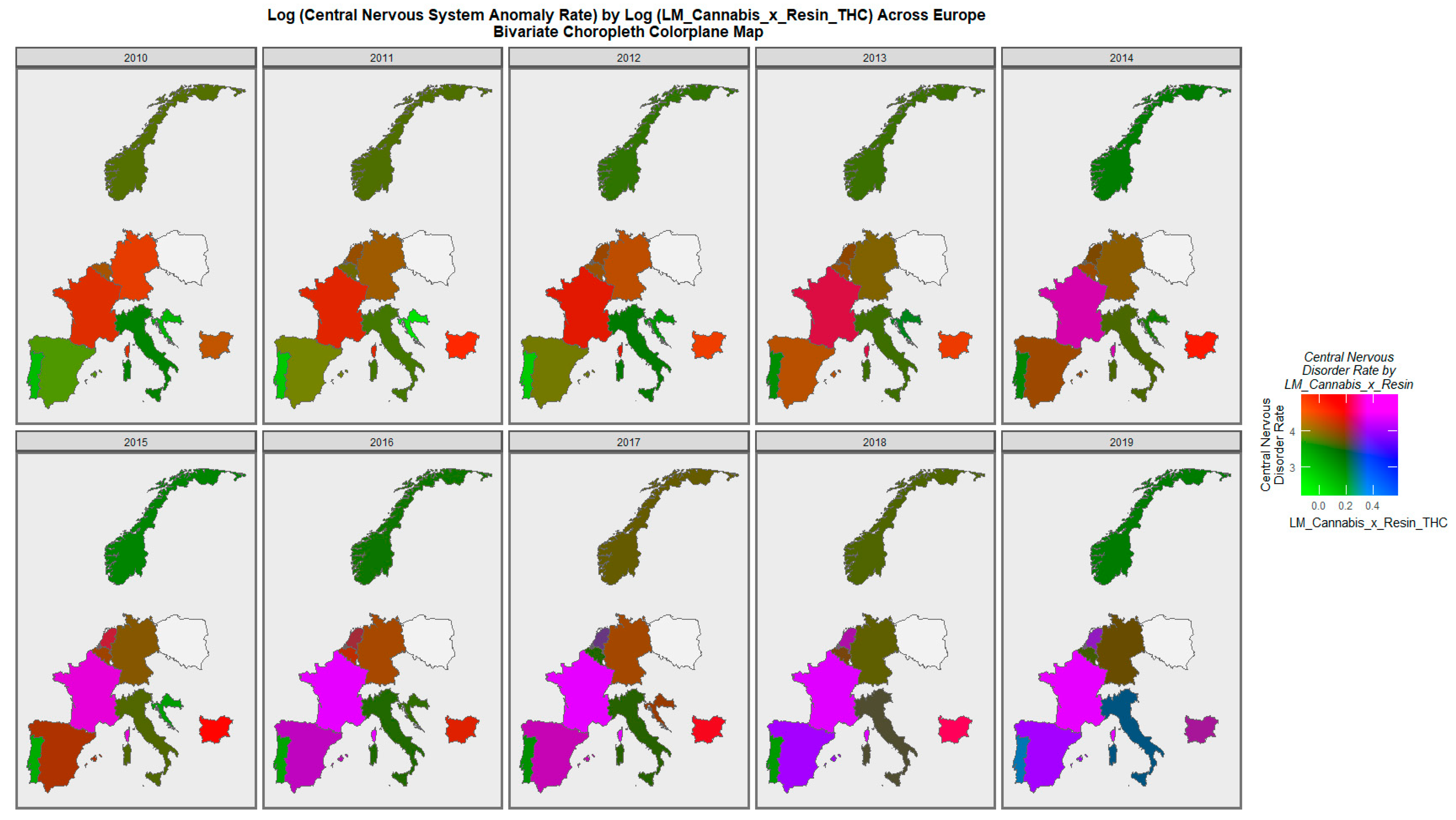

| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Resin.THC + Amphetamines + Cocaine + Income) | |||||
| Tobacco | 0.05 (0.02, 0.09) | 0.0010 | psi | 0.8032 | <2.2 × 10−16 |
| LM.Cannabis_x_Herb.THC | 5.23 (1.82, 8.64) | 0.0027 | rho | 0.62462 | 4.19 × 10−10 |
| Income | 0 (0, 0) | 0.0005 | lambda | −0.7094 | 8.38 × 10−15 |
| Interactive | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Herb.THC_x_Daily.Interpol. * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Herb.THC + Alcohol + Amphetamines + Cocaine + Income | |||||
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | −15.7 (−27.56, −3.84) | 0.0094 | psi | 0.79839 | <2.2 × 10−16 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 5.18 (0.18, 10.18) | 0.04239 | rho | 0.70837 | <2.2 × 10−16 |
| Alcohol | 0.12 (0.07, 0.18) | 4.14 × 10−5 | lambda | −0.72814 | <2.2 × 10−16 |
| Cocaine | 0.15 (0.02, 0.28) | 0.0261 | |||
| Income | 0 (0, 0) | 0.0011 | |||
| Tobacco: LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 0.68 (0.21, 1.16) | 0.0050 | |||
| Tobacco: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | −0.22 (−0.42, −0.02) | 0.0314 | |||
| 1 Lag | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. * LM.Cannabis_x_Herb.THC + Alcohol + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.06 (0.03, 0.1) | 0.0008 | psi | 0.8217 | <2.2 × 10−16 |
| Income | 0 (0, 0) | 0.0029 | rho | 0.6356 | 2.40 × 10−9 |
| lambda | −0.71467 | 1.39 × 10−13 | |||
| 2 Lags | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. * LM.Cannabis_x_Herb.THC + Alcohol + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.08 (0.04, 0.12) | 3.23 × 10−5 | psi | 0.75493 | <2.2 × 10−16 |
| Income | 0 (0, 0) | 0.0004 | rho | 0.41765 | 0.0521 |
| lambda | −0.4742 | 0.0225 | |||
| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Resin.THC + Amphetamines + Cocaine + Income) | |||||
| Tobacco | 0.06 (0.02, 0.11) | 0.0050 | psi | 0.68084 | <2.2 × 10−16 |
| Income | 0 (0, 0) | 0.0001 | rho | 0.65932 | 8.56 × 10−12 |
| lambda | −0.77182 | <2.2 × 10−16 | |||
| Interactive | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Resin.THC + Resin + Alcohol + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.09 (0.04, 0.13) | 0.0002 | psi | 0.5578 | 7.07 × 10−6 |
| LM.Cannabis_x_Resin.THC | 2.7 (0.66, 4.74) | 0.0092 | rho | −0.6788 | 9.48 × 10−8 |
| Income | 0 (0, 0) | 0.0002 | lambda | 0.5255 | 0.000365 |
| Tobacco: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | −0.07 (−0.15, 0) | 0.0466 | |||
