Investigation of Antimicrobial Resistance Genes in Listeria monocytogenes from 2010 through to 2021
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antimicrobial Resistance Data from the NCBI Pathogen Detection Isolate Browser
- Organism group = Listeria monocytogenes;
- Collection date = from: 31 December 2009, to: 31 December 2021.
2.2. Principal Component Analysis and Hierarchical Clustering
3. Results
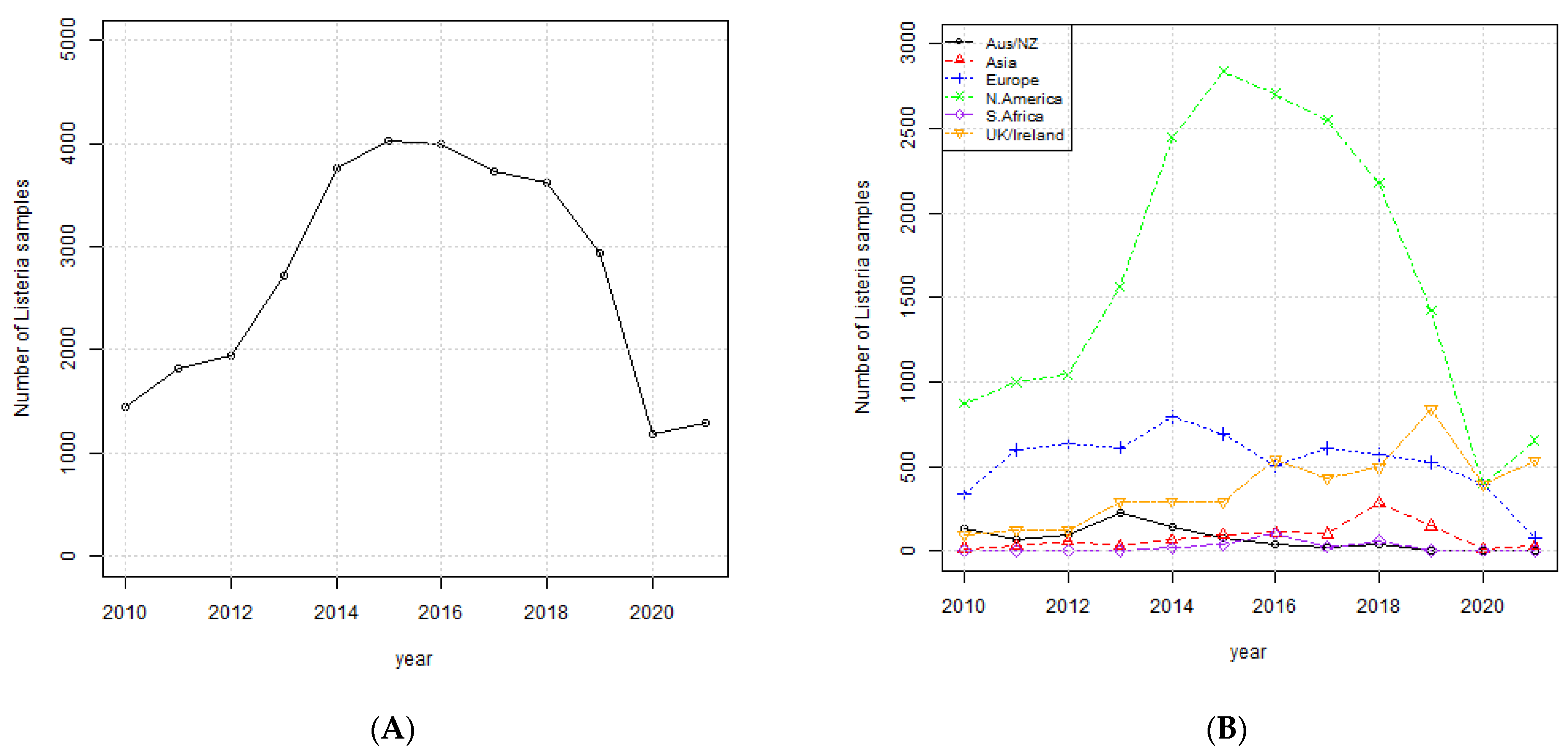
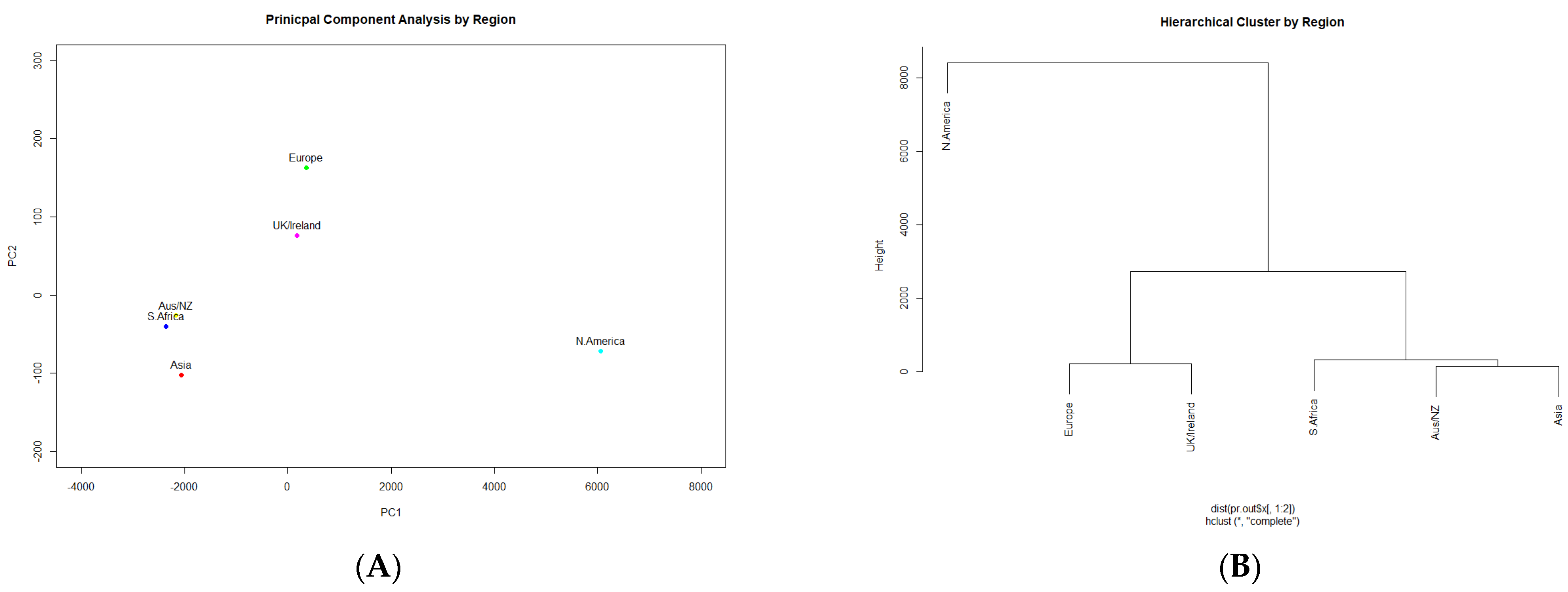
3.1. Occurrence of Listeria monocytogenes
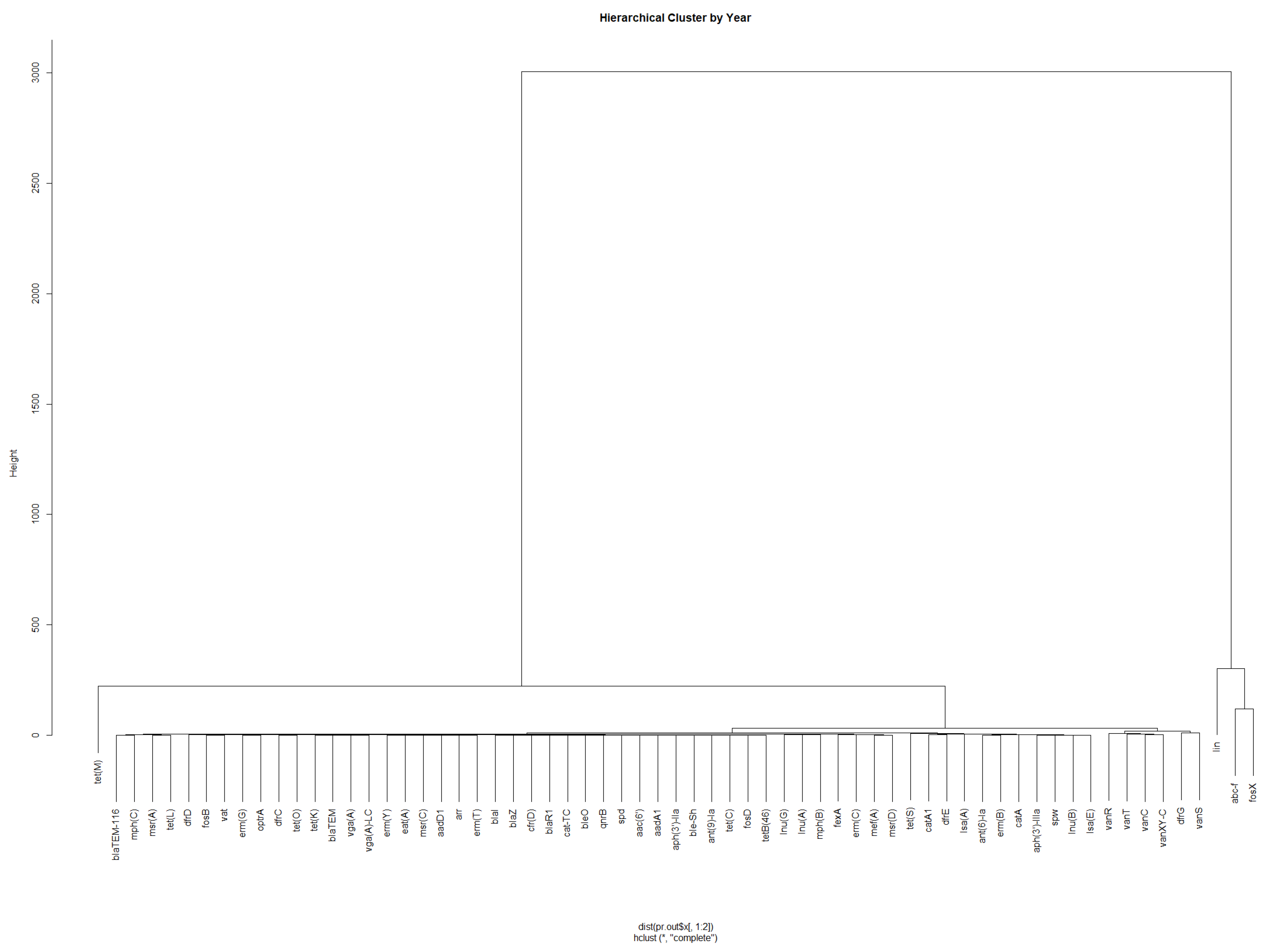
3.2. Presence of Antimicrobial Resistance Genes
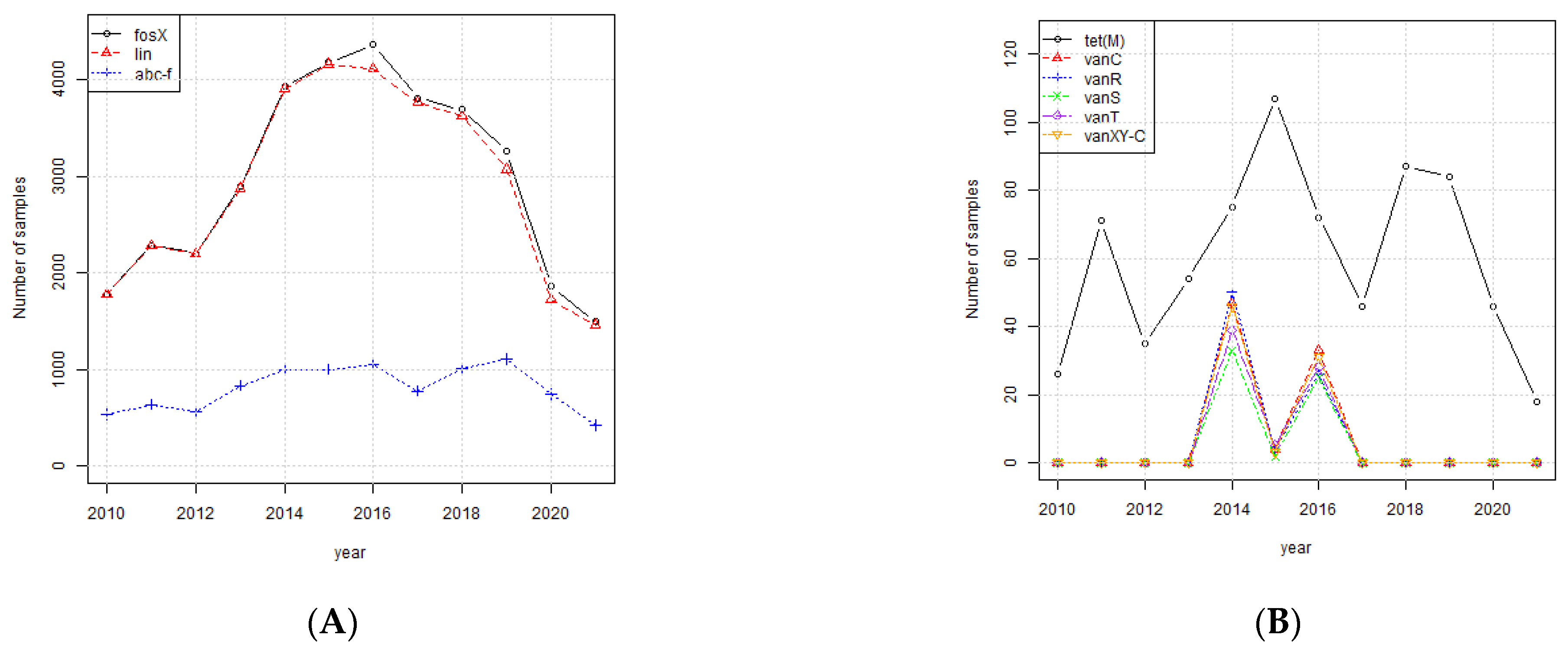
3.3. Investigation of Highly Occurring AMR Genes
3.4. The Biological Functions of the Highly Occurring Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 12, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial resistance in bacteria: Mechanisms, evolution, and persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Mühlberg, E.; Umstätter, F.; Kleist, C.; Domhan, C.; Mier, W.; Uhl, P. Renaissance of vancomycin: Approaches for breaking antibiotic resistance in multidrug-resistant bacteria. Can. J. Microbiol. 2020, 66, 11–16. [Google Scholar] [CrossRef] [Green Version]
- Bowler, J. The 3rd Leading Global Cause of Death Is Likely Not What You Think, New Study Reveals. Available online: https://www.sciencealert.com/the-third-leading-cause-of-death-globally-in-2019-was-antibiotic-resistant-bacterial-infection (accessed on 24 February 2022).
