Factors Driving Microbial Community Dynamics and Potential Health Effects of Bacterial Pathogen on Landscape Lakes with Reclaimed Water Replenishment in Beijing, PR China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Water Samples
2.2. Water Quality Analysis
2.3. DNA Extraction and High-Throughput Sequencing
2.4. Data Processing and Statistical Analysis
3. Results and Discussion
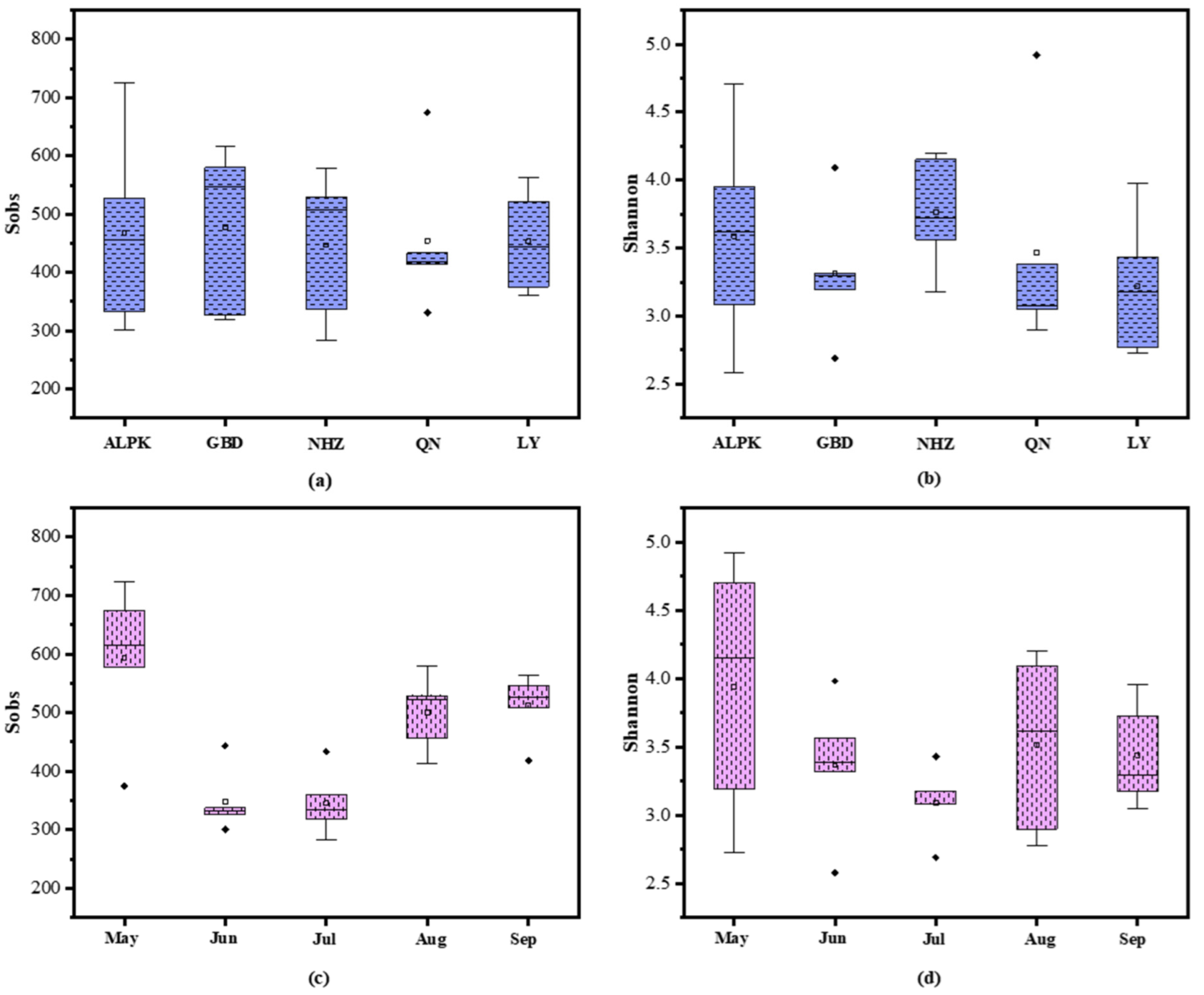
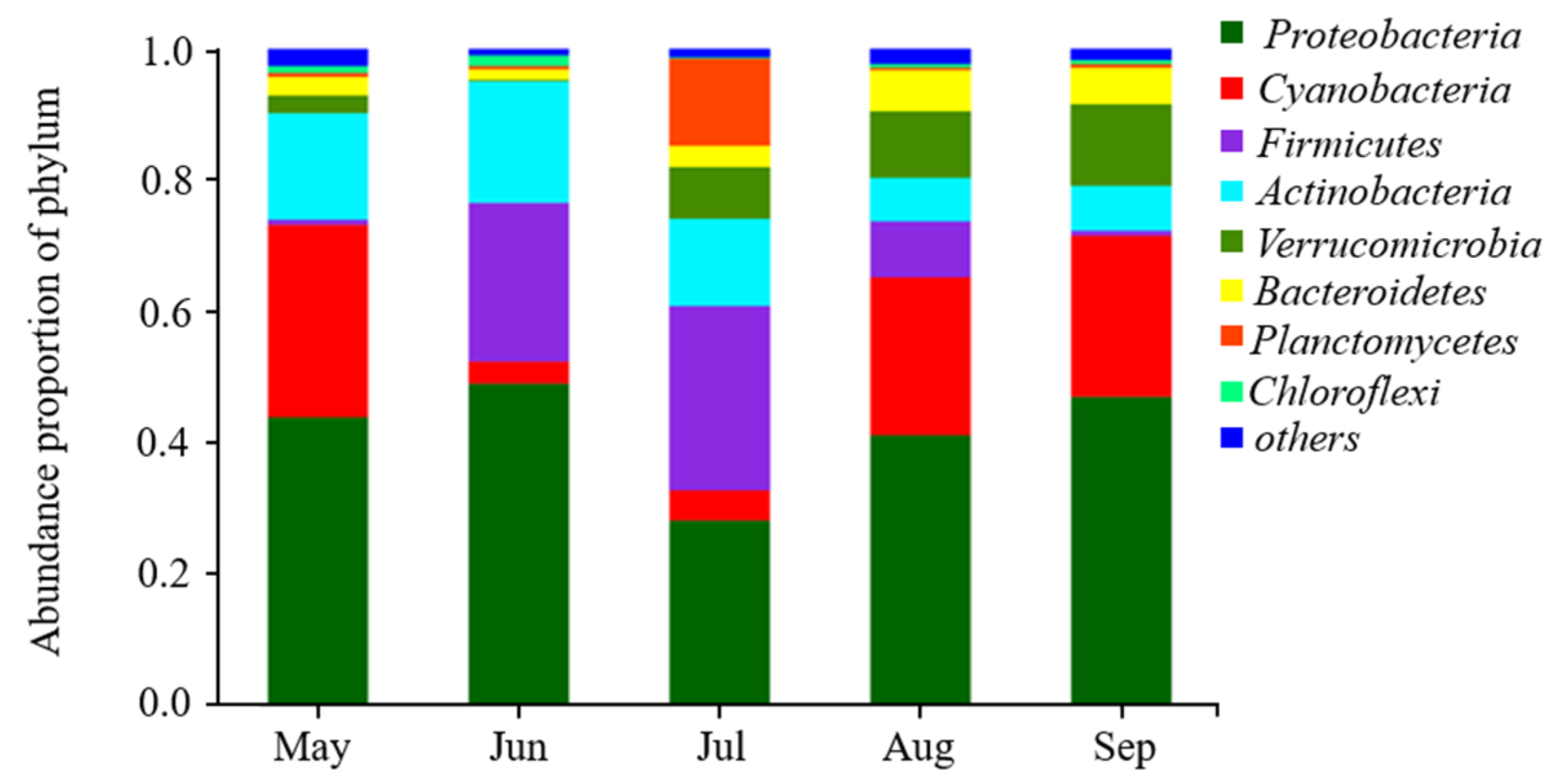
3.1. Microbial Community of Examined Lakes
3.2. Microbial Community Diversity and Potential Bacterial Pathogens
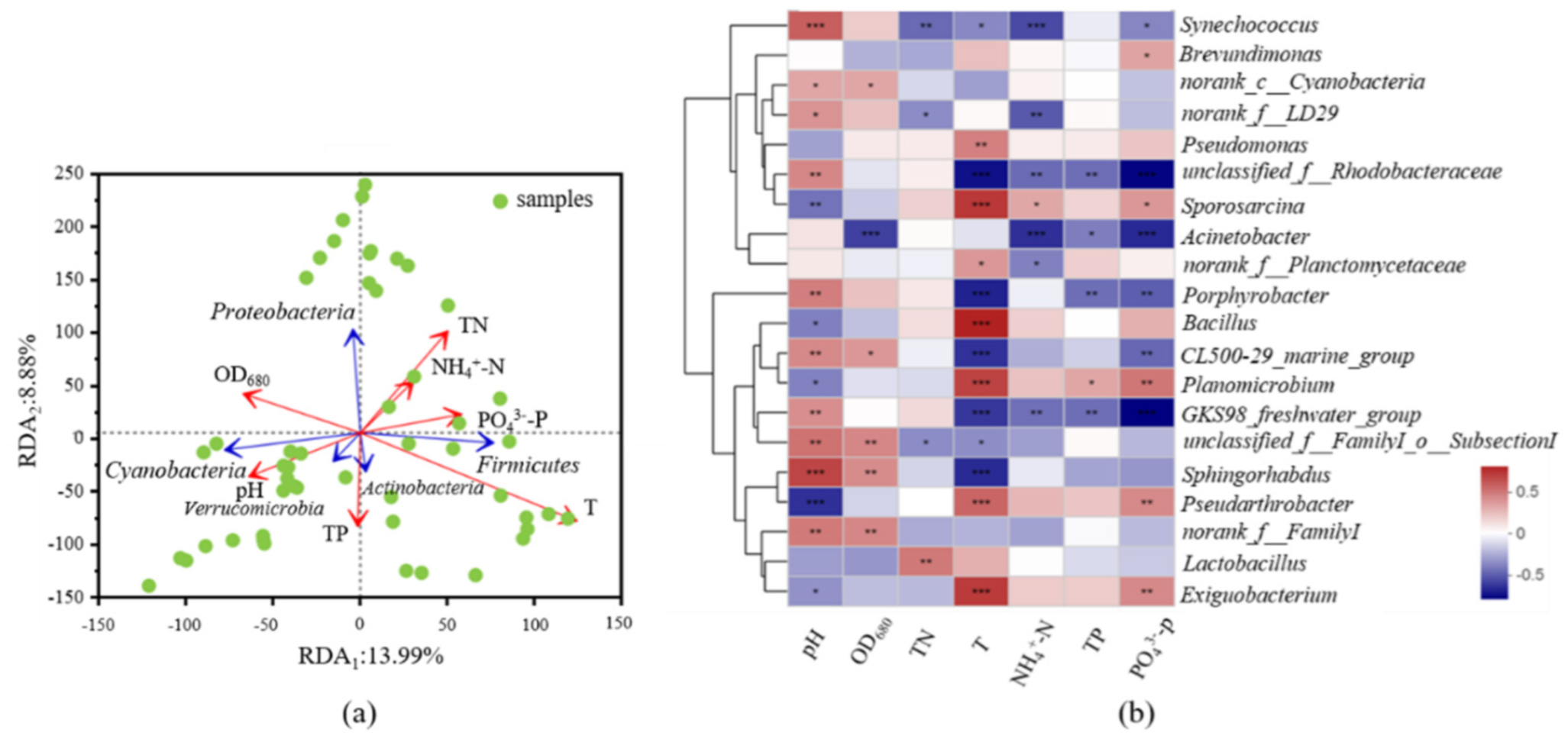
3.3. Responses of Community Changes and Potential Bacterial Pathogens to Key Environmental Factors
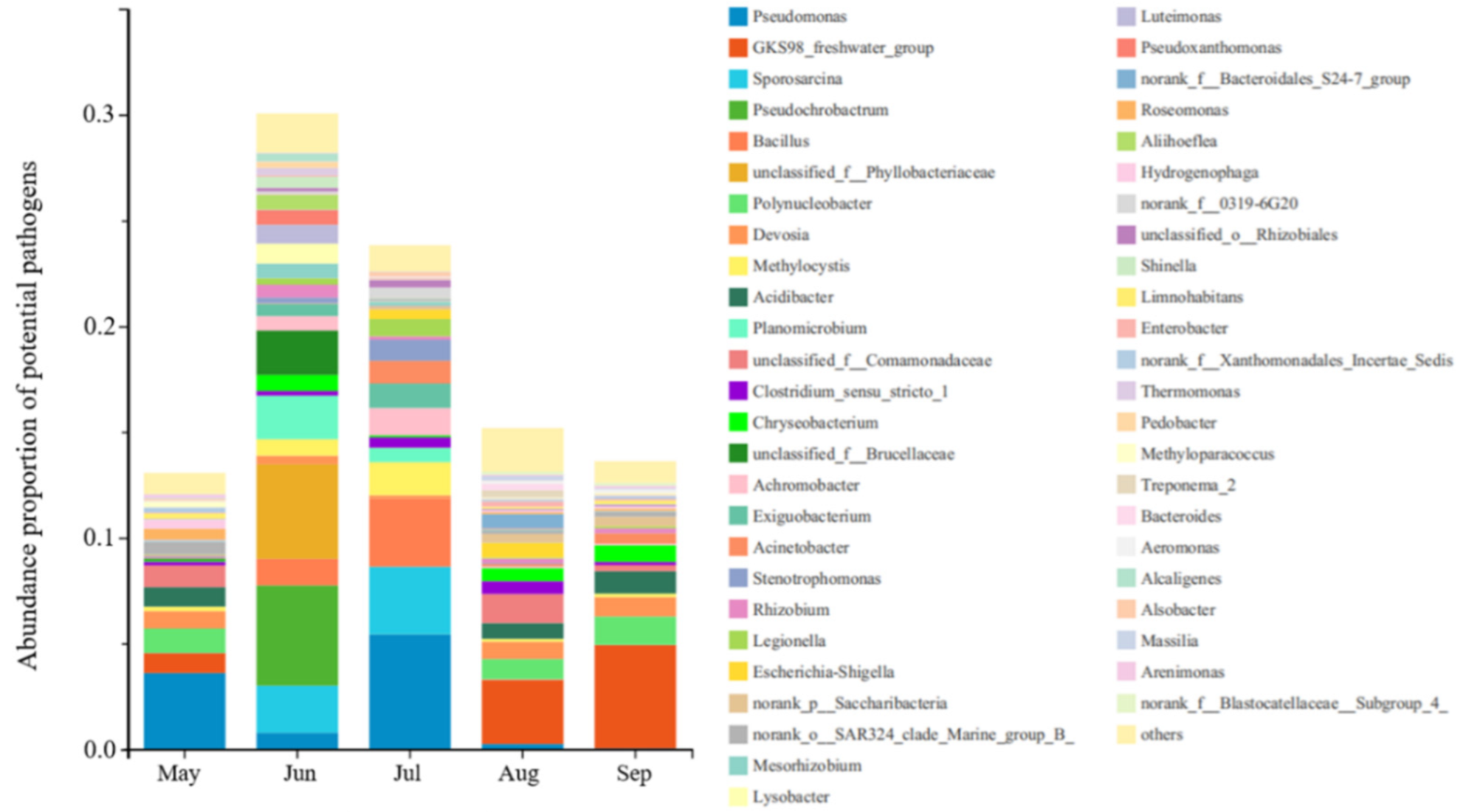
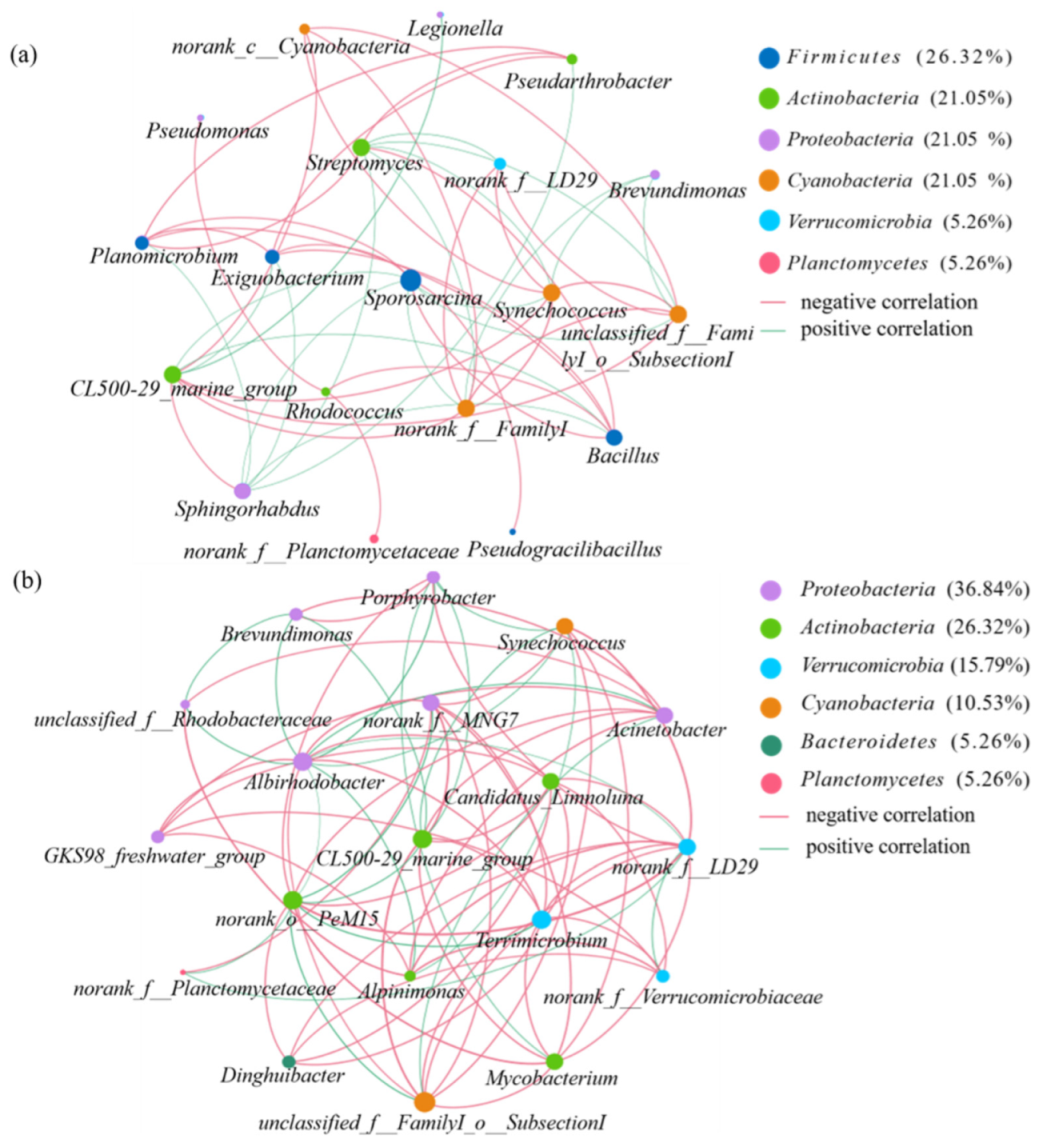
3.4. Health Effects of Potential Pathogens and Microbial Co-Occurrence
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Z.; Tong, Y.; Ngo, H.H.; Yun, L.; Li, G.Q.; Wu, Q.Y.; Li, K.X.; Bai, Y.; Liu, S.M.; Hu, H.Y. Assimilable organic carbon (AOC) variation in reclaimed water: Insight on biological stability evaluation and control for sustainable water reuse. Bioresour. Technol. 2018, 254, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.L.; Chen, H.B. Effect of residual chlorine on the interaction between bacterial growth and assimilable organic carbon and biodegradable organic carbon in reclaimed water. Sci. Total Environ. 2021, 752, 141223. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Q.; Huang, D.; Guo, C.F.; Guo, X.Y. Multivariate statistical analysis of temporal–spatial variations in water quality of a constructed wetland purification system in a typical park in Beijing, China. Environ. Monit. Assess. 2014, 187, 4219. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xu, Z.Q.; Long, Y.J.; Feng, M.Q. Replenishment of urban landscape ponds with reclaimed water: Spatiotemporal variations of water quality and mechanism of algal inhibition with alum sludge. Sci. Total Environ. 2021, 790, 148052. [Google Scholar] [CrossRef] [PubMed]
- Marks, J.S. Taking the public seriously: The case of potable and non-potable reuse. Desalination 2006, 187, 137–147. [Google Scholar] [CrossRef]