| 1 Lag | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Resin + Daily.Interpol. + Alcohol + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.07 (0.03, 0.12) | 0.0004 | psi | 0.6289 | 2.34 × 10−14 |
| LM.Cannabis_x_Resin.THC | 5.5 (3.44, 7.56) | 1.53 × 10−7 | rho | 0.6326 | 1.57 × 10−8 |
| Income | 0 (0, 0) | 0.0005 | lambda | −0.635 | 1.69 × 10−8 |
| Tobacco: LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | −0.08 (−0.13, −0.02) | 0.0036 | |||
| 2 Lags | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC * Resin + Daily.Interpol. + Alcohol + Amphetamines + Cocaine + Income | |||||
| Resin | 3.48 (2.17, 4.79) | 1.97 × 10−7 | psi | 0.88545 | <2.2 × 10−16 |
| rho | 0.59616 | 2.10 × 10−6 | |||
| lambda | −0.6442 | 1.76 × 10−7 | |||
| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Resin.THC + Amphetamines + Cocaine + Income) | |||||
| Alcohol | 0.22 (0.14, 0.3) | 2.14 × 10−8 | psi | 0.32056 | 0.00128 |
| Daily.Interpol. | 37.8 (21.83, 53.77) | 3.65 × 10−6 | rho | 0.53742 | 1.16 × 10−5 |
| Amphetamines | −0.25 (−0.43, −0.06) | 0.01052 | lambda | −0.4586 | 0.000461 |
| Income | 0 (0, 0) | 0.00335 | |||
| Interactive | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Resin.THC + Herb + Alcohol + Herb + Amphetamines + Cocaine + Income | |||||
| Resin | 37.8 (21.83, 53.77) | 3.65 × 10−6 | psi | 0.32056 | 0.00128 |
| Alcohol | 0.22 (0.14, 0.3) | 2.14 × 10−8 | rho | 0.53742 | 1.17 × 10−5 |
| Amphetamines | −0.25 (−0.43, −0.06) | 0.01052 | lambda | −0.4586 | 0.000463 |
| Income | 0 (0, 0) | 0.00335 | |||
| 2 Lags | |||||
| Rate ~ Tobacco * LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Herb.THC + Alcohol + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.15 (0.09, 0.21) | 1.98 × 10−6 | psi | 0.238 | 0.0374 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 45.4 (17.96, 72.84) | 0.00117 | rho | −0.2833 | 0.2029 |
| Alcohol | 0.14 (0.06, 0.23) | 0.00132 | lambda | 0.2726 | 0.145 |
| Amphetamines | −0.36 (−0.58, −0.14) | 0.00149 | |||
| Income | 0 (0, 0) | 3.26 × 10−7 | |||
| Tobacco: LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | −1.68 (−2.76, −0.6) | 0.00234 | |||
| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Resin.THC + Amphetamines + Cocaine + Income) | |||||
| Tobacco | 0.03 (0.01, 0.06) | 0.01993 | psi | 0.1805 | 0.085341 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 1.32 (0.11, 2.53) | 0.03192 | rho | 0.5289 | 0.000136 |
| Income | 0 (0, 0) | 0.00654 | lambda | −0.51 | 0.000565 |
| Interactive | |||||
