Salinity as a Determinant Structuring Microbial Communities in Coastal Lakes
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling
2.3. Physico-Chemical Analyses of Water
2.4. Microbial Community Analyses
2.5. Statistical Analyses
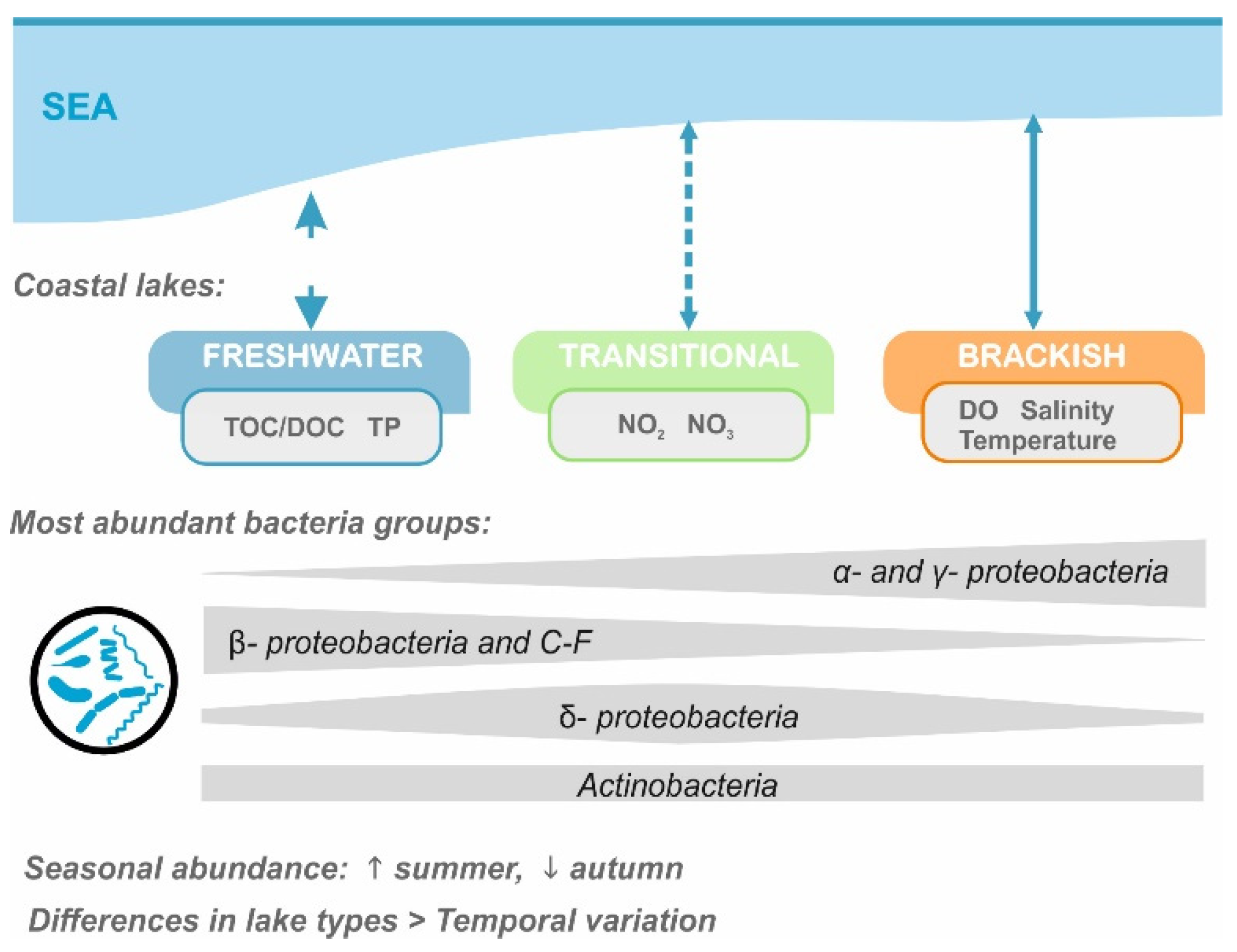
3. Results
3.1. Physico-Chemical Parameters of Water
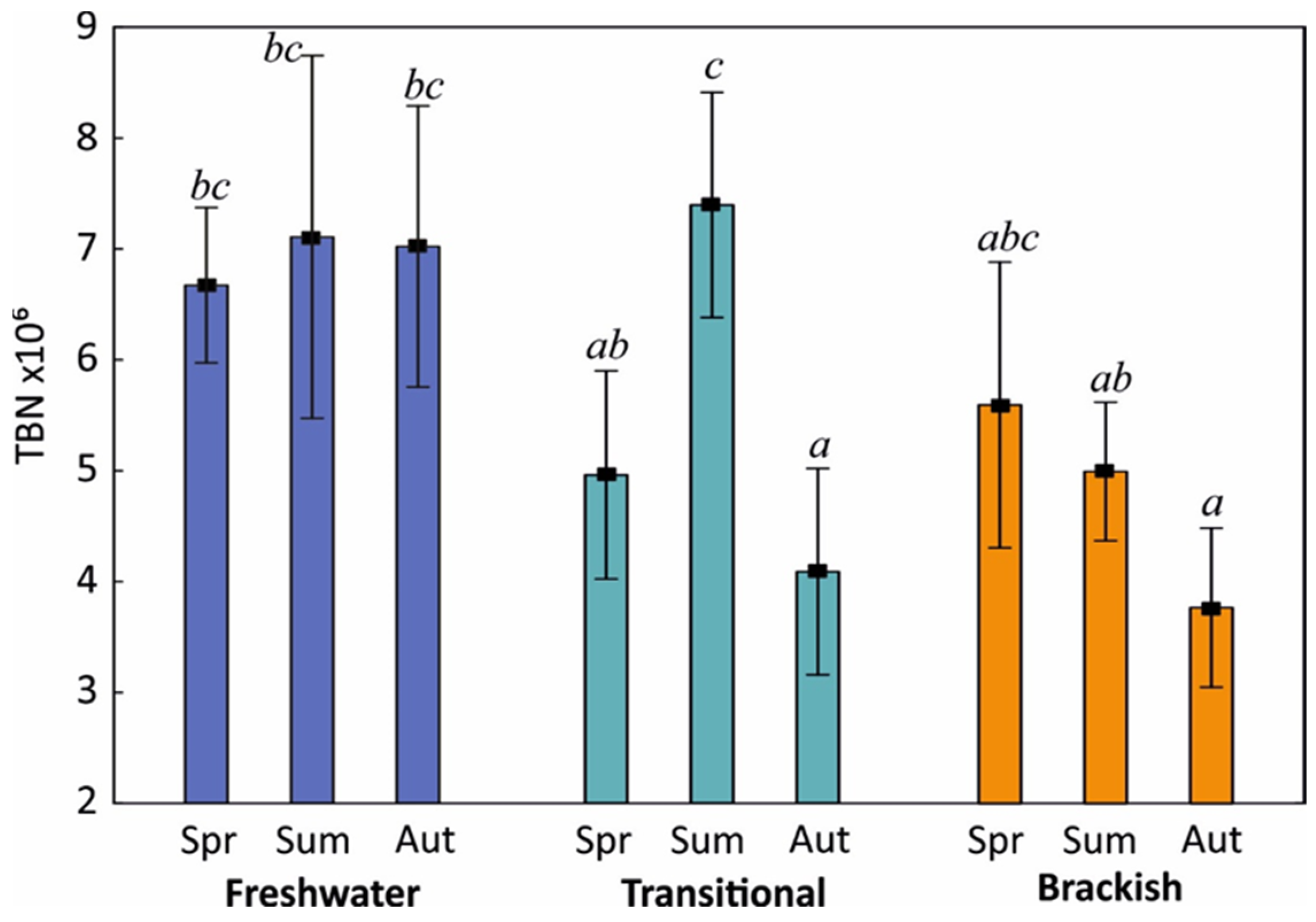
3.2. Bacterioplankton Characteristics in Coastal Lakes
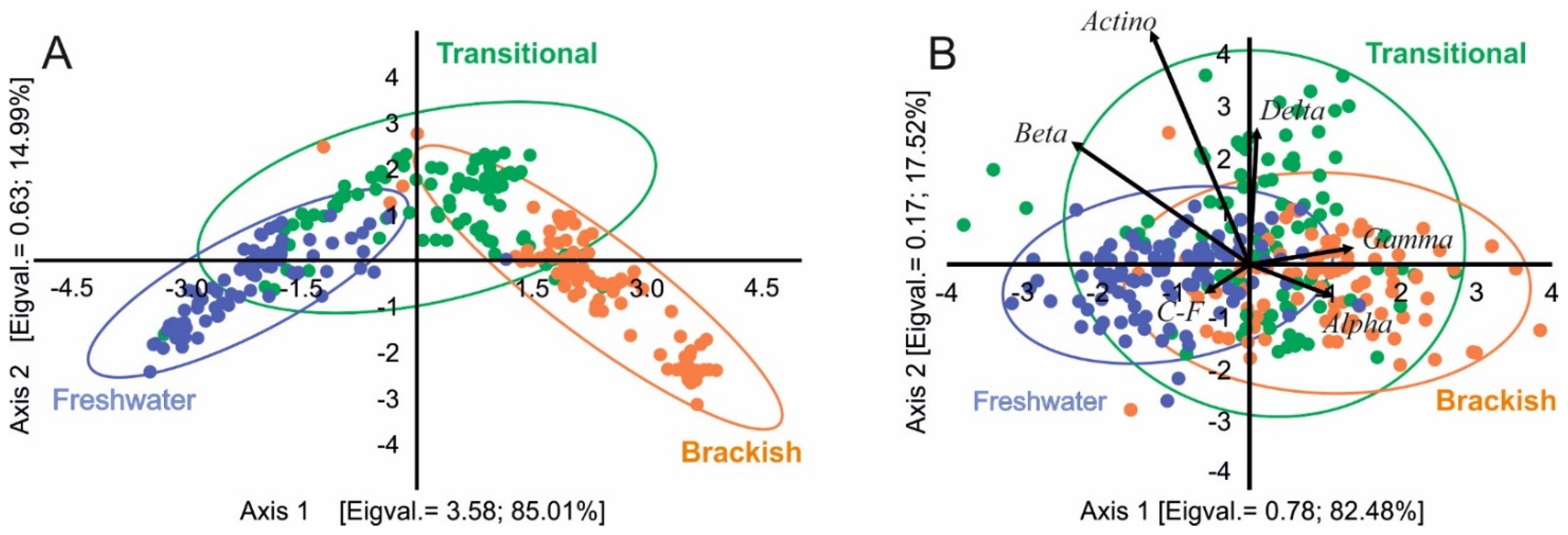
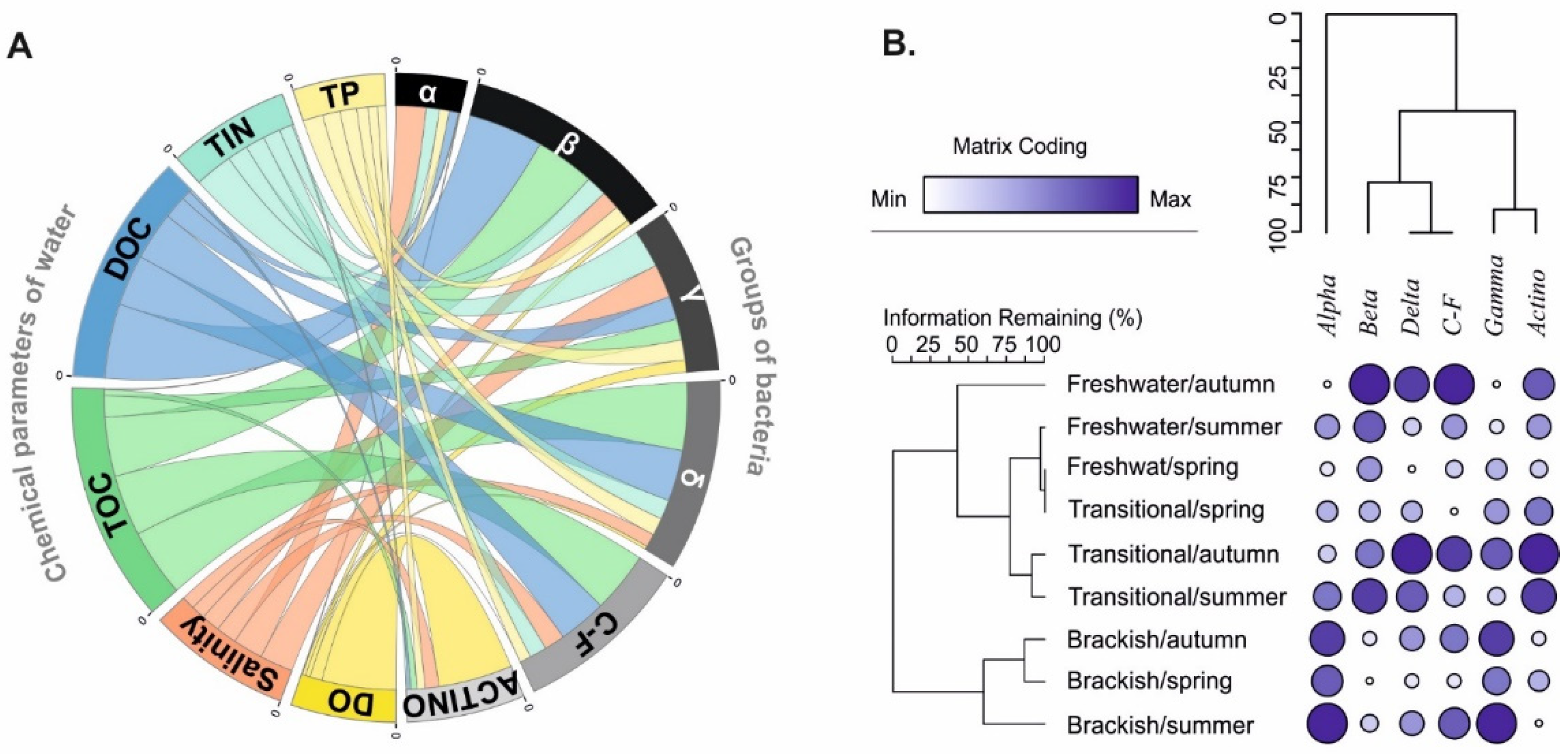
3.3. Environmental Determinants of Bacterial Communities Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jeffries, T.C.; Schmitz Fontes, M.L.; Harrison, D.P.; Van-Dongen-Vogels, V.; Eyre, B.D.; Ralph, P.J.; Seymour, J.R. Bacterioplankton dynamics within a large anthropogenically impacted urban estuary. Front. Microbiol. 2016, 6, 1438. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Vincx, M.; Herman, P.M.J.; Heip, C. Monitoring meiobenthos using cm-, m- and km-scales in the Southern Bight of the North Sea. Mar. Environ. Res. 1997, 43, 265–278. [Google Scholar] [CrossRef]
- Wang, K.; Ye, X.; Chen, H.; Zhao, Q.; Hu, C.; He, J.; Qian, Y.; Xiong, J.; Zhu, J.; Zhang, D. Bacterial biogeography in the coastal waters of Northern Zhejiang, East China Sea is highly controlled by spatially structured environmental gradients. Environ. Microbiol. 2015, 17, 3898–3913. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pan, J.; Yang, J.; Zhou, Z.; Pan, Y.; Li, M. Patterns and processes of free-living and particle-associated bacterioplankton and archaeaplankton communities in a subtropical river-bay system in South China. Limnol. Oceanogr. 2020, 65, 161–179. [Google Scholar] [CrossRef]
- Morrow, K.M.; Fiore, C.L.; Lesser, M.P. Environmental drivers of microbial community shifts in the giant barrel sponge, Xestospongia Muta, over a shallow to mesophotic depth gradient. Environ. Microbiol. 2016, 18, 2025–2038. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shen, T.; Cheng, Y.; Zhao, T.; Li, L.; Qi, P. Temporal and spatial variations in the bacterial community composition in Lake Bosten, a large, brackish lake in China. Sci. Rep. 2020, 10, 304. [Google Scholar] [CrossRef]
- Rath, K.M.; Fierer, N.; Murphy, D.V.; Rousk, J. Linking Bacterial Community Composition to Soil Salinity along Environmental Gradients. ISME J. 2019, 13, 836–846. [Google Scholar] [CrossRef]
- de Souza Silva, C.M.M.; Francisconi, E. Effect of salinity on soil microorganisms. In Soil Health and Land Use Management; Hernandez Soriano, M.C., Ed.; InTech Press: Rijeka, Croatia, 2012; pp. 177–198. [Google Scholar] [CrossRef]
- Andronov, E.E.; Petrova, S.N.; Pinaev, A.G.; Pershina, E.V.; Rakhimgalieva, S.Z.; Akhmedenov, K.M.; Gorobets, A.V.; Sergaliev, N.K. Analysis of the structure of microbial community in soils with different degrees of salinization using T-RFLP and Real-Time PCR techniques. Eurasian Soil Sci. 2012, 45, 147–156. [Google Scholar] [CrossRef]