- Zhu, L.B.; Torres, M.; Betancourt, W.Q.; Sharma, M.; Shirley, A.; Micallef, S.A.; Gerba, C.; Sapkota, A.R.; Sapkota, A.; Parveen, S.; et al. Incidence of fecal indicator and pathogenic bacteria in reclaimed and return flow waters in Arizona, USA. Environ. Res. 2019, 170, 122–127. [Google Scholar] [CrossRef]
- Yue, L.Y.; Kong, W.D.; Ji, M.K.; Liu, J.B.; Morgan-Kiss, R.M. Community response of microbial primary producers to salinity is primarily driven by nutrients in lakes. Sci. Total Environ. 2019, 696, 134001. [Google Scholar] [CrossRef]
- Malayil, L.; Ramachandran, P.; Chattopadhyay, S.; Cagle, R.; Hittle, L.; Ottesen, A.; Mongodin, E.F.; Sapkota, A.R. Metabolically-active bacteria in reclaimed water and ponds revealed using bromodeoxyuridine DNA labeling coupled with 16S rRNA and shotgun sequencing. Water Res. 2020, 184, 116185. [Google Scholar] [CrossRef]
- Caicedo, C.; Rosenwinkel, K.H.; Exner, M.; Verstraete, W.; Suchenwirth, R.; Hartemann, P.; Nogueira, R. Legionella occurrence in municipal and industrial wastewater treatment plants and risks of reclaimed wastewater reuse: Review. Water Res. 2019, 149, 21–34. [Google Scholar] [CrossRef]
- Kulkarni, P.; Olson, N.D.; Paulson, J.N.; Pop, M.; Maddox, C.; Claye, E.; Rosenberg Goldstein, R.E.; Sharma, M.; Gibbs, S.G.; Mongodin, E.F.; et al. Conventional wastewater treatment and reuse site practices modify bacterial community structure but do not eliminate some opportunistic pathogens in reclaimed water. Sci. Total Environ. 2018, 639, 1126–1137. [Google Scholar] [CrossRef]
- Solaiman, S.; Micallef, S.A. Aeromonas spp. diversity in U.S. mid-Atlantic surface and reclaimed water, seasonal dynamics, virulence gene patterns and attachment to lettuce. Sci. Total Environ. 2021, 779, 146472. [Google Scholar] [CrossRef] [PubMed]
- Pepper, I.L.; Gerba, C.P. Risk of infection from Legionella associated with spray irrigation of reclaimed water. Water Res. 2018, 139, 101–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, K.A.; Hamilton, M.T.; Johnson, W.; Jjemba, P.; Bukhari, Z.; LeChevallier, M.; Haas, C.N. Health risks from exposure to Legionella in reclaimed water aerosols: Toilet flushing, spray irrigation, and cooling towers. Water Res. 2018, 134, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Banda, J.F.; Zhang, Q.; Ma, L.Q.; Pei, L.X.; Du, Z.R.; Hao, C.B.; Dong, H.L. Both pH and salinity shape the microbial communities of the lakes in Badain Jaran Desert, NW China. Sci. Total Environ. 2021, 791, 148108. [Google Scholar] [CrossRef]
