Molecular Epidemiology of Antibiotic Resistance Genes and Virulence Factors in Multidrug-Resistant Escherichia coli Isolated from Rodents, Humans, Chicken, and Household Soils in Karatu, Northern Tanzania
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Bacterial Isolates
2.3. DNA Extraction
2.4. Detection of Antimicrobial Resistance and Virulence Genes
2.5. Statistical Analysis
3. Results
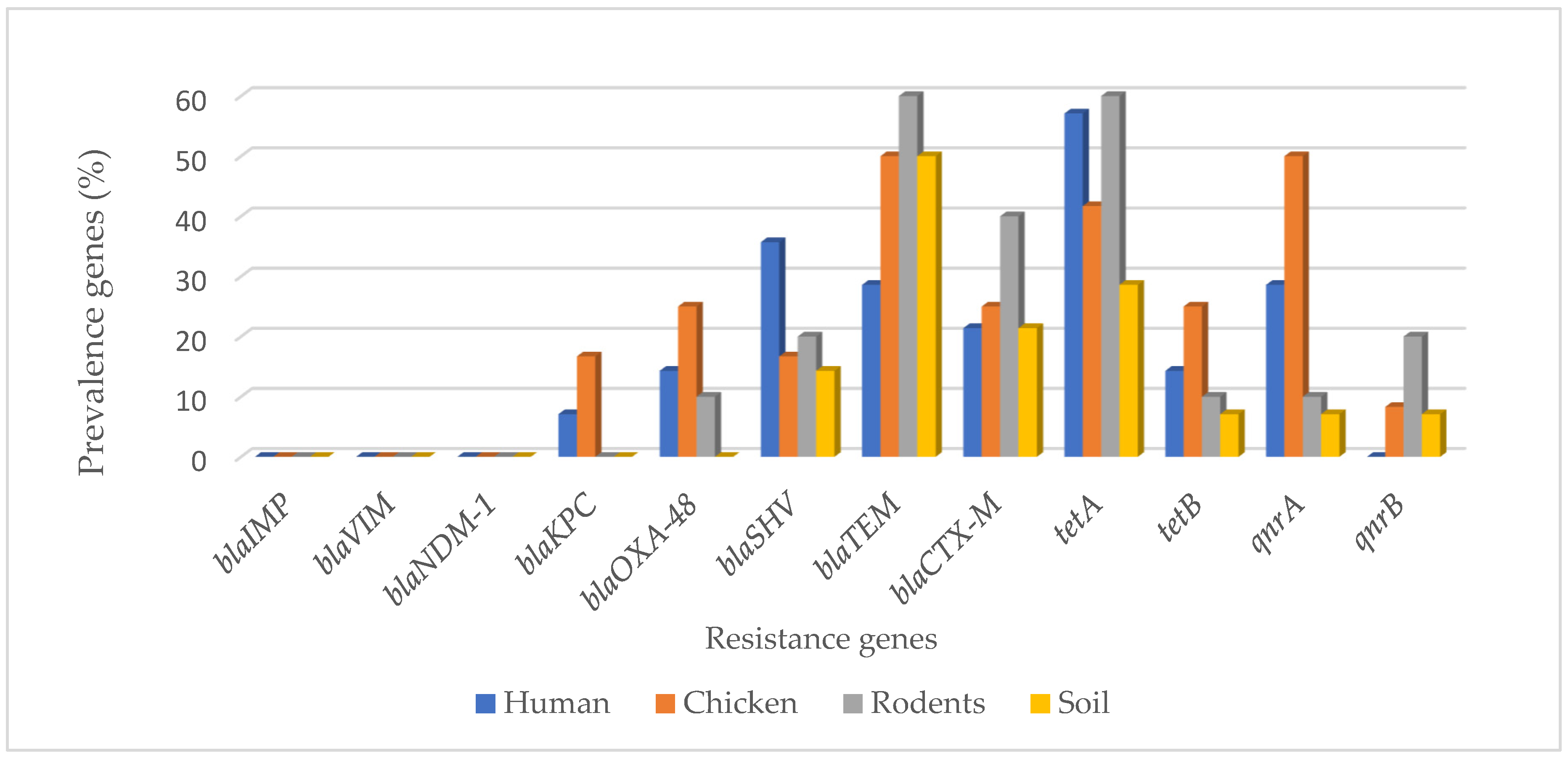
3.1. Carbapenems, ESBL, Tetracycline, and Quinolones Resistance Genes in MDR E. coli Isolates from Different Sample Sources
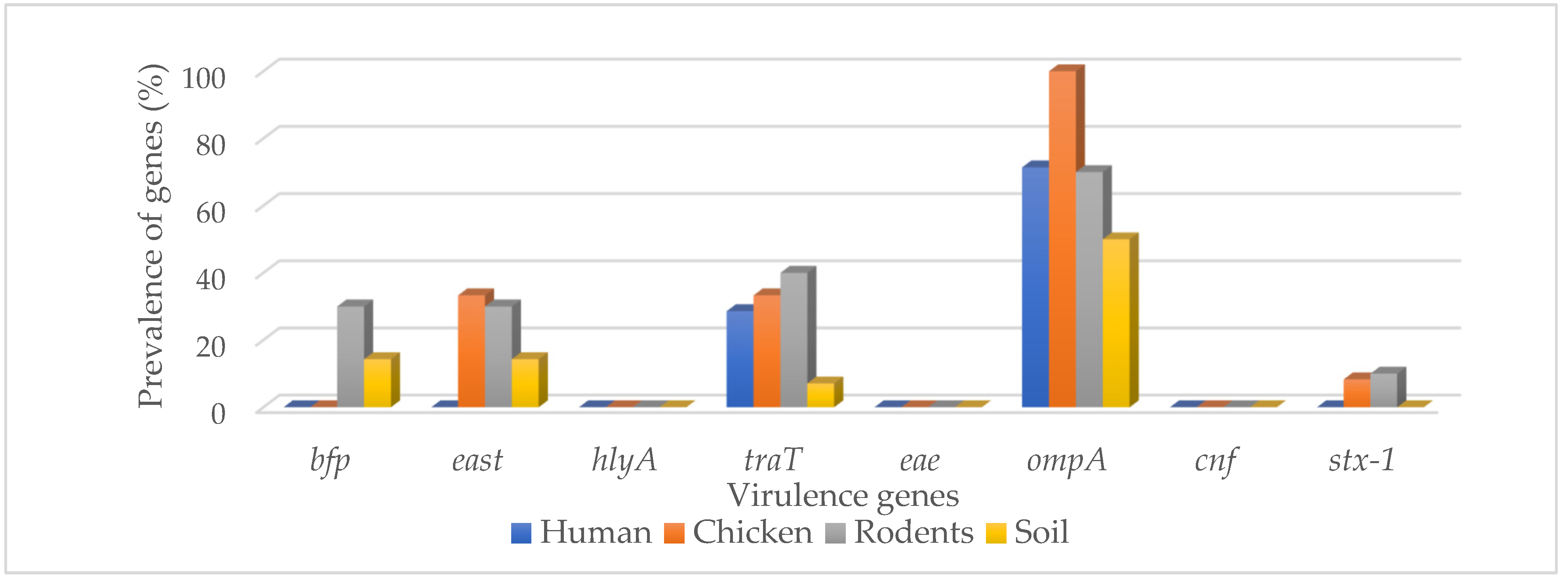
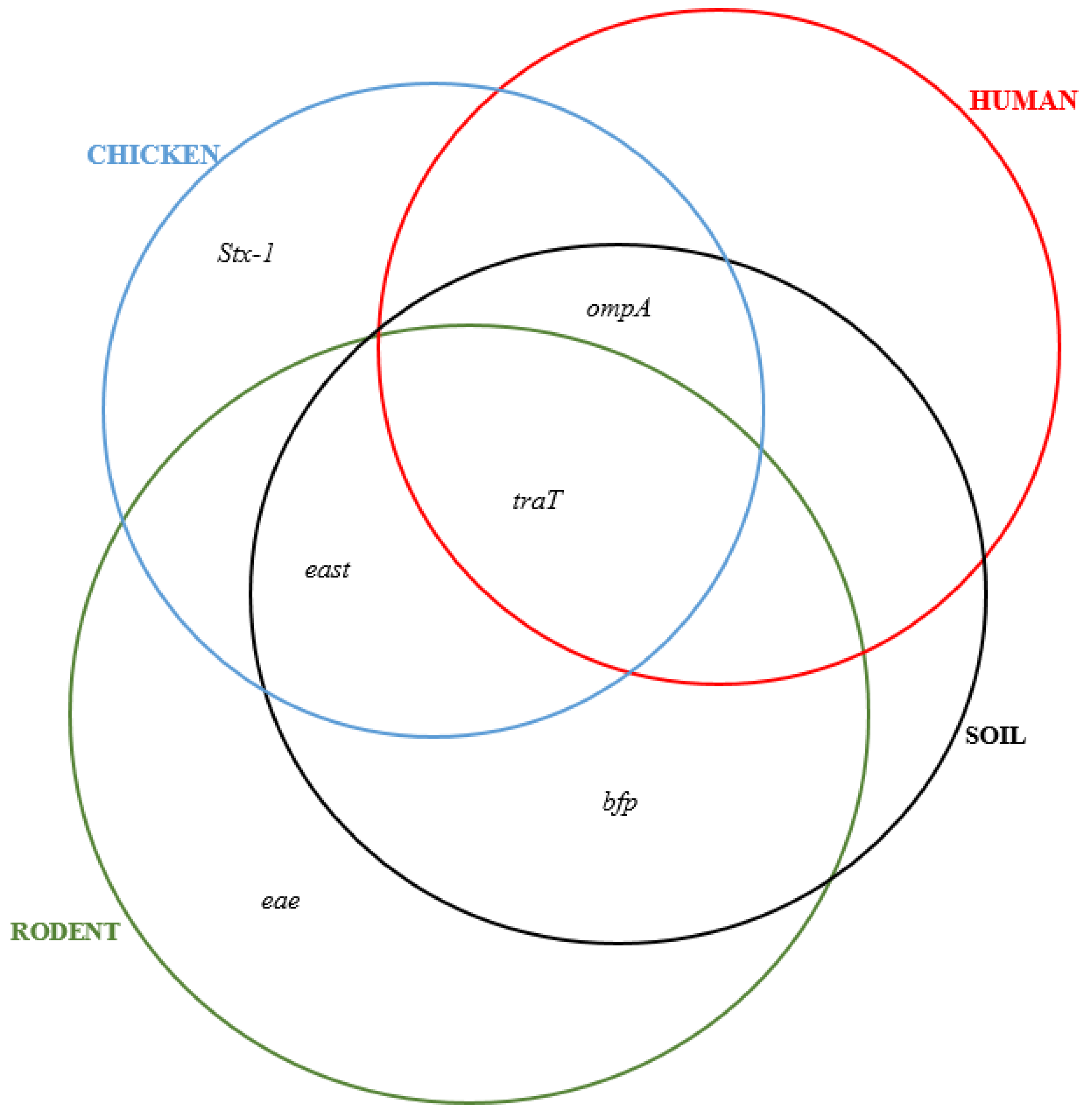
3.2. Detection of Virulence Genes in MDR E. coli Isolates from Different Sample Sources
3.3. Comparison between Phenotypic and Genotypic Antibiotic Resistance
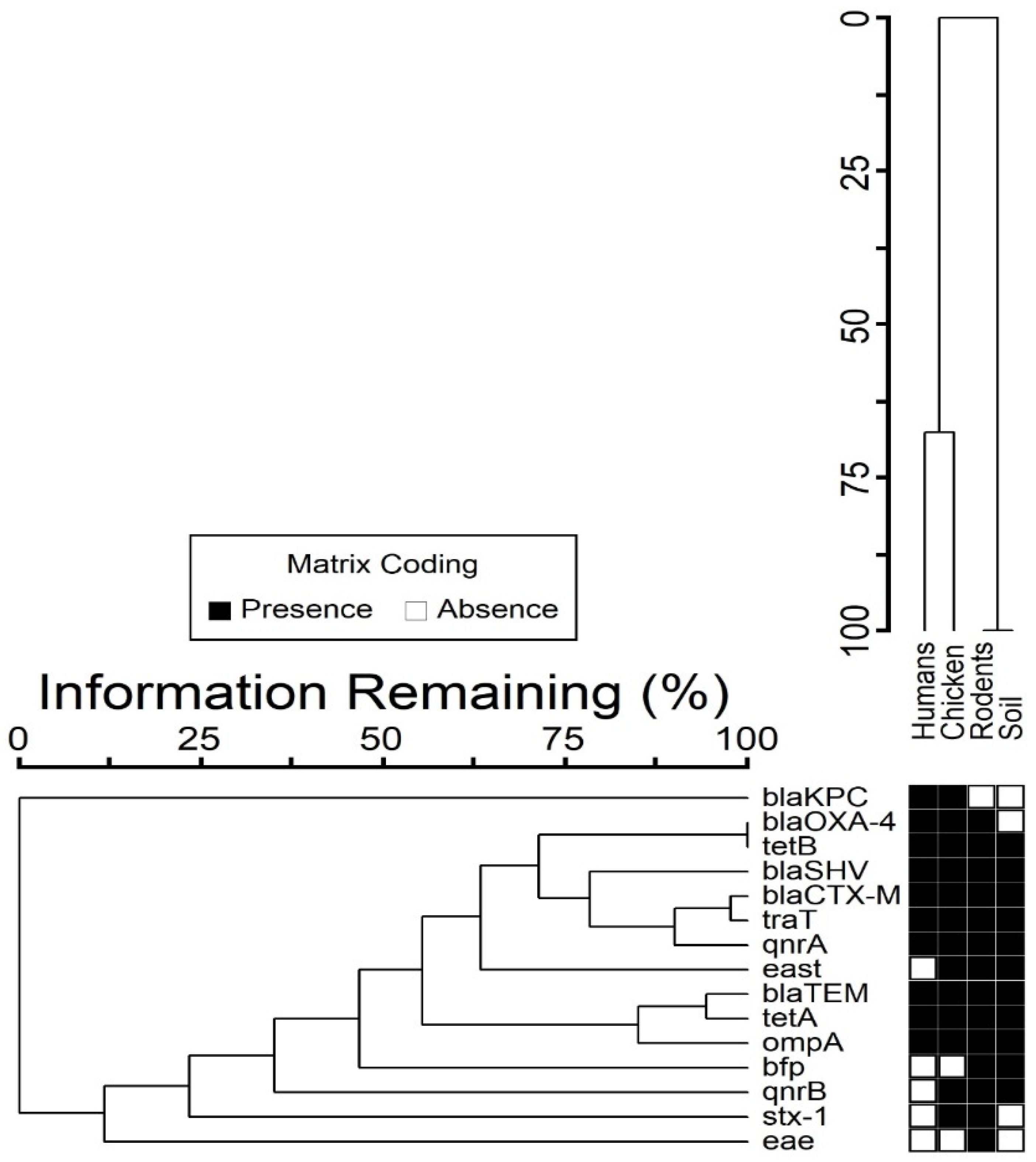
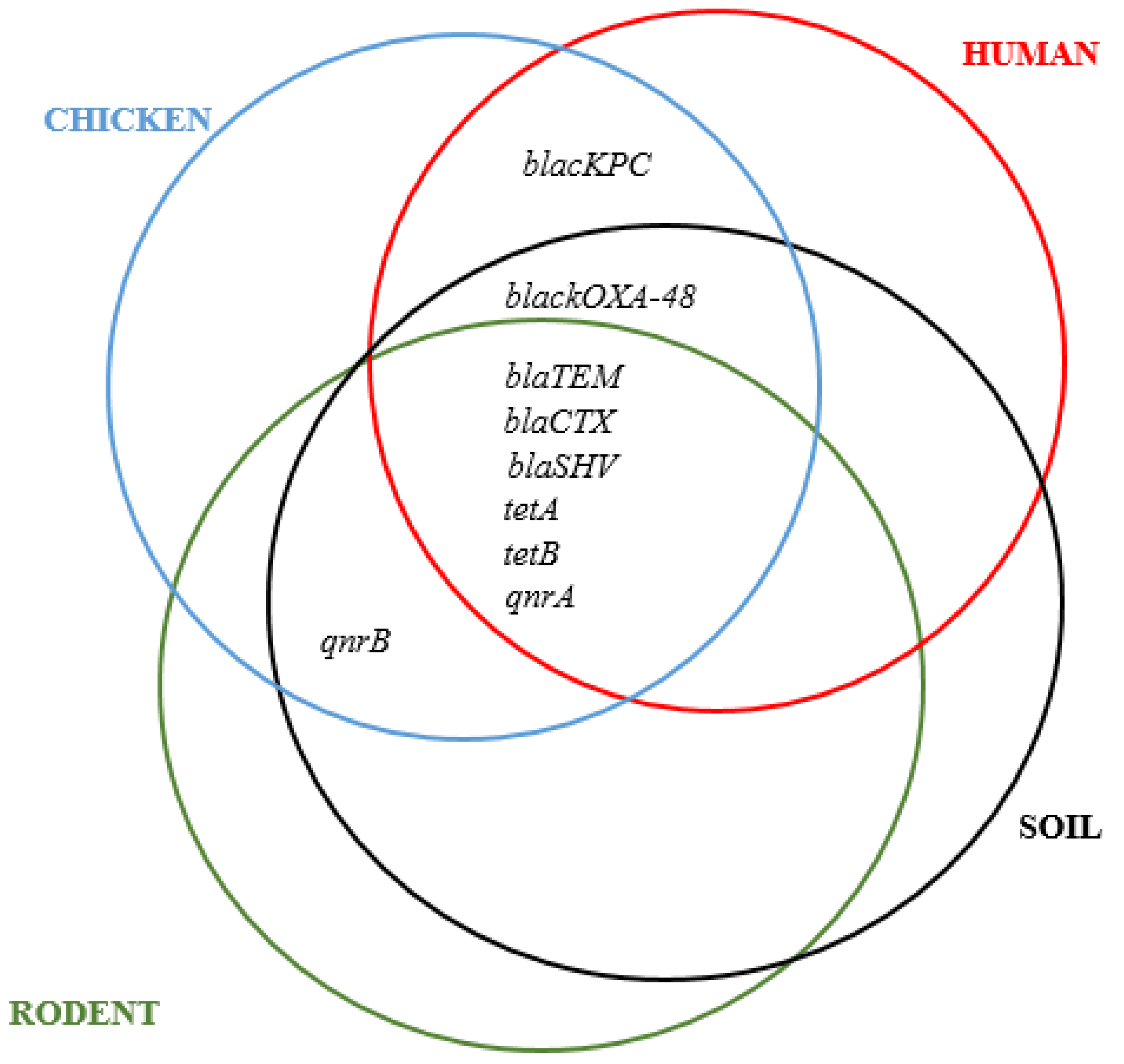
3.4. Co-Occurrence between Resistance and Virulence Genes
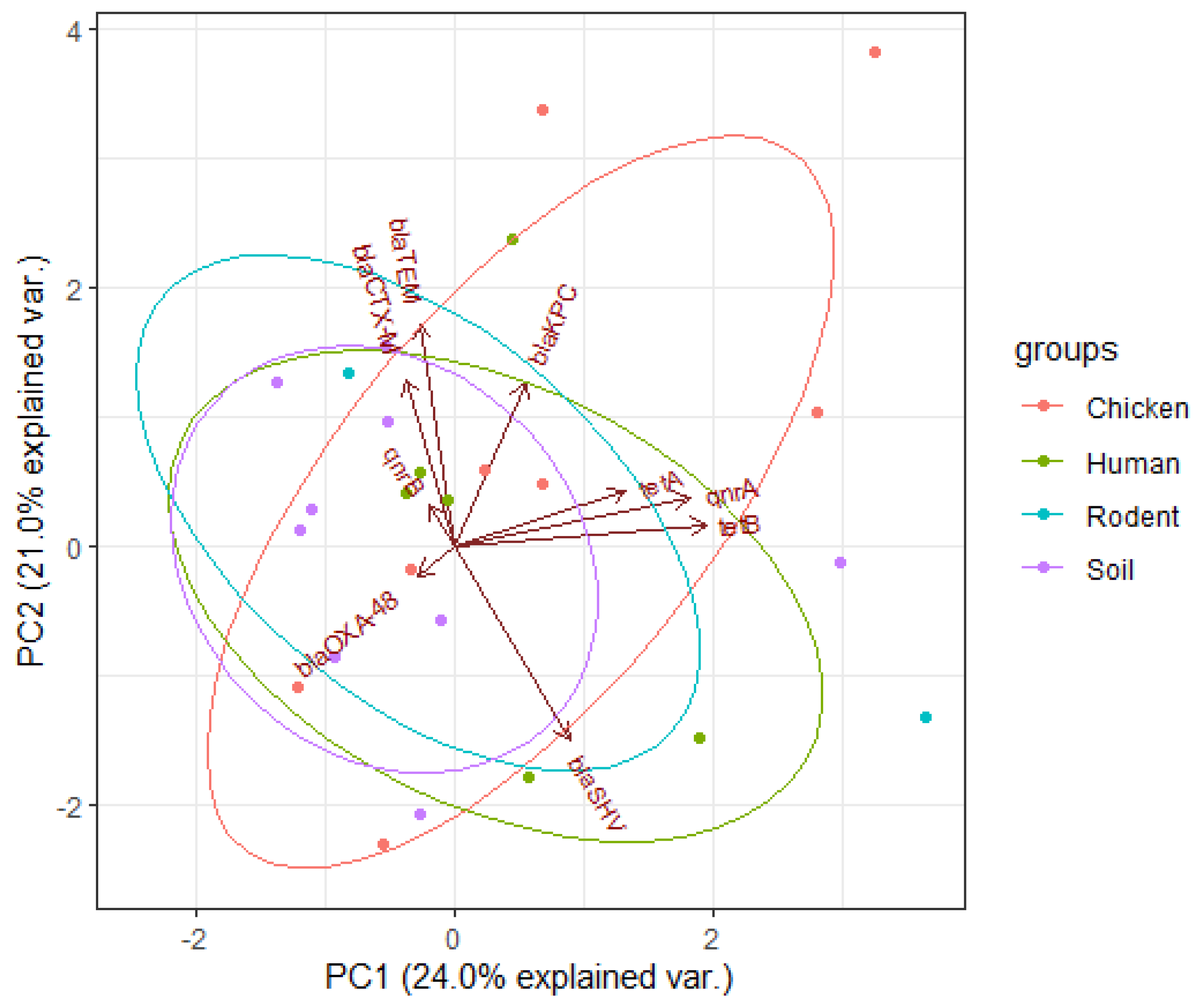
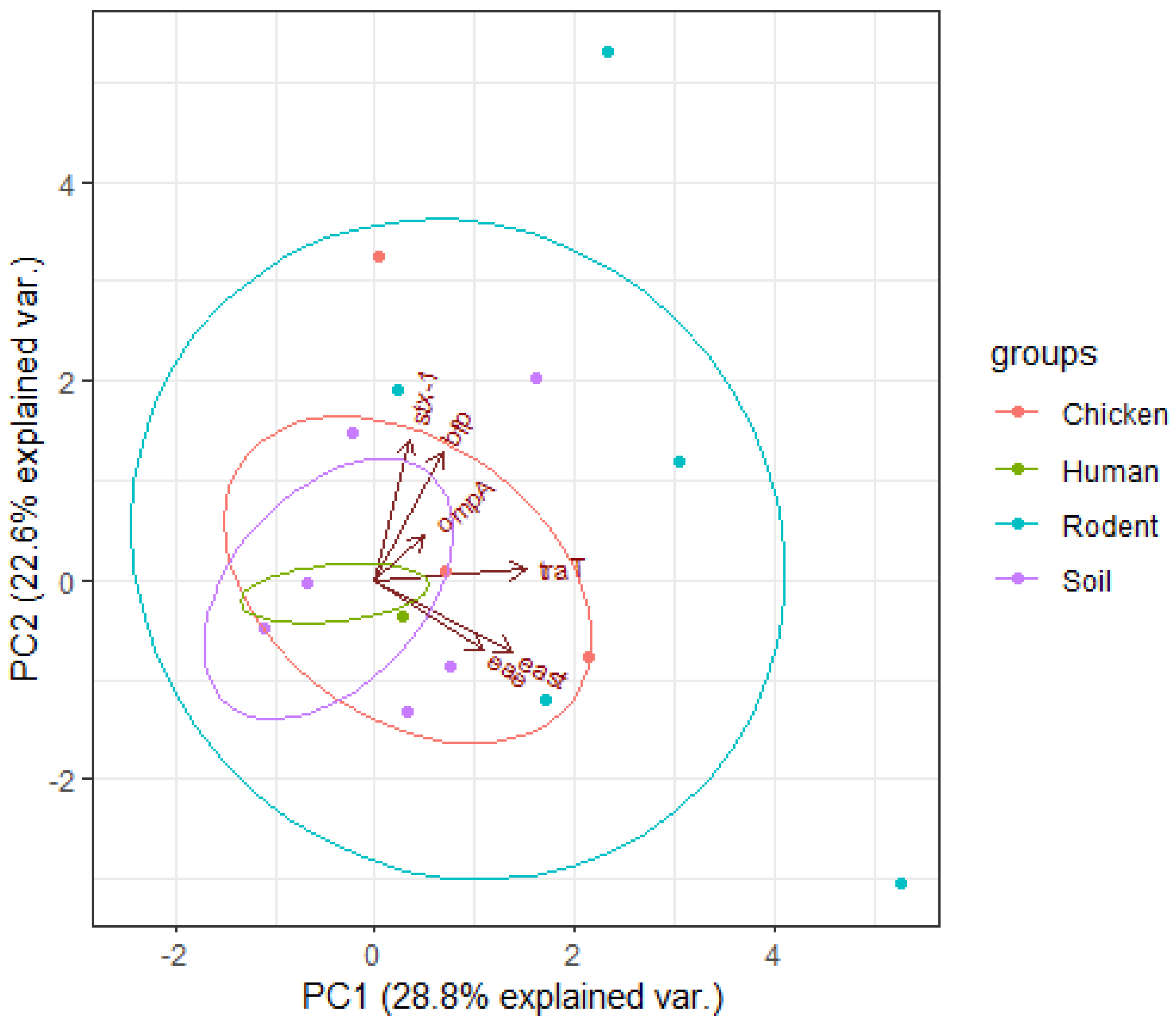