- Obolewski, K.; Glińska-Lewczuk, K.; Bąkowska, M.; Szymańska, M.; Mrozińska, N. Patterns of phytoplankton composition in coastal lakes differed by connectivity with the Baltic Sea. Sci. Total Environ. 2018, 631–632, 951–961. [Google Scholar] [CrossRef]
- Ghosh, A.; Bhadury, P. Exploring Biogeographic patterns of bacterioplankton communities across global estuaries. Microbiologyopen 2019, 8, e00741. [Google Scholar] [CrossRef]
- Grizzetti, B.; Liquete, C.; Pistocchi, A.; Vigiak, O.; Zulian, G.; Bouraoui, F.; de Roo, A.; Cardoso, A.C. Relationship between ecological condition and ecosystem services in european rivers, lakes and coastal waters. Sci. Total Environ. 2019, 671, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Intergovernmental Panel on Climate Change. IPCC (2001a) Special Report on the Regional Impacts of Climate Change. Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2001; Available online: https://www.ipcc.ch (accessed on 10 January 2022).
- Intergovernmental Panel on Climate Change. IPCC (2001b) Third Report of the Working Group of the Intergovernmental Panel on Climate Change. Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2001; Available online: https://www.ipcc.ch (accessed on 10 January 2022).
- Anonymous, J. Symposium on the Classification of Brackish Waters. Venice, 8–14 April 1958. The Venice System for the classification of marine waters according to salinity. Limnol. Oceanogr. 1958, 3, 346–347. [Google Scholar] [CrossRef]