- Aping, N.; Song, L.Y.; Xiong, Y.H.; Lu, C.J.; Junaid, M.; Pei, D.S. Impact of water quality on the microbial diversity in the surface water along the three Gorge reservoir (Tgr), China. Ecotoxicol. Environ. Saf. 2019, 181, 412–418. [Google Scholar]
- Zhang, Y.T.; Shen, H.; He, X.H.; Thomas, B.W.; Lupwayi, N.Z.; Hao, X.Y.; Thomas, M.C.; Shi, X.J. Fertilization shapes bacterial community structure by alteration of soil pH. Front. Microbiol. 2017, 8, 1325. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.B.; Liu, H.; Wan, S.X.; Hua, K.K.; Wang, D.Z.; Guo, X.S.; He, C.L. Fertilisation practice changes rhizosphere microbial community structure in the agroecosystem. Ann. Appl. Biol. 2019, 174, 123–132. [Google Scholar] [CrossRef]
- Yang, Y.Y.; Chen, J.F.; Chen, X.L.; Jiang, Q.S.; Liu, Y.; Xie, S.G. Cyanobacterial Bloom Induces Structural and Functional Succession of Microbial Communities in Eutrophic Lake Sediments. Environ. Pollut. 2021, 284, 117157. [Google Scholar] [CrossRef]
- Kevin, P.; Wang, J.; Fernando, S.C.; Thompson, J.R. Secondary metabolite gene expression and interplay of bacterial functions in a tropical freshwater cyanobacterial bloom. ISME J. 2014, 8, 1866–1878. [Google Scholar]
- Wang, K.; Razzano, M.; Mou, X.Z. Cyanobacterial blooms alter the relative importance of neutral and selective processes in assembling freshwater bacterioplankton community. Sci. Total Environ. 2020, 706, 135724. [Google Scholar] [CrossRef]
- Zhang, H.H.; Jia, J.Y.; Chen, S.N.; Huang, T.L.; Wang, Y.; Zhao, Z.F.; Feng, J.; Hao, H.Y.; Li, S.L.; Ma, X.X. Dynamics of Bacterial and Fungal Communities during the Outbreak and Decline of an Algal Bloom in a Drinking Water Reservoir. Int. J. Environ. Res. Public Health 2018, 15, 361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerba, C.P.; Betancourt, W.Q.; Kitajima, M.; Rock, C.M. Reducing uncertainty in estimating virus reduction by advanced water treatment processes. Water Res. 2018, 133, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Costán-Longares, A.; Montemayor, M.; Payán, A.; Méndez, J.; Jofre, J.; Mujeriego, R.; Lucena, F. Microbial indicators and pathogens: Removal, relationships and predictive capabilities in water reclamation facilities. Water Res. 2008, 42, 4439–4448. [Google Scholar] [CrossRef] [PubMed]