| Rate ~ Tobacco * Resin + Daily.Interpol. + Herb + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + Alcohol + Amphetamines + Cocaine + Income | |||||
| Resin | −6.17 (−9.17, −3.17) | 5.26 × 10−5 | rho | −0.5311 | 0.000340 |
| Cocaine | 0.23 (0.09, 0.36) | 0.00111 | lambda | 0.4738 | 0.000591 |
| Income | 0 (0, 0) | 0.00197 | |||
| Tobacco: Resin | 0.24 (0.13, 0.35) | 3.76 × 10−5 | |||
| 1 Lag | |||||
| Rate ~ Tobacco * Resin + Daily.Interpol. + Herb + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + Alcohol + Amphetamines + Cocaine + Income | |||||
| Resin | −11.7 (−16.25, −7.15) | 4.47 × 10−7 | psi | −0.03769 | 0.748 |
| LM.Cannabis_x_Herb.THC_x_Daily.Interpol. | 2.71 (1.02, 4.4) | 0.001741 | rho | 0.65087 | 2.06 × 10−11 |
| Alcohol | −0.12 (−0.18, −0.05) | 0.000918 | lambda | −0.64586 | 2.57 × 10−11 |
| Cocaine | −0.2 (−0.4, 0) | 0.047988 | |||
| Income | 0 (0, 0) | 1.55 × 10−6 | |||
| Tobacco: Resin | 0.41 (0.25, 0.57) | 5.18 × 10−7 | |||
| 2 Lags | |||||
| Rate ~ Tobacco * Resin + Daily.Interpol. + Herb + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + Alcohol + Amphetamines + Cocaine + Income | |||||
| Resin | −11.7 (−15.82, −7.58) | 2.73 × 10−8 | rho | 0.5539 | 3.46 × 10−5 |
| Alcohol | −0.1 (−0.17, −0.03) | 0.00318 | lambda | −0.6037 | 1.21 × 10−6 |
| Income | 0 (0, 0) | 9.17 × 10−7 | |||
| Tobacco: Resin | 0.45 (0.29, 0.61) | 2.13 × 10−8 | |||
| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Resin.THC + Amphetamines + Cocaine + Income) | |||||
| Tobacco | 0.07 (0.04, 0.11) | 1.26 × 10−4 | psi | 0.6859 | <2 × 10−16 |
| Herb | 4.26 (1.26, 7.26) | 0.0054 | rho | 0.5583 | 2.33 × 10−5 |
| Income | 0 (0, 0) | 0.0007 | lambda | −0.6312 | 1.70 × 10−7 |
| Interactive | |||||
| Rate ~ Tobacco + Daily.Interpol. * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Resin + Alcohol + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.08 (0.04, 0.12) | 2.27 × 10−5 | psi | 0.6508 | <2 × 10−16 |
| Income | 0 (0, 0) | 5.01 × 10−5 | rho | 0.5338 | 0.0003 |
| lambda | −0.6369 | 2.75 × 10−6 | |||
| 1 Lag | |||||
| Rate ~ Tobacco + Daily.Interpol. * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Alcohol + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.07 (0.03, 0.11) | 0.0002 | psi | 0.5946 | 8.78 × 10−12 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | −1.15 (−2.29, −0.01) | 0.0485 | rho | 0.4815 | 0.0063 |
| LM.Cannabis_x_Resin.THC | 3.59 (1.73, 5.45) | 0.0002 | lambda | −0.5681 | 0.0003 |
| Income | 0 (0, 0) | 0.0007 | |||
| 2 Lags | |||||