- CDC. Antibiotic Resistance Threats in the United States; U.S Department of Health and Human Services: Washington, DC, USA, 2019; pp. 1–113. [CrossRef] [Green Version]
- Friedrich, A.W. Control of hospital acquired infections and antimicrobial resistance in Europe: The way to go. Wien. Med. Wochenschr. 2019, 169, 25–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FDA. Common Foodborne Disease Causes. Available online: https://www.fda.gov/files/food/published/Most-Common-Foodborne-Illnesses-%28PDF%29.pdf (accessed on 13 February 2022).
- CDC. National Enteric Disease Surveillance: The Listeria Initiative; National Center for Emerging and Zoonotic Infectious Diseases: Atlanta, GA, USA, 2016; pp. 1–2. [Google Scholar]
- Lotfollahi, L. Prevalence and antimicrobial resistance profiles of Listeria monocytogenes in spontaneous abortions in humans. Afr. J. Microbiol. Res. 2011, 5, 1990–1993. [Google Scholar] [CrossRef]
- Hurley, D.; Luque-Sastre, L.; Parker, C.T.; Huynh, S.; Eshwar, A.K.; Nguyen, S.V.; Andrews, N.; Moura, A.; Fox, E.M.; Jordan, K.; et al. Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. Appl. Environ. Sci. 2019, 4, e00252-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Wang, A.; Fu, M.; Wang, A.; Chen, K.; Jia, Q.; Huang, Z. Investigation of incidents and trends of antimicrobial resistance in foodborne pathogens in eight countries from historical sample data. Int. J. Environ. Res. Public Health 2020, 17, 472. [Google Scholar] [CrossRef] [Green Version]
- Abd Al-Mayahi, F.S.; Jaber, S.M. Multiple drug resistance of Listeria monocytogenes isolated from aborted women by using serological and molecular techniques in Diwaniyah city/Iraq. Iran. J. Microbiol. 2020, 12, 305–312. [Google Scholar] [CrossRef]
- Charpentier, E.; Courvalin, P. Antibiotic resistance in Listeria spp. Antimicrob. Agents Chem. 1999, 43, 2103–2108. [Google Scholar] [CrossRef] [Green Version]
- Morvan, A.; Moubareck, C.; Leclercq, A.; Hervé-Bazin, M.; Bremont, S.; Lecuit, M.; Courvalin, P.; Le Monnier, A. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob. Agents Chem. 2010, 54, 2728–2731. [Google Scholar] [CrossRef] [Green Version]
- Walsh, D.; Duffy, G.; Sheridan, J.J.; Blair, I.S.; McDowell, D.A. Antibiotic resistance among Listeria, including Listeria monocytogenes, in retail foods. J. Appl. Microbiol. 2001, 90, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.; Fraccalvieri, R.; Pasquali, F.; Santagada, G.; Latorre, L.M.; Difato, L.M.; Miccolupo, A.; Normanno, G.; Parisi, A. Antimicrobial susceptibility and multilocus sequence typing of Listeria monocytogenes isolated over 11 years from food, humans, and the environment in Italy. Foodborne Pathog. Dis. 2020, 17, 284–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gómez, D.; Azón, E.; Marco, N.; Carramiñana, J.J.; Rota, C.; Ariño, A.; Yangüela, J. Antimicrobial resistance of Listeria monocytogenes and Listeria innocua from meat products and meat-processing environment. Food Microbiol. 2014, 42, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Mota, M.I.; Vázquez, S.; Cornejo, C.; D’Alessandro, B.; Braga, V.; Caetano, A.; Betancor, L.; Varela, G. Does shiga toxin-producing Escherichia coli and Listeria monocytogenes contribute significantly to the burden of antimicrobial resistance in Uruguay? Front. Vet. Sci. 2020, 7, 903. [Google Scholar] [CrossRef] [PubMed]
- Hailu, W.; Helmy, Y.A.; Carney-Knisely, G.; Kauffman, M.; Fraga, D.; Rajashekara, G. Prevalence and antimicrobial resistance profiles of foodborne pathogens isolated from dairy cattle and poultry manure amended farms in northeastern Ohio, the United States. Antibiotics 2021, 10, 1450. [Google Scholar] [CrossRef] [PubMed]
- National Database of Antibiotic Resistant Organisms. Available online: https://www.ncbi.nlm.nih.gov/pathogens/antimicrobial-resistance/ (accessed on 13 February 2022).