- Langenheder, S.; Kisand, V.; Wikner, J.; Tranvik, L.J. Salinity as a structuring factor for the composition and performance of bacterioplankton degrading riverine DOC. FEMS Microbiol. Ecol. 2003, 45, 189–202. [Google Scholar] [CrossRef]
- Ward, C.S.; Yung, C.-M.; Davis, K.M.; Blinebry, S.K.; Williams, T.C.; Johnson, Z.I.; Hunt, D.E. Annual community patterns are driven by seasonal switching between closely related marine bacteria. ISME J. 2017, 11, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, C.S.; Herfort, L.; Zuber, P.; Baptista, A.M.; Crump, B.C. Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. ISME J. 2012, 6, 554–563. [Google Scholar] [CrossRef]
- Joshi, P.; Pande, V.; Joshi, P. Microbial diversity of aquatic ecosystem and its industrial potential. J. Bacteriol. Mycol. Open Access 2016, 3, 177–179. [Google Scholar] [CrossRef]
- Piwosz, K.; Salcher, M.M.; Zeder, M.; Ameryk, A.; Pernthaler, J. Seasonal dynamics and activity of typical freshwater bacteria in brackish waters of the Gulf of Gdańsk. Limnol. Oceanogr. 2013, 58, 817–826. [Google Scholar] [CrossRef]
- Reissmann, J.H.; Burchard, H.; Feistel, R.; Hagen, E.; Lass, H.U.; Mohrholz, V.; Nausch, G.; UmLauf, L.; Wieczorek, G. Vertical mixing in the Baltic Sea and consequences for eutrophication—A review. Prog. Oceanogr. 