| Rate ~ Tobacco + Daily.Interpol. * LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Alcohol + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.1 (0.05, 0.15) | 3.49 × 10−5 | psi | 0.6627 | 1.03 × 10−14 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 2.28 (0.35, 4.21) | 0.0208 | rho | 0.5557 | 9.20 × 10−5 |
| LM.Cannabis_x_Resin.THC | −2.54 (−4.74, −0.34) | 0.0239 | lambda | −0.6187 | 6.85 × 10−6 |
| Income | 0 (0, 0) | 4.71 × 10−5 | |||
| Parameter Values | Model Parameters | ||||
|---|---|---|---|---|---|
| Parameter | Estimate (C.I.) | p-Value | Parameter | Value | Significance |
| Additive | |||||
| Rate ~ Tobacco + Alcohol + LM.Cannabis_x_Herb.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Daily.Interpol. + LM.Cannabis_x_Resin.THC + Amphetamines + Cocaine + Income) | |||||
| Tobacco | 0.08 (0.05, 0.11) | 2.21 × 10−8 | psi | 0.3329 | 0.0006 |
| Herb | 3.28 (1.18, 5.38) | 0.0021 | rho | −0.7510 | <2.2 × 10−16 |
| Amphetamines | 0.13 (0.02, 0.24) | 0.0201 | lambda | 0.5524 | 5.98 × 10−8 |
| Income | 0 (0, 0) | 2.44 × 10−5 | |||
| Interactive | |||||
| Rate ~ Tobacco + Herb * Resin + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + LM.Cannabis_x_Resin.THC + Alcohol + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.09 (0.07, 0.11) | 1.06 × 10−15 | rho | −0.7547 | |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | −0.42 (−0.75, −0.09) | 0.0131 | lambda | 0.4883 | <2.2 × 10−16 |
| Income | 0 (0, 0) | 5.11 × 10−11 | 4.70 × 10−6 | ||
| Herb: Resin | 17.1 (11.12, 23.08) | 1.92 × 10−8 | |||
| 1 Lag | |||||
| Rate ~ Tobacco + Herb * Resin + LM.Cannabis_x_Resin.THC + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Alcohol + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.09 (0.06, 0.11) | 5.79 × 10−15 | rho | −0.7561 | <2.2 × 10−16 |
| LM.Cannabis_x_Resin.THC | 4.01 (2.86, 5.16) | 9.76 × 10−12 | lambda | 0.4818 | 2.44 × 10−6 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | −2.15 (−2.95, −1.35) | 1.39 × 10−7 | |||
| Cocaine | −0.16 (−0.29, −0.02) | 0.0238 | |||
| Income | 0 (0, 0) | 7.63 × 10−15 | |||
| 2 Lags | |||||
| Rate ~ Tobacco + Herb * Resin + LM.Cannabis_x_Resin.THC + LM.Cannabis_x_Resin.THC_x_Daily.Interpol. + Alcohol + Daily.Interpol. + Amphetamines + Cocaine + Income | |||||
| Tobacco | 0.06 (0.03, 0.09) | 0.0002 | psi | 0.2973 | 0.00632 |
| Resin | −3.51 (−6.18, −0.84) | 0.0096 | rho | 0.6272 | 3.52 × 10−8 |
| LM.Cannabis_x_Resin.THC_x_Daily.Interpol. | 1.58 (0.37, 2.79) | 0.0102 | lambda | −0.6951 | 8.93 × 10−10 |
| Daily.Interpol. | −23.4 (−41.55, −5.25) | 0.0115 | |||
| Income | 0 (0, 0) | 0.0004 | |||