3.5. Principal Component Analysis Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Song, Y.; Yu, L.; Zhang, Y.; Dai, Y.; Wang, P.; Feng, C.; Liu, M.; Sun, S.; Xie, Z.; Wang, F. Prevalence and characteristics of multidrug-resistant mcr-1-positive Escherichia coli isolates from broiler chickens in Tai’an, China. Poult. Sci. 2020, 99, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Yoon, M.Y.; Ha, J.S.; Seo, K.W.; Noh, E.B.; Son, S.H.; Lee, Y.J. Molecular characterization of avian pathogenic Escherichia coli from broiler chickens with colibacillosis. Poult. Sci. 2020, 99, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.R.; Shrivastava, P.S.; Ramasamy, J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018, 32, 76–77. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-Ploskonska, G.; Choroszy-Krol, I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: Recent reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Baky, R.M.; Ibrahim, R.A.; Mohamed, D.S.; Ahmed, E.F.; Hashem, Z.S. Prevalence of virulence genes and their association with antimicrobial resistance among pathogenic E. coli isolated from Egyptian patients with different clinical infections. Infect. Drug. Resist. 2020, 13, 1221–1236. [Google Scholar] [CrossRef]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef]
- Masoud, S.M.; El-Baky, A.; Mahmoud, R.; Aly, S.A.; Ibrahem, R.A. Co-Existence of Certain ESBLs, MBLs and Plasmid Mediated Quinolone Resistance Genes among MDR E. coli Isolated from Different Clinical Specimens in Egypt. Antibiotics 2021, 10, 835. [Google Scholar] [CrossRef]
- Ibrahim, M.E.; Bilal, N.E.; Magzoub, M.A.; Hamid, M.E. Prevalence of extended-spectrum β-lactamases-producing Escherichia coli from Hospitals in Khartoum State, Sudan. Oman Med. J. 2013, 28, 116–120. [Google Scholar] [CrossRef]
- Yano, H.; Uemura, M.; Endo, S.; Kanamori, H.; Inomata, S.; Kakuta, R.; Ichimura, S.; Ogawa, M.; Shimojima, M.; Ishibashi, N. Molecular characteristics of extended-spectrum β-lactamases in clinical isolates from Escherichia coli at a Japanese tertiary hospital. PLoS ONE 2013, 8, e64359. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile β-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Livermore, D.M. The impact of carbapenemases on antimicrobial development and therapy. Curr. Opin. Investig. Drugs (Lond. Engl. 2000) 2002, 3, 218–224. [Google Scholar]
- Bryan, A.; Shapir, N.; Sadowsky, M.J. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains isolated from diverse human and animal sources. Appl. Environ. Microbiol. 2004, 70, 2503–2507. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Cattoir, V.; Nordmann, P. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front. Microbiol. 2012, 3, 24. [Google Scholar] [CrossRef] [PubMed]
- Allocati, N.; Masulli, M.; Alexeyev, M.F.; Di Ilio, C. Escherichia coli in Europe: An overview. Int. J. Environ. Res. Public Health 2013, 10, 6235–6254. [Google Scholar] [CrossRef]
- Robins-Browne, R.M.; Holt, K.E.; Ingle, D.J.; Hocking, D.M.; Yang, J.; Tauschek, M. Are Escherichia coli pathotypes still relevant in the era of whole-genome sequencing? Front. Cell. Infect. Microbiol. 2016, 6, 141. [Google Scholar] [CrossRef]
- Gomes, T.A.; Elias, W.P.; Scaletsky, I.C.; Guth, B.E.; Rodrigues, J.F.; Piazza, R.M.; Ferreira, L.; Martinez, M.B. Diarrheagenic Escherichia coli. Braz. J. Microbiol. 2016, 47, 3–30. [Google Scholar] [CrossRef]
- White, A.; Hughes, J.M. Critical importance of a one health approach to antimicrobial resistance. EcoHealth 2019, 16, 404–409. [Google Scholar] [CrossRef] [PubMed]
- Kilonzo, B.; Mbise, T.; Mwalimu, D.; Kindamba, L. Observations on the endemicity of plague in Karatu and Ngorongoro, northern Tanzania. Tanzan. J. Health Res. 2006, 8, 1–6. [Google Scholar] [CrossRef]
- Makundi, R.H.; Massawe, A.W.; Mulungu, L.S.; Katakweba, A.; Mbise, T.J.; Mgode, G. Potential mammalian reservoirs in a bubonic plague outbreak focus in Mbulu District, northern Tanzania, in 2007. Available online: https://www.degruyter.com/document/doi/10.1515/MAMM.2008.038/pdf (accessed on 2 February 2022).