- Pathogen Detection. Available online: https://www.ncbi.nlm.nih.gov/pathogens/ (accessed on 16 January 2022).
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Jollife, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Stogios, P.J. R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 20 February 2022).
- Sharkey, L.K.R.; Edwards, T.A.; O’Neill, A.J. ABC-F proteins mediate antibiotic resistance through ribosomal protection. mBio 2016, 7, e01975-15. [Google Scholar] [CrossRef] [Green Version]
- Haubert, L.; Mendonça, M.; Lopes, G.V.; de Itapema Cardoso, M.R.; da Silva, W.P. Listeria monocytogenes isolates from food and food environment harbouring tetM and ermB resistance genes. Lett. Appl. Microbiol. 2016, 62, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Brenciani, A.; Bacciaglia, A.; Vecchi, M.; Vitali, L.A.; Varaldo, P.E.; Giovanetti, E. Genetic elements carrying erm(B) in Streptococcus pyogenes and association with tet(M) tetracycline resistance gene. Antimicrob. Agents Chem. 2007, 51, 1209–1216. [Google Scholar] [CrossRef] [Green Version]
- Hua, M.; Huang, W.; Chen, A.; Rehmet, M.; Jin, C.; Huang, Z. Comparison of antimicrobial resistance detected in environmental and clinical isolates from historical data for the US. BioMed Res. Int. 2020, 51, 1209–1216. [Google Scholar] [CrossRef] [PubMed]
- Almahdawy, O.; Shalal, O.S.; Najee, H. Description of vancomycin resistance genes in enterococcus sp. clinical strains isolated from Bucharest, Romania. Rom. Biotechnol. Lett. 2019, 24, 395–399. [Google Scholar] [CrossRef]
- Evers, S.; Courvalin, P. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in enterococcus faecalis V583. J. Bacteriol. 1996, 178, 1302–1309. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, P.E.; Arias, C.A.; Courvalin, P. Gene vanXY C encodes D,D-dipeptidase (VanX) and D,D-carboxypeptidase (VanY) activities in vancomycin-resistant Enterococcus gallinarum BM4174. Mol. Microbiol. 2002, 34, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Stogios, P.J.; Savchenko, A. Molecular mechanisms of vancomycin resistance. Protein Sci. 2020, 29, 654–669. [Google Scholar] [CrossRef]
- Gómez-Sanz, E.; Haro-Moreno, J.M.; Jensen, S.O.; Roda-García, J.J.; López-Pérez, M. The resistome and mobilome of multidrug-resistant Staphylococcus sciuri C2865 unveil a transferable trimethoprim resistance gene, designated dfrE, spread unnoticed. mSystems 2021, 6, e00511-21. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chem. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Zhu, X.Q.; Wang, X.M.; Li, H.; Shang, Y.H.; Pan, Y.S.; Wu, C.M.; Wang, Y.; Du, X.D.; Shen, J.Z. Novel lnu(G) gene conferring resistance to lincomycin by nucleotidylation, located on Tn6260 from enterococcus faecalis E531. J. Antimicrob. Chem. 2017, 72, 993–997. [Google Scholar] [CrossRef] [Green Version]
- Bokaeian, M.; Zahedani, S.S.; Bajgiran, M.S.; Moghaddam, A.A. Frequency of PER, VEB, SHV, TEM and CTX-M genes in resistant strains of Pseudomonas aeruginosa producing extended spectrum β-Lactamases. Jundishapur J. Microbiol. 2015, 8, 13783. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Fuentes, M.A.; Abriouel, H.; Ortega Morente, E.; Pérez Pulido, R.; Gálvez, A. Genetic determinants of antimicrobial resistance in gram positive bacteria from organic foods. Int. J. Food Microbiol. 2014, 172, 49–56. [Google Scholar] [CrossRef]
- Nair, S.; Ashton, P.; Doumith, M.; Connell, S.; Painset, A.; Mwaigwisya, S.; Langridge, G.; De Pinna, E.; Godbole, G.; Day, M. WGS for surveillance of antimicrobial resistance: A pilot study to detect the prevalence and mechanism of resistance to azithromycin in a UK population of non-typhoidal Salmonella. J. Antimicrob. Chem. 2016, 71, 3400–3408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- FoodNet Fast. Available online: https://wwwn.cdc.gov/foodnetfast/ (accessed on 13 February 2022).