| Herb: Resin | 26.5 (3.96, 49.04) | 0.0209 | |||
| No. | E-Value Estimate | Lower Bound E-Value |
|---|---|---|
| 1 | 1.89 × 1035 | 1.04 × 1029 |
| 2 | 1.89 × 1035 | 1.04 × 1029 |
| 3 | 1.89 × 1035 | 1.04 × 1029 |
| 4 | 1.89 × 1035 | 1.04 × 1029 |
| 5 | 6.54 × 1034 | 3.32 × 1025 |
| 6 | 6.54 × 1034 | 3.32 × 1025 |
| 7 | 5.85 × 1029 | 9.39 × 1020 |
| 8 | 5.85 × 1029 | 9.39 × 1020 |
| 1 | 1.56 × 1022 | 2.92 × 109 |
| 9 | 1.56 × 1022 | 2.92 × 109 |
| 10 | 2.71 × 1017 | 1.45 × 106 |
| 11 | 2.71 × 1017 | 1.45 × 106 |
| 12 | 9.28 × 1011 | 1.11 × 106 |
| 13 | 9.28 × 1011 | 1.11 × 106 |
| 14 | 1.82 × 107 | 7.92 × 104 |
| 15 | 1.82 × 107 | 7.92 × 104 |
| 16 | 5.31 × 105 | 2.43 × 104 |
| 17 | 5.31 × 105 | 2.43 × 104 |
| 18 | 4.84 × 105 | 868.15 |
| 19 | 4.84 × 105 | 868.15 |
| 20 | 1.35 × 104 | 455.47 |
| 21 | 1.35 × 104 | 455.47 |
| 22 | 4.98 × 103 | 81.79 |
| 23 | 4.98 × 103 | 81.79 |
| 24 | 727.65 | 44.80 |
| 25 | 727.65 | 44.80 |
| 26 | 727.65 | 38.03 |
| 27 | 727.65 | 38.03 |
| 28 | 272.41 | 28.95 |
| 29 | 272.41 | 28.95 |
| 30 | 184.48 | 28.95 |
| 31 | 184.48 | 28.95 |
| 32 | 49.08 | 22.02 |
| 33 | 49.08 | 22.02 |
| 34 | 49.08 | 22.02 |
| 35 | 49.08 | 22.02 |
| 36 | 35.92 | 8.79 |
| 37 | 35.92 | 8.79 |
| 38 | 35.92 | 8.79 |
| 39 | 35.92 | 8.79 |
| 40 | 23.23 | 1.64 |
| 41 | 23.23 | 1.64 |
| 42 | 23.23 | 1.60 |
| 43 | 23.23 | 1.60 |
| 44 | 2.17 | 1.60 |
| 45 | 2.17 | 1.60 |
| 46 | 2.11 | 1.44 |
| 47 | 2.11 | 1.44 |
| 48 | 1.81 | 1.19 |
| 49 | 1.81 | 1.19 |
| Anomaly | Number | Mean Minimum E-Value | Median Minimum E-Value | Min Minimum E-Value | Max Minimum E-Value | Mean E-Value Estimate | Median E-Value Estimate | Min E-Value Estimate | Max E-Value Estimate |
|---|---|---|---|---|---|---|---|---|---|
| Severe Microcephalus | 6 | 6.93 × 1028 | 1.04 × 1029 | 1.11 × 106 | 1.04 × 1029 | 1.26 × 1035 | 1.89 × 1035 | 1.82 × 107 | 1.89 × 1035 |
| Microphthalmos | 8 | 8.30 × 1024 | 4.70 × 1020 | 1.44 | 3.32 × 1025 | 1.64 × 1034 | 2.93 × 1029 | 2.11 | 6.54 × 1034 |
| Nervous | 10 | 5.84 × 108 | 7.92 × 104 | 38.03 | 2.92 × 109 | 3.12 × 1021 | 9.28 × 1011 | 272.41 | 1.56 × 1022 |
| Neural Tube Defects | 8 | 231.81 | 28.95 | 1.19 | 868.15 | 1269.9925 | 49.08 | 1.81 | 4980.00 |
| Anencephalus | 6 | 41.94 | 22.02 | 22.02 | 81.79 | 85.44 | 35.92 | 35.92 | 184.48 |
| Spina Bifida | 12 | 86.84 | 8.79 | 1.6 | 455.47 | 9.10 × 104 | 727.65 | 23.23 | 5.31 × 105 |
| Nearest Gene Name | Number of Annotations | Page | Chromosome | Status | Nearest Gene | Distance to Nearest Gene | Relative Location | p-Value | P-Adjust |