- Steele, M.; Odumeru, J. Irrigation Water as Source of Foodborne Pathogens on Fruit and Vegetables. J. Food Prot. 2004, 67, 2839–2849. [Google Scholar] [CrossRef]
- Deviller, G.; Lundy, L.; Despo, F.K. Recommendations to derive quality standards for chemical pollutants in reclaimed water intended for reuse in agricultural irrigation. Chemosphere 2020, 240, 124911. [Google Scholar] [CrossRef]
- Hartemann, P.; Hautemaniere, A. Legionellosis prevention in France. Bundesgesundheitsblatt 2011, 54, 724–727. [Google Scholar] [CrossRef]
- Nogueira, R.; Utecht, K.U.; Exner, M.; Verstraete, W.; Rosenwinkel, K.H. Strategies for the reduction of Legionella in biological treatment systems. Water Sci. Technol. 2016, 74, 816–823. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency (USEPA). Risk Assessment Guidance for Superfund. Human Health Evaluation Manual, Part A; Office of Emergency and Remedial Response: Washington, DC, USA, 1989.
- Zhang, J.Z.; Zhang, H.X.; Li, L.W.; Wang, Q.; Yu, J.W.; Chen, Y.S. Microbial community analysis and correlation with 2-methylisoborneol occurrence in landscape lakes of Beijing. Environ. Res. 2020, 183, 109217. [Google Scholar] [CrossRef]
- Chen, Z.C.; Zhang, X.L.; Yang, Y.; Zhou, K.; Wragg, N.; Liu, Y.; Lewis, M.; Liu, C.Q. Fabrication and characterization of 3D complex hydroxyapatite scaffolds with hierarchical porosity of different features for optimal bioactive performance. Ceram. Int. 2017, 43, 336–344. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi, J.; Chaganti, S.R.; Shahraki, A.H.; Heath, D.D. Microbial community and abiotic effects on aquatic bacterial communities in north temperate lakes. Sci. Total Environ. 2021, 781, 146771. [Google Scholar] [CrossRef]