- National Outbreak Reporting System. Available online: https://wwwn.cdc.gov/norsdashboard/ (accessed on 13 February 2022).
- Scortti, M.; Han, L.; Alvarez, S.; Leclercq, A.; Moura, A.; Lecuit, M.; Vazquez-Boland, J. Epistatic control of intrinsic resistance by virulence genes in Listeria. PLoS Genet. 2018, 14, e1007525. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Category | Description |
|---|---|
| Scientific name | Listeria monocytogenes |
| Collection date | Date the sample was collected |
| Location | Location from which the sample was collected |
| Isolation type | Clinical or environmental/other |
| Serovar | Serovar |
| AMR genotype | List of the AMR genotypes identified in sample |
| Category | Abbreviated Name | Entries | Comments |
|---|---|---|---|
| Scientific name | Sci_name | 1 = Listeria monocytogenes | |
| Collection date | Year | 2010 through to 2021 | |
| Location | Region | 1 = Australia/New Zealand 2 = Asia 3 = Europe 4 = North America 5 = South Africa 6 = United Kingdom/Ireland | |
| Isolation type | Epi_type | 1 = clinical 2 = environmental | |
| Serovar | Serovar | 1 = 1/2a 2 = 1/2b 3 = 4b | |
| AMR gene | Gene name, e.g., fosX | 0 = not found in sample 1 = found in sample | There is 1 column for each gene |
| Aus/NZ | Asia | Europe | N. America | S. Africa | UK/Ireland |
|---|---|---|---|---|---|
| fosX | fosX | fosX | fosX | fosX | fosX |
| abc-f | lin | abc-f | abc-f | lin | abc-f |
| lin | abc-f | lin | lin | abc-f | lin |
| erm(G) | tet(M) | tet(M) | tet(M) | fexA | tet(M) |
| tet(M) | dfrG | vanC | tet(M) | ||
| tet(S) | vanR | dfrG | |||
| ant(6)-Ia | vanXY-C | tet(S) | |||
| erm(B) | vanT | ||||
| aph(3′)-IIIa | vanS | ||||
| lnu(B) | tet(S) | ||||
| spw | catA1 | ||||
| catA | |||||
| catA1 | |||||
| mef(A) | |||||
| msr(D) | |||||
| erm(C) | |||||
| fexA | |||||
| lnu(A) |
| Clinical | Environmental/Other |
|---|---|
| fosX | fosX |
| lin | abc-f |
| abc-f | lin |
| tet(M) | tet(M) |
| catA1 | vanC |
| catA | vanR |
| mef(A) | vanXY-C |
| msr(D) | vanT |
| dfrG | vanS |
| fexA | tet(S) |
| dfrE | |
| fexA | |
| dfrG | |
| erm(B) | |
| lnu(G) | |
| blaTEM-116 | |
| erm(C) | |
| lsa(A) | |
| mph(B) | |
| tet(L) |
| AMR Gene | Biological Function | References |
|---|---|---|
| fosX | Catalyzes hydration of fosfomycin breaking the oxirane ring | [18] |
| abc-f | ATP-binding cassette protein that mediates resistance to a broad array of antibiotic classes that target the ribosome of Gram-positive pathogens | [25] |
| lin | Ribosomal protection protein, lincomycin | [18] |
| tet(M) | Tetracycline resistance (ribosome protection), class M | [27,28] |
| vanC | Glycopeptide resistance gene; vancomycin, class C | [29] |