2009, 82, 47–80. [Google Scholar] [CrossRef]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Herlemann, D.P.R.; Lundin, D.; Andersson, A.F.; Labrenz, M.; Jürgens, K. Phylogenetic signals of salinity and season in bacterial community composition across the salinity gradient of the Baltic Sea. Front. Microbiol. 2016, 7, 1883. [Google Scholar] [CrossRef]
- Pawlak, J.F.; Laamanen, M.; Andersen, J.H. HELCOM 2009 Eutrophication in the Baltic Sea—An Integrated Thematic Assessment of the Effects of Nutrient Enrichment and Eutrophication in the Baltic Sea Region: Executive Summary. Baltic Sea Environment Proceedings No. 115A; Helsinki Commission: Helsinki, Finland, 2009. [CrossRef]
- Devictor, V.; Clavel, J.; Julliard, R.; Lavergne, S.; Mouillot, D.; Thuiller, W.; Venail, P.; Villéger, S.; Mouquet, N. Defining and Measuring Ecological Specialization. J. Appl. Ecol. 2010, 47, 15–25. [Google Scholar] [CrossRef]
- Shade, A.; Peter, H.; Allison, S.D.; Baho, D.L.; Berga, M.; Bürgmann, H.; Huber, D.H.; Langenheder, S.; Lennon, J.T.; Martiny, J.B.H.; et al. Fundamentals of microbial community resistance and resilience. Front. Microbiol. 2012, 3, 417. [Google Scholar] [CrossRef] [PubMed]
- Shen, D. Aquatic Bacterioplankton Communities along Salinity Gradients: Insights into Composition, Community Assembly and Functional Traits. University in Rostock, Germany. 2018. Available online: https://rosdok.uni-rostock.de/file/rosdok_disshab_0000001953/rosdok_derivate_0000057192/Dissertation_Shen_2018.pdf (accessed on 10 January 2022).