- Ziwa, M.H.; Matee, M.I.; Hangombe, B.M.; Lyamuya, E.F.; Kilonzo, B.S. Plague in Tanzania: An overview. Tanzan. J. Health Res. 2013, 15, 4. [Google Scholar] [CrossRef]
- Guenther, S.; Bethe, A.; Fruth, A.; Semmler, T.; Ulrich, R.G.; Wieler, L.H.; Ewers, C. Frequent combination of antimicrobial multiresistance and extraintestinal pathogenicity in Escherichia coli isolates from urban rats (Rattus norvegicus) in Berlin, Germany. PLoS ONE 2012, 7, e50331. [Google Scholar] [CrossRef]
- Feng, A.Y.; Himsworth, C.G. The secret life of the city rat: A review of the ecology of urban Norway and black rats (Rattus norvegicus and Rattus rattus). Urban Ecosyst. 2014, 17, 149–162. [Google Scholar] [CrossRef]
- Hassell, J.M.; Ward, M.J.; Muloi, D.; Bettridge, J.M.; Robinson, T.P.; Kariuki, S.; Ogendo, A.; Kiiru, J.; Imboma, T.; Kangethe, E.K. Clinically relevant antimicrobial resistance at the wildlife–livestock–human interface in Nairobi: An epidemiological study. Lancet Planet. Health 2019, 3, e259–e269. [Google Scholar] [CrossRef]
- Desvars-Larrive, A.; Ruppitsch, W.; Lepuschitz, S.; Szostak, M.P.; Spergser, J.; Feßler, A.T.; Schwarz, S.; Monecke, S.; Ehricht, R.; Walzer, C. Urban brown rats (Rattus norvegicus) as possible source of multidrug-resistant Enterobacteriaceae and meticillin-resistant Staphylococcus spp., Vienna, Austria, 2016 and 2017. Eurosurveillance 2019, 24, 1900149. [Google Scholar] [CrossRef]
- Sonola, V.S.; Katakweba, A.S.; Misinzo, G.; Matee, M.I. Occurrence of Multi-Drug-Resistant Escherichia coli in Chickens, Humans, Rodents and Household Soil in Karatu, Northern Tanzania. Antibiotics 2021, 10, 1137. [Google Scholar] [CrossRef] [PubMed]
- Cepas, V.; Soto, S.M. Relationship between Virulence and Resistance among Gram-Negative Bacteria. Antibiotics 2020, 9, 719. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Zhang, X.; Gao, Y.; Song, S.; Xu, D.; Yan, L. Development and application of multiplex PCR method for simultaneous detection of seven viruses in ducks. BMC Vet. Res. 2019, 15, 103. [Google Scholar] [CrossRef]
- United Republic of Tanzania, 2012 Population and Housing Census-Population Distribution by Administrative Areas. Available online: http://tanzania.countrystat.org/fileadmin/user_upload/countrystat_fenix/congo/docs/Census%20General%20Report-2012PHC.pdf (accessed on 2 February 2022).
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- R: A Language and Environment for Statistical Computing. Available online: https://www.eea.europa.eu/data-and-maps/indicators/oxygen-consuming-substances-in-rivers/r-development-core-team-2006 (accessed on 2 February 2022).
- Gakuya, F.; Kyule, M.; Gathura, P.; Kariuki, S. Antimicrobial susceptibility and plasmids from Escherichia coli isolated from rats. East Afr. Med. J. 2001, 78, 518–522. [Google Scholar] [CrossRef][Green Version]
- Wang, H.; Sangwan, N.; Li, H.Y.; Su, J.Q.; Oyang, W.Y.; Zhang, Z.J.; Gilbert, J.A.; Zhu, Y.G.; Ping, F.; Zhang, H.L. The antibiotic resistome of swine manure is significantly altered by association with the Musca domestica larvae gut microbiome. ISME J. 2017, 11, 100–111. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef]
- Comparison of Antibiotic Resistant Escherichia coli Obtained from Drinking Water Sources in Northern Tanzania: A Cross-Sectional Study. Available online: https://www.researchgate.net/publication/309765672_Comparison_of_antibiotic_resistant_Escherichia_coli_obtained_from_drinking_water_sources_in_northern_Tanzania_a_cross-sectional_study (accessed on 2 February 2022).
- Antimicrobial Resistance Profiles of Escherichia coli Isolated from Broiler and Layer Chickens in Arusha and Mwanza, Tanzania. Available online: https://www.hindawi.com/journals/ijmicro/2021/6759046/ (accessed on 2 February 2022).
- Sale of Fluoroquinolones in Northern Tanzania: A Potential Threat for Fluoroquinolone Use in Tuberculosis Treatment. Available online: https://academic.oup.com/jac/article/65/1/145/727526?login=false# (accessed on 2 February 2022).
- Comparison of the Prevalence of Antibiotic-Resistant Escherichia coli Isolates from Commercial-Layer and Free-Range Chickens in Arusha District, Tanzania. Available online: https://www.researchgate.net/publication/308594993_Comparison_of_the_prevalence_of_antibiotic-resistant_Escherichia_coli_isolates_from_commercial-layer_and_free-range_chickens_in_Arusha_district_Tanzania (accessed on 2 February 2022).
- Subbiah, M.; Caudell, M.A.; Mair, C.; Davis, M.A.; Matthews, L.; Quinlan, R.J.; Quinlan, M.B.; Lyimo, B.; Buza, J.; Keyyu, J. Antimicrobial resistant enteric bacteria are widely distributed amongst people, animals and the environment in Tanzania. Nat. Commun. 2020, 11, 228. [Google Scholar] [CrossRef] [PubMed]
- Genetic Relatedness of Multidrug Resistant Escherichia coli Isolated from Humans, Chickens and Poultry Environments. Available online: https://aricjournal.biomedcentral.com/articles/10.1186/s13756-021-00930-x (accessed on 2 February 2022).