| vanR | Glycopeptide resistance gene; vancomycin, class R | [30] |
| vanXY-C | Glycopeptide resistance gene; vancomycin | [31] |
| vanT | Glycopeptide resistance gene; vancomycin, class T | [32] |
| vanS | Glycopeptide resistance gene; vancomycin, class S | [30] |
| tet(S) | Tetracycline resistance (ribosome protection), class S | [27,28] |
| dfrE | Trimethoprim resistance | [33] |
| fexA | Active efflux, phenicols | [28] |
| dfrG | Trimethoprim resistance | [34] |
| erm(B) | Ribosome modification-mediated resistance; macrolide, lincosamide, and streptogramin B | [27] |
| lnu(G) | Enzymatic inactivation by nucleotidylation, lincomycin | [35] |
| blaTEM-116 | Β-lactamase, broad-spectrum cephalosporin | [36] |
| erm(C) | Ribosome modification-mediated resistance; macrolide, lincosamide, and streptogramin B | [28] |
| lsa(A) | Lincosamide and streptogramin A resistance | [37] |
| mph(B) | Encode phosphotransferases conferring macrolide resistance | [38] |
| tet(L) | Tetracycline resistance (active efflux), class L | [27,28] |
| Year | Infections (Incidence Per 100,000 Population) |
|---|---|
| 2010 | 0.26 |
| 2011 | 0.28 |
| 2012 | 0.26 |
| 2013 | 0.25 |
| 2014 | 0.24 |
| 2015 | 0.25 |
| 2016 | 0.26 |
| 2017 | 0.32 |
| 2018 | 0.26 |
| 2019 | 0.27 |
| 2020 | 0.2 |
| Year | Outbreaks | Illnesses | Hospitalizations | Deaths |
|---|---|---|---|---|
| 2009 | 4 | 35 | 18 | 0 |
| 2010 | 5 | 32 | 29 | 9 |
| 2011 | 6 | 209 | 184 | 39 |
| 2012 | 5 | 41 | 38 | 6 |
| 2013 | 10 | 86 | 77 | 16 |
| 2014 | 14 | 84 | 79 | 20 |
| 2015 | 6 | 75 | 61 | 7 |
| 2016 | 6 | 77 | 69 | 10 |
| 2017 | 11 | 54 | 47 | 7 |
| 2018 | 4 | 43 | 38 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanes, R.M.; Huang, Z. Investigation of Antimicrobial Resistance Genes in Listeria monocytogenes from 2010 through to 2021. Int. J. Environ. Res. Public Health 2022, 19, 5506. https://doi.org/10.3390/ijerph19095506
Hanes RM, Huang Z. Investigation of Antimicrobial Resistance Genes in Listeria monocytogenes from 2010 through to 2021. International Journal of Environmental Research and Public Health. 2022; 19(9):5506. https://doi.org/10.3390/ijerph19095506
Chicago/Turabian StyleHanes, Robert M., and Zuyi Huang. 2022. "Investigation of Antimicrobial Resistance Genes in Listeria monocytogenes from 2010 through to 2021" International Journal of Environmental Research and Public Health 19, no. 9: 5506. https://doi.org/10.3390/ijerph19095506
APA StyleHanes, R. M., & Huang, Z. (2022). Investigation of Antimicrobial Resistance Genes in Listeria monocytogenes from 2010 through to 2021. International Journal of Environmental Research and Public Health, 19(9), 5506. https://doi.org/10.3390/ijerph19095506