- Becerra-Castro, C.; Gonçalo, M.; Silva, A.M.T.; Manaia, C.M.; Nunes, O.C. Proteobacteria become predominant during regrowth after water disinfection. Sci. Total Environ. 2016, 573, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. Guide to the Natural History of Freshwater Lake Bacteria. Microbiol. Mol. Biol. 2011, 75, 14–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondol, M.A.; Shin, S.J.; Islam, M.T. Diversity of secondary metabolites from marine bacillus species: Chemistry and biological activity. Mar. Drugs 2013, 11, 2846–2872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Yang, J.; Amalfitano, S.; Yu, X.Q.; Liu, L.M. Effects of water stratification and mixing on microbial community structure in a subtropical deep reservoir. Sci. Rep. 2014, 4, 5821. [Google Scholar]
- Woodhouse, J.N.; Ongley, S.E.; Brown, M.V.; Neilan, B.A. Microbial diversity and diazotrophy associated with the freshwater non-heterocyst forming cyanobacterium lyngbya robusta. J. Appl. Phycol. 2013, 25, 1039–1045. [Google Scholar] [CrossRef]
- Sepa. Environmental Quality Standard for Surface Water (Eqssw), Gb3838–2002; Inspection and Quarantine of PR China: Beijing, China, 2002. (In Chinese) [Google Scholar]
- Jing, S.; Ji, D.F.; Lin, M.; Chen, Y.Q.; Sun, Y.Y.; Huo, L.L.; Zhu, J.C.; Xi, B.D. Developing Surface Water Quality Standards in China. Resour. Conserv. Recycl. 2017, 117, 294–303. [Google Scholar]
- Theodorou, C.M.; Stokes, S.C.; Hegazi, M.S.; Brown, E.G.; Saadai, P. Is pseudomonas infection associated with worse outcomes in pediatric perforated appendicitis? J. Pediatr. Surg. 2020, 56, 1826–1830. [Google Scholar] [CrossRef]
- Dong, P.Y.; Cui, Q.J.; Fang, T.T.; Huang, Y.; Wang, H. Occurrence of antibiotic resistance genes and bacterial pathogens in water and sediment in urban recreational water. J. Environ. Sci. 2019, 77, 65–74. [Google Scholar] [CrossRef]
- Cui, Q.J.; Fang, T.T.; Huang, Y.; Dong, P.Y.; Wang, H. Evaluation of bacterial pathogen diversity, abundance and health risks in urban recreational water by amplicon next-generation sequencing and quantitative PCR. J. Environ. Sci. 2017, 57, 137–149. [Google Scholar] [CrossRef]
- Ranjbar, M.; Behrouz, B.; Norouzi, F.; Gargari, S.L.M. Anti-Pcrv Igy antibodies protect against pseudomonas aeruginosa infection in both acute pneumonia and burn wound models. Mol. Immunol. 2019, 116, 98–105. [Google Scholar] [CrossRef]