|---|---|---|---|---|---|---|---|---|---|
| Glutamate/GABA Receptors | |||||||||
| GRIA | 132 | 8 | 11 | Dependence | ENSG00000152578 | 0 | Intron | 1.96 × 10−8 | 0.000888 |
| GRIK | 165 | 24 | 1 | Dependence | ENSG00000163873 | 0 | Intron | 5.18 × 10−7 | 0.005036 |
| GRIN | 26 | 14 | 16 | Dependence | ENSG00000183454 | 0 | Intron | 1.18 × 10−7 | 0.002371 |
| GRID | 11 | 7 | 4 | Dependence | ENSG00000152208 | 0 | Intron | 1.10 × 10−8 | 0.000694 |
| GRM | 122 | 10 | 3 | Dependence | ENSG00000196277 | 0 | Intron | 4.74 × 10−8 | 0.001439 |
| GABR | 143 | 18 | 4 | Dependence | ENSG00000151834 | 0 | Intron | 2.72 × 10−7 | 0.003664 |
| GABRA | 142 | 52 | 4 | Dependence | ENSG00000151834 | 0 | Intron | 2.74 × 10−6 | 0.011395 |
| GABRB | 22 | 18 | 4 | Dependence | ENSG00000163288 | 0 | Intron | 2.72 × 10−7 | 0.003664 |
| Other Receptors | |||||||||
| HTR | 85 | 36 | X | Dependence | ENSG00000147246 | 0 | Intron | 1.31 × 10−6 | 0.007988 |
| DRD | 17 | 33 | 5 | Dependence | ENSG00000184845 | 0 | Intron | 1.06 × 10−6 | 0.007226 |
| DAT | 52 | 214 | 4 | Withdrawal | ENSG00000109576 | 0 | Intron | 1.18 × 10−5 | 0.024539 |
| MOR | 379 | 68 | X | Dependence | ENSG00000231154 | 0 | Intron | 4.87 × 10−6 | 0.015091 |
| DOR/KOR | 0 | - | - | - | - | - | - | - | - |
| OXTR | 7 | 55 | 3 | Dependence | ENSG00000180914 | 0 | Intron | 3.10 × 10−6 | 0.012043 |
| Synaptic Scaffolding | |||||||||
| NXN | 10 | 32 | 1 | Dependence | ENSG00000226693 | 0 | Intron | 1.03 × 10−6 | 0.007159 |
| NLG | 10 | 48 | 3 | Dependence | ENSG00000169760 | 0 | Intron | 2.35 × 10−6 | 0.010632 |
| Nearest Gene Name | Page | Chromosome | Status | Nearest Gene | Distance to Nearest Gene | Relative Location | p-Value | p-Adjust |
| ROBO1 | ||||||||
| ROBO1 | 22 | 3 | Dependence | ENCG00000169855 | 0 | Intron | 4.08 × 10−7 | 0.004487 |
| ROBO1 | 34 | 3 | Dependence | ENCG00000169855 | 0 | Intron | 1.10 × 10−6 | 0.007347 |
| ROBO1 | 179 | 3 | Withdrawal | ENCG00000169855 | 0 | Intron | 4.75 × 10−6 | 0.016077 |
| ROBO1 | 75 | 3 | Dependence | ENCG00000169855 | 0 | Intron | 5.83 × 10−6 | 0.016430 |
| ROBO1 | 82 | 3 | Dependence | ENCG00000169855 | 1176 | Downstream | 7.09 × 10−6 | 0.017986 |
| ROBO1 | 103 | 3 | Dependence | ENCG00000169855 | 0 | Intron | 1.10 × 10−5 | 0.022061 |
| ROBO1 | 118 | 3 | Dependence | ENCG00000169855 | 90264 | Upstream | 1.46 × 10−5 | 0.025364 |
| ROBO2 | ||||||||
| ROBO2 | 8 | 3 | Dependence | ENCG00000185008 | 0 | Intron | 1.72 × 10−8 | 0.000855 |
| ROBO2 | 17 | 3 | Dependence | ENCG00000185008 | 0 | Intron | 2.30 × 10−7 | 0.003353 |