- Székely, A.J.; Berga, M.; Langenheder, S. Mechanisms determining the fate of dispersed bacterial communities in new environments. ISME J. 2013, 7, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.Y.; Dafforn, K.A.; Brown, M.V.; Johnston, E.L. Bacterial communities are sensitive indicators of contaminant stress. Mar. Pollut. Bull. 2012, 64, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Lew, S.; Glińska-Lewczuk, K.; Ziembińska-Buczyńska, A. Prokaryotic community composition affected by seasonal changes in physicochemical properties of water in peat bog lakes. Water 2018, 10, 485. [Google Scholar] [CrossRef]
- Selak, L.; Osterholz, H.; Stanković, I.; Hanžek, N.; Udovič, M.G.; Dittmar, T.; Orlić, S. Adaptations of microbial communities and dissolved organics to seasonal pressures in a mesotrophic coastal Mediterranean lake. Environ. Microbiol. 2022; online version. [Google Scholar] [CrossRef]
- Laas, P.; Ugarelli, K.; Travieso, R.; Stumpf, S.; Gaiser, E.E.; Kominoski, J.S.; Stingl, U. Water column microbial communities vary along salinity gradients in the Florida Coastal Everglades wetlands. Microorganisms 2022, 10, 215. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Amann, R.; Alfreider, A.; Pernthaler, J.; Psenner, R.; Trebesius, K.; Schleifer, K.-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 1996, 19, 403–406. [Google Scholar] [CrossRef]
- Pernthaler, J.; Glöckner, F.-O.; Schönhuber, W.; Amann, R. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. J. Microbiol. Methods 2001, 30, 207–226. [Google Scholar] [CrossRef]
- Stoecker, K.; Dorninger, C.; Daims, H.; Wagner, M. Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl. Environ. Microbiol. 2010, 76, 922–926. [Google Scholar] [CrossRef]
- Loy, A.; Lehner, A.; Lee, N.; Adamczyk, J.; Meier, H.; Ernst, J.; Schleifer, K.-H.; Wagner, M. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 2002, 68, 5064–5081. [Google Scholar] [CrossRef]
- Manz, W.; Amann, R.; Ludwig, W.; Wagner, M.; Schleifer, K.-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: Problems and solutions. Syst. Appl. Microbiol. 1992, 15, 593–600. [Google Scholar] [CrossRef]
- Roller, C.; Wagner, M.; Amann, R.; Ludwig, W.; Schleifer, K.-H. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 1995, 141, 1267. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lew, S.; Lew, M.; Mieszczyński, T.; Szarek, J. Selected fluorescent techniques for identification of the physiological state of individual water and soil bacterial cells—Review. Folia Microbiol. 2010, 55, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Hammer, D.A.T.; Ryan, P.D.; Hammer, Ø.; Harper, D.A.T. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- McCune, B.; Mefford, M.J. PC-ORD. Multivariate Analysis of Ecological Data, Version 6; MjM Software: Gleneden Beach, OR, USA, 2011.