- El-Saed, A.; Balkhy, H.H.; Al-Dorzi, H.M.; Khan, R.; Rishu, A.H.; Arabi, Y.M. Acinetobacter is the most common pathogen associated with late-onset and recurrent ventilator-associated pneumonia in an adult intensive care unit in saudi arabia. Int. J. Infect. Dis. 2013, 17, e696–e701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebringer, A.; Rashid, T.; Wilson, C. Chapter 29—Multiple sclerosis and creutzfeldt-jakob disease are autoimmune diseases probably caused by exposure to the nasal microbe Acinetobacter. In Infection and Autoimmunity, 2nd ed.; Shoenfeld, Y., Agmon-Levin, N., Rose, N.R., Eds.; Academic Press: Amsterdam, The Netherlands, 2015; pp. 505–517. [Google Scholar]
- Te, S.H.; Tan, B.F.; Thompson, J.R.; Gin, K.Y.H. Relationship of microbiota and cyanobacterial secondary metabolites in planktothricoides-dominated bloom. Environ. Sci. Technol. 2017, 51, 4199–4209. [Google Scholar] [CrossRef] [PubMed]
- Cuzman, O.A.; Rescic, S.; Richter, K.; Wittig, L.; Tiano, P. Sporosarcina pasteurii use in extreme alkaline conditions for recycling solid industrial wastes. J. Biotechnol. 2015, 214, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, X.; Wang, Y.; Zhang, S.; Zhang, G.; Han, Y.; Li, M.; Liu, L. Spatial and temporal dynamics of microbial community composition and factors influencing the surface water and sediments of urban rivers. J. Environ. Sci. 2023, 124, 187–197. [Google Scholar] [CrossRef]
- Bunse, C.; Bertos-Fortis, M.; Sassenhagen, I.; Sildever, S.; Sjöqvist, C.; Godhe, A.; Gross, S.; Kremp, A.; Lips, I.; Lundholm, N.; et al. Spatio-temporal interdependence of bacteria and phytoplankton during a baltic sea spring bloom. Front. Microbiol. 2016, 7, 517. [Google Scholar] [CrossRef] [Green Version]
- Bleves, S.; Viarre, V.; Salacha, R.; Michel, G.P.F.; Filloux, A.; Voulhoux, R. Protein secretion systems in pseudomonas aeruginosa: A wealth of pathogenic weapons. Int. J. Med. Microbiol. 2010, 300, 534–543. [Google Scholar] [CrossRef]
- Curran, C.S.; Bolig, T.; Torabi-Parizi, P. Mechanisms and targeted therapies for Pseudomonas aeruginosa lung infection. Am. J. Respir. Crit. Care Med. 2018, 197, 708–727. [Google Scholar] [CrossRef]
- Casciaro, B.; d’Angelo, I.; Zhang, X.; Loffredo, M.R.; Conte, G.; Cappiello, F.; Quaglia, F.; Di, Y.P.P.; Ungaro, F.; Mangoni, M.L. Poly (Lactide-Co-Glycolide) Nanoparticles for prolonged therapeutic efficacy of esculentin-1a-derived antimicrobial peptides against pseudomonas aeruginosa lung infection: In vitro and in vivo studies. Biomacromolecules 2019, 20, 1876–1888. [Google Scholar] [CrossRef]