| ROBO2 | 139 | 3 | Withdrawal | ENCG00000185008 | 0 | Intron | 4.68 × 10−7 | 0.005298 |
| ROBO2 | 149 | 3 | Withdrawal | ENCG00000185008 | 0 | Intron | 1.20 × 10−6 | 0.008443 |
| ROBO2 | 167 | 3 | Withdrawal | ENCG00000185008 | 0 | Intron | 3.15 × 10−6 | 0.013257 |
| ROBO2 | 66 | 3 | Dependence | ENCG00000185008 | 0 | Intron | 4.52 × 10−6 | 0.014551 |
| ROBO2 | 200 | 3 | Withdrawal | ENCG00000185008 | 0 | Intron | 8.80 × 10−6 | 0.021641 |
| ROBO2 | 204 | 3 | Withdrawal | ENCG00000185008 | 0 | Intron | 9.48 × 10−6 | 0.022315 |
| ROBO2 | 106 | 3 | Dependence | ENCG00000185008 | 0 | Intron | 1.16 × 10−5 | 0.022679 |
| ROBO2 | 223 | 3 | Withdrawal | ENCG00000185008 | 0 | Intron | 1.40 × 10−5 | 0.026641 |
| ROBO2 | 149 | 3 | Withdrawal | ENCG00000185008 | 0 | Intron | 1.20 × 10−6 | 0.084427 |
| ROBO4 | ||||||||
| ROBO4 | 230 | 11 | Withdrawal | ENCG00000154133 | 0 | Intron | 1.59 × 10−5 | 0.028295 |
| Nearest Gene Name | Page | Chromosome | Status | Nearest Gene | Fold Enrichment | p-Value | P-Bonferroni | P-FDR |
| ROBO1 | 237 | 3 | Dependence | ENCG00000169855 | 1.7082 | 1.12 × 10−2 | 0.9504 | 0.001219 |
| ROBO2 | 237 | 3 | Dependence | ENCG00000185008 | 1.7082 | 1.12 × 10−2 | 0.9504 | 0.001219 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reece, A.S.; Hulse, G.K. European Epidemiological Patterns of Cannabis- and Substance-Related Congenital Neurological Anomalies: Geospatiotemporal and Causal Inferential Study. Int. J. Environ. Res. Public Health 2023, 20, 441. https://doi.org/10.3390/ijerph20010441
Reece AS, Hulse GK. European Epidemiological Patterns of Cannabis- and Substance-Related Congenital Neurological Anomalies: Geospatiotemporal and Causal Inferential Study. International Journal of Environmental Research and Public Health. 2023; 20(1):441. https://doi.org/10.3390/ijerph20010441
Chicago/Turabian StyleReece, Albert Stuart, and Gary Kenneth Hulse. 2023. "European Epidemiological Patterns of Cannabis- and Substance-Related Congenital Neurological Anomalies: Geospatiotemporal and Causal Inferential Study" International Journal of Environmental Research and Public Health 20, no. 1: 441. https://doi.org/10.3390/ijerph20010441
APA StyleReece, A. S., & Hulse, G. K. (2023). European Epidemiological Patterns of Cannabis- and Substance-Related Congenital Neurological Anomalies: Geospatiotemporal and Causal Inferential Study. International Journal of Environmental Research and Public Health, 20(1), 441. https://doi.org/10.3390/ijerph20010441