- Mahmoud, N.E.; Altayb, H.N.; Gurashi, R.M. Detection of Carbapenem-Resistant Genes in Escherichia coli Isolated from Drinking Water in Khartoum, Sudan. Available online: https://pubmed.ncbi.nlm.nih.gov/32612664/ (accessed on 2 February 2022).
- Schages, L.; Wichern, F.; Kalscheuer, R.; Bockmühl, D. Winter is coming–Impact of temperature on the variation of beta-lactamase and mcr genes in a wastewater treatment plant. Sci. Total Environ. 2020, 712, 136499. [Google Scholar] [CrossRef] [PubMed]
- Gutkind, G.O.; Di Conza, J.; Power, P.; Radice, M. β-Lactamase-mediated resistance: A biochemical, epidemiological and genetic overview. Curr. Pharm. Des. 2013, 19, 164–208. [Google Scholar] [CrossRef] [PubMed]
- Evans, B.A.; Amyes, S.G. OXA β-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef]
- Hamilton, M.J.; Hadi, A.Z.; Griffith, J.F.; Ishii, S.; Sadowsky, M.J. Large scale analysis of virulence genes in Escherichia coli strains isolated from Avalon Bay, CA. Water Res. 2010, 44, 5463–5473. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Yin, H.B.; Bauchan, G.; Mowery, J. Inhibition of Escherichia coli O157: H7 and Salmonella enterica virulence factors by benzyl isothiocyanate. Food Microbiol. 2020, 86, 103303. [Google Scholar] [CrossRef]
- Pan, Z.; Chen, Y.; McAllister, T.A.; Gänzle, M.; Plastow, G.; Guan, L.L. Abundance and Expression of Shiga Toxin Genes in Escherichia coli at the Recto-Anal Junction Relates to Host Immune Genes. Front. Cell. Infect. Microbiol. 2021, 11, 148. [Google Scholar] [CrossRef]
- Ménard, L.-P.; Dubreuil, J.D. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): A new toxin with an old twist. Crit. Rev. Microbiol. 2002, 28, 43–60. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, K.S. Role of OmpA and IbeB in Escherichia coli K1 invasion of brain microvascular endothelial cells in vitro and in vivo. Pediatr. Res. 2002, 51, 559–563. [Google Scholar] [CrossRef]
- Firoozeh, F.; Saffari, M.; Neamati, F.; Zibaei, M. Detection of virulence genes in Escherichia coli isolated from patients with cystitis and pyelonephritis. Int. J. Infect. Dis. 2014, 29, 219–222. [Google Scholar] [CrossRef]
- Guan, J.; Liu, S.; Lin, Z.; Li, W.; Liu, X.; Chen, D. Severe sepsis facilitates intestinal colonization by extended-spectrum-β-lactamase-producing Klebsiella pneumoniae and transfer of the SHV-18 resistance gene to Escherichia coli during antimicrobial treatment. Antimicrob. Agents Chemother. 2014, 58, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Duffitt, A.D.; Reber, R.T.; Whipple, A.; Chauret, C. Gene expression during survival of Escherichia coli O157: H7 in soil and water. Int. J. Microbiol. 2011, 2011, 340506. [Google Scholar] [CrossRef] [PubMed]
- Cooley, M.B.; Jay-Russell, M.; Atwill, E.R.; Carychao, D.; Nguyen, K.; Quiñones, B.; Patel, R.; Walker, S.; Swimley, M.; Pierre-Jerome, E. Development of a robust method for isolation of Shiga toxin-positive Escherichia coli (STEC) from fecal, plant, soil and water samples from a leafy greens production region in California. PLoS ONE 2013, 8, e65716. [Google Scholar] [CrossRef]
- Le Huy, H.; Koizumi, N.; Ung, T.T.H.; Le, T.T.; Nguyen, H.L.K.; Hoang, P.V.M.; Nguyen, C.N.; Khong, T.M.; Hasebe, F.; Haga, T.; et al. Antibiotic-resistant Escherichia coli isolated from urban rodents in Hanoi, Vietnam. J. Vet. Med. Sci. 2020, 82, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Himsworth, C.G.; Zabek, E.; Desruisseau, A.; Parmley, E.J.; Reid-Smith, R.; Jardine, C.M.; Tang, P.; Patrick, D.M. Prevalence and characteristics of Escherichia coli and Salmonella spp. in the feces of wild urban Norway and black rats (Rattus norvegicus and Rattus rattus) from an inner-city neighborhood of Vancouver, Canada. J. Wildl. Dis. 2015, 51, 589–600. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Hammaren, R.; Pal, C.; Östman, M.; Björlenius, B.; Flach, C.-F.; Fick, J.; Kristiansson, E.; Tysklind, M.; Larsson, D.J. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 2016, 572, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Soto, S.; Zuniga, S.; Ulleryd, P.; Vila, J. Acquisition of a pathogenicity island in an Escherichia coli clinical isolate causing febrile urinary tract infection. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Hacker, J.; Blum-Oehler, G.; Mühldorfer, I.; Tschäpe, H. Pathogenicity islands of virulent bacteria: Structure, function and impact on microbial evolution. Mol. Microbiol. 1997, 23, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
| Genes | Different Sample Sources n (%) | ||||
|---|---|---|---|---|---|
| Humans (n = 14) | Chickens (n = 12) | Rodents (n = 10) | Soil (n = 14) | Total (n = 50) | |