- Savini, V.; Fazii, P.; Favaro, M.; Astolfi, D.; Polilli, E.; Pompilio, A.; Vannucci, M.; D’Amario, C.; Bonaventura, G.D.; Fontana, C.; et al. Tuberculosis-Like pneumonias by the aerobic actinomycetes Rhodococcus, Tsukamurella and Gordonia. Microbes Infect. 2012, 14, 401–410. [Google Scholar] [CrossRef]
- Zhang, H.H.; Wang, Y.; Chen, S.N.; Zhao, Z.F.; Feng, J.; Zhang, Z.H.; Lu, K.Y.; Jia, J.Y. Water Bacterial and Fungal Community Compositions Associated with Urban Lakes, Xi’an, China. Int. J. Environ. Res. Public Health 2018, 15, 469. [Google Scholar] [CrossRef] [Green Version]
- Fang, T.T.; Cui, Q.J.; Huang, Y.; Dong, P.Y.; Wang, H.; Liu, W.-T.; Ye, Q.H. Distribution Comparison and Risk Assessment of Free-Floating and Particle-Attached Bacterial Pathogens in Urban Recreational Water: Implications for Water Quality Management. Sci. Total Environ. 2018, 613–614, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Perkins, S.D.; Shaw, M.; Nichols, T.L. Evaluation of exposure to brevundimonas diminuta and Pseudomonas aeruginosa during showering. J. Aerosol. Sci. 2017, 114, 77–93. [Google Scholar] [CrossRef] [PubMed]
- Montoya, J.E.M.; Moran, M.A.M.; Ardila, J.A.B.; Henao, P.G.C.; Rodriguez, E.E.M.; Meza, G.A.C. Brain abscess by Kocuria Rosea: Case report and literature review. Interdiscip. Neurosurg. 2017, 7, 59–61. [Google Scholar] [CrossRef]
- Moreira, J.S.; Riccetto, A.G.L.; Silva, M.T.N.D.; Vilela, M.M.D.S. Endocarditis by Kocuria Rosea in an immunocompetent child. Braz. J. Infect. Dis. 2015, 19, 82–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baruah, M.; Lyngdoh, C.; Lyngdoh, W.V.; Talukdar, R. Noncatheter-related bacteraemia due to Chryseobacterium Indologenes in an immunocompetent patient. Indian J. Med. Microbiol. 2016, 34, 380–381. [Google Scholar] [CrossRef]
- Bhuyar, G.; Jain, S.; Shah, H.; Mehta, V.K. Urinary tract infection by Chryseobacterium Indologenes. Indian J. Med. Microbiol. 2012, 30, 370–372. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Loria, R. Virulence mechanisms of Gram-Positive plant pathogenic bacteria. Curr. Opin. Plant Biol. 2008, 11, 449–456. [Google Scholar] [CrossRef]
- Loria, R.; Kers, J.; Madhumita, J. Evolution of plant pathogenicity in Streptomyces. Annu. Rev. Phytopathol. 2006, 44, 469–487. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, B.; Ning, D.L.; Zhang, Y.; Dai, T.J.; Wu, L.W.; Li, T.L.; Liu, W.; Zhou, J.Z.; Wen, X.H. Seasonal dynamics of the microbial community in two full-scale wastewater treatment plants: Diversity, composition, phylogenetic group based assembly and co-occurrence pattern. Water Res. 2021, 200, 117295. [Google Scholar]

| Parameters | Value | Parameters | Value |
|---|---|---|---|
| T (°C) | 23.0~30.0 | TN (mg/L) | 3.70 ± 0.87 |
| pH | 7.21~8.85 | TP (mg/L) | 0.024 ± 0.009 |
| NH4+-N (mg/L) | 0.0851 ± 0.0012 | PO43−-P (mg/L) | 0.0177 ± 0.0003 |
| NO3− (mg/L) | 5.31 ± 0.55 | OD680 (cm−1) | 0.457 ± 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; He, X.; Zhang, H.; Liao, Y.; Wang, Q.; Li, L.; Yu, J. Factors Driving Microbial Community Dynamics and Potential Health Effects of Bacterial Pathogen on Landscape Lakes with Reclaimed Water Replenishment in Beijing, PR China. Int. J. Environ. Res. Public Health 2022, 19, 5127. https://doi.org/10.3390/ijerph19095127
Zhang J, He X, Zhang H, Liao Y, Wang Q, Li L, Yu J. Factors Driving Microbial Community Dynamics and Potential Health Effects of Bacterial Pathogen on Landscape Lakes with Reclaimed Water Replenishment in Beijing, PR China. International Journal of Environmental Research and Public Health. 2022; 19(9):5127. https://doi.org/10.3390/ijerph19095127
Chicago/Turabian StyleZhang, Junzhi, Xiao He, Huixin Zhang, Yu Liao, Qi Wang, Luwei Li, and Jianwei Yu. 2022. "Factors Driving Microbial Community Dynamics and Potential Health Effects of Bacterial Pathogen on Landscape Lakes with Reclaimed Water Replenishment in Beijing, PR China" International Journal of Environmental Research and Public Health 19, no. 9: 5127. https://doi.org/10.3390/ijerph19095127
APA StyleZhang, J., He, X., Zhang, H., Liao, Y., Wang, Q., Li, L., & Yu, J. (2022). Factors Driving Microbial Community Dynamics and Potential Health Effects of Bacterial Pathogen on Landscape Lakes with Reclaimed Water Replenishment in Beijing, PR China. International Journal of Environmental Research and Public Health, 19(9), 5127. https://doi.org/10.3390/ijerph19095127