| Bfp | 0 (0.0) | 0 (0.0) | 3 (30.0) | 2 (14.3) | 5 (10.0) |
| East | 0 (0.0) | 4 (33.3) | 3 (30.0) | 2 (14.3) | 9 (18.0) |
| hlyA | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| traT | 4 (28.6) | 4 (33.3) | 4 (40.0) | 1 (7.1) | 13 (26.0) |
| eae | 0 (0.0) | 0 (0.0) | 1 (10.0) | 0 (0.0) | 1 (2.0) |
| ompA | 10 (71.4) | 12 (100.0) | 7 (70.0) | 7 (50.0) | 36 (72.0) |
| cnf | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| stx-1 | 0 (0.0) | 1 (8.3) | 1 (10.0) | 0 (0.0) | 2 (4.0) |
| Total | 2 (14.3) | 4 (33.3) | 6 (60.0) | 4 (33.3) | 16 (32.0) |
| χ2-square | 52.29 | 46.43 | 2.00 | 26.67 | |
| p-value | 0.001 | 0.001 | 0.0188 | 0.0004 | |
| Genes | Different Types of Sample Sources n (%) | ||||
|---|---|---|---|---|---|
| Human (n = 14) | Chicken (n = 12) | Rodents (n = 10) | Soil (n = 14) | Total Isolates (n = 50) | |
| blaIMP | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| blaVIM | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| blaNDM-1 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| blaKPC | 1 (7.1) | 2 (16.7) | 0 (0.0) | 0 (0.0) | 3 (6.0) |
| blaOXA-48 | 2 (14.3) | 3 (25.0) | 1 (10.0) | 0 (0.0) | 6 (12.0) |
| blaSHV | 5 (35.7) | 2 (16.7) | 2 (20.0) | 2 (14.3) | 11 (22.0) |
| blaTEM | 4 (28.6) | 6 (50.0) | 6 (60.0) | 7 (50.0) | 23 (46.0) |
| blaCTX-M | 3 (21.4) | 3 (25.0) | 4 (40.0) | 3 (21.4) | 13 (26.0) |
| tetA | 8 (57.1) | 5 (41.7) | 6 (60.0) | 4 (28.6) | 23 (46.0) |
| tetB | 2 (14.3) | 3 (25.0) | 1 (10.0) | 1 (7.1) | 7 (14.0) |
| qnrA | 4 (28.6) | 6 (50.0) | 1 (10.0) | 1 (7.1) | 12 (24.0) |
| qnrB | 0 (0.0) | 1 (8.3) | 2 (20.0) | 1 (7.1) | 4 (8.0) |
| Total | 8 (57.1) | 9 (75.0) | 8 (80.0) | 7 (50.0%) | 32 (64.0) |
| χ2-square | 52.29 | 46.43 | 2.00 | 26.67 | |
| p-value | 0.001 | 0.001 | 0.0188 | 0.0004 | |
| Genotypes of Isolates | Phenotypic Resistance of Isolates | |||
|---|---|---|---|---|
| Correlation Coefficients (r) | ||||
| Tetracycline | Imipenem | Ciprofloxacin | Cefotaxime | |
| tetA | 0.53 | 0.51 | 0.62 | 0.43 |
| tetB | 0.90 | 0.90 | 0.86 | 0.93 |
| blaKPC | 0.90 | 0.90 | 0.86 | 0.93 |
| blaOXA-48 | 0.91 | 0.89 | 0.90 | 0.90 |
| qnrA | 0.94 | 0.94 | 0.90 | 0.96 |
| qnrB | −0.69 | −0.71 | −0.67 | −0.69 |
| blaCTX-M | −0.54 | −0.58 | −0.45 | −0.61 |
| blaTEM | −0.71 | −0.69 | −0.78 | −0.63 |
| blaSHV | 0.43 | 0.41 | 0.51 | 0.33 |
| ABR Genes | Virulence Genes | |||||
|---|---|---|---|---|---|---|
| Correlation Coefficients (r) | ||||||
| bfp | east | traT | eae | ompA | stx-1 | |
| blaKPC | −0.11 | 0.30 | 0.41 | −0.05 | 0.11 | −0.07 |
| blaOXA-48 | −0.16 | −0.06 | 0.15 | 0.38 | −0.05 | 0.22 |
| blaSHV | −0.24 | −0.34 | −0.44 | −0.10 | 0.24 | −0.15 |
| blaTEM | 0.39 | 0.44 | 0.51 | 0.17 | −0.39 | 0.01 |
| blaCTX-M | 0.05 | 0.39 | 0.31 | 0.23 | −0.22 | 0.08 |
| tetA | 0.36 | −0.10 | 0.11 | 0.15 | 0.26 | 0.22 |
| tetB | −0.18 | 0.06 | −0.05 | −0.08 | 0.18 | −0.11 |
| qnrA | −0.26 | 0.03 | 0.00 | −0.11 | 0.09 | 0.10 |
| qnrB | 0.63 | −0.19 | 0.12 | −0.05 | 0.13 | 0.30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sonola, V.S.; Katakweba, A.; Misinzo, G.; Matee, M.I. Molecular Epidemiology of Antibiotic Resistance Genes and Virulence Factors in Multidrug-Resistant Escherichia coli Isolated from Rodents, Humans, Chicken, and Household Soils in Karatu, Northern Tanzania. Int. J. Environ. Res. Public Health 2022, 19, 5388. https://doi.org/10.3390/ijerph19095388
Sonola VS, Katakweba A, Misinzo G, Matee MI. Molecular Epidemiology of Antibiotic Resistance Genes and Virulence Factors in Multidrug-Resistant Escherichia coli Isolated from Rodents, Humans, Chicken, and Household Soils in Karatu, Northern Tanzania. International Journal of Environmental Research and Public Health. 2022; 19(9):5388. https://doi.org/10.3390/ijerph19095388
Chicago/Turabian StyleSonola, Valery Silvery, Abdul Katakweba, Gerald Misinzo, and Mecky Isaac Matee. 2022. "Molecular Epidemiology of Antibiotic Resistance Genes and Virulence Factors in Multidrug-Resistant Escherichia coli Isolated from Rodents, Humans, Chicken, and Household Soils in Karatu, Northern Tanzania" International Journal of Environmental Research and Public Health 19, no. 9: 5388. https://doi.org/10.3390/ijerph19095388
APA StyleSonola, V. S., Katakweba, A., Misinzo, G., & Matee, M. I. (2022). Molecular Epidemiology of Antibiotic Resistance Genes and Virulence Factors in Multidrug-Resistant Escherichia coli Isolated from Rodents, Humans, Chicken, and Household Soils in Karatu, Northern Tanzania. International Journal of Environmental Research and Public Health, 19(9), 5388. https://doi.org/10.3390/ijerph19095388