- Dufrene, M.; Legendre, P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol. Monogr. 1997, 67, 345. [Google Scholar] [CrossRef]
- Obolewski, K.; Glińska-Lewczuk, K.; Sidoruk, M.; Szymańska, M.M. Response of benthic fauna to habitat heterogeneity in a shallow temperate lake. Animals 2021, 11, 2488. [Google Scholar] [CrossRef]
- Obolewski, K.; Glińska-Lewczuk, K. Connectivity and complexity of coastal lakes as determinants for their restoration—A case study of the Southern Baltic Sea. Ecol. Eng. 2020, 155, 105948. [Google Scholar] [CrossRef]
- Euler, S.; Jeffrey, L.C.; Maher, D.T.; Mackenzie, D.; Tait, D.R. Shifts in methanogenic archaea communities and methane dynamics along a subtropical estuarine land use gradient. PLoS ONE 2020, 15, e0242339. [Google Scholar] [CrossRef]
- Crump, B.C.; Hopkinson, C.S.; Sogin, M.L.; Hobbie, J.E. Microbial biogeography along an estuarine salinity gradient: Combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 2004, 70, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Kirchman, D.L.; Dittel, A.I.; Malmstrom, R.R.; Cottrell, M.T. Biogeography of major bacterial groups in the Delaware estuary. Limnol. Oceanogr. 2005, 50, 1697–1706. [Google Scholar] [CrossRef]
- Lew, S.; Glińska-Lewczuk, K. Environmental controls on the abundance of methanotrophs and methanogens in peat bog lakes. Sci. Total Environ. 2018, 645, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Nold, S.C.; Zwart, G. Patterns and governing forces in aquatic microbial communities. Aquat. Ecol. 1998, 32, 17–35. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Fuchs, B.M.; Amann, R. Bacterioplankton compositions of lakes and oceans: A first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 1999, 65, 3721–3726. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, M.; Huang, T.; Miao, Y.; Li, H.; Liu, K.; Yang, W.; Ma, B. Spatial and temporal dynamics of actinobacteria in drinking water reservoirs: Novel sights into abundance, community structure, and co-existence model. Sci. Total Environ. 2022, 814, 152804. [Google Scholar] [CrossRef]
- Newton, R.J.; Jones, S.E.; Eiler, A.; McMahon, K.D.; Bertilsson, S. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 2011, 75, 14–49. [Google Scholar] [CrossRef]
- Weyman-Kaczmarkowa, W. Interdependencies between oligotrophic and copiotrophic bacteria in soils of different mechanical structure. Pol. J. Soil Sci. 1996, 29, 62–72. [Google Scholar]
- Glöckner, F.O.; Zaichikov, E.; Belkova, N.; Denissova, L.; Pernthaler, J.; Pernthaler, A.; Amann, R. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 2000, 66, 5053–5065. [Google Scholar] [CrossRef]
- Holmfeldt, K.; Dziallas, C.; Titelman, J.; Pohlmann, K.; Grossart, H.-P.; Riemann, L. Diversity and abundance of freshwater Actinobacteria along environmental gradients in the brackish northern Baltic Sea. Environ. Microbiol. 2009, 11, 2042–2054. [Google Scholar] [CrossRef]
- Mohapatra, M.; Behera, P.; Kim, J.Y.; Rastogi, G. Seasonal and spatial dynamics of bacterioplankton communities in a brackish water coastal lagoon. Sci. Total Environ. 2020, 705, 134729. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, T.C.; del Giorgio, P.A. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 2002, 47, 453–470. [Google Scholar] [CrossRef]
- Eiler, A.; Bertilsson, S. Flavobacteria blooms in four eutrophic lakes: Linking population dynamics of freshwater bacterioplankton to resource availability. Appl. Environ. Microbiol. 2007, 73, 3511–3518. [Google Scholar] [CrossRef] [PubMed]
- Rissanen, A.J.; Saarenheimo, J.; Tiirola, M.; Peura, S.; Aalto, S.L.; Karvinen, A.; Nykänen, H. Gammaproteobacterial methanotrophs dominate methanotrophy in aerobic and anaerobic layers of boreal lake waters. Aquat. Microb. Ecol. 2018, 81, 257–276. [Google Scholar] [CrossRef]
- Hu, Y.O.; Karlson, B.; Charvet, S.; Andersson, A.F. Diversity of pico to mesoplankton along the 2000 km salinity gradient of the Baltic Sea. Front. Microbiol. 2016, 7, 679. [Google Scholar] [CrossRef]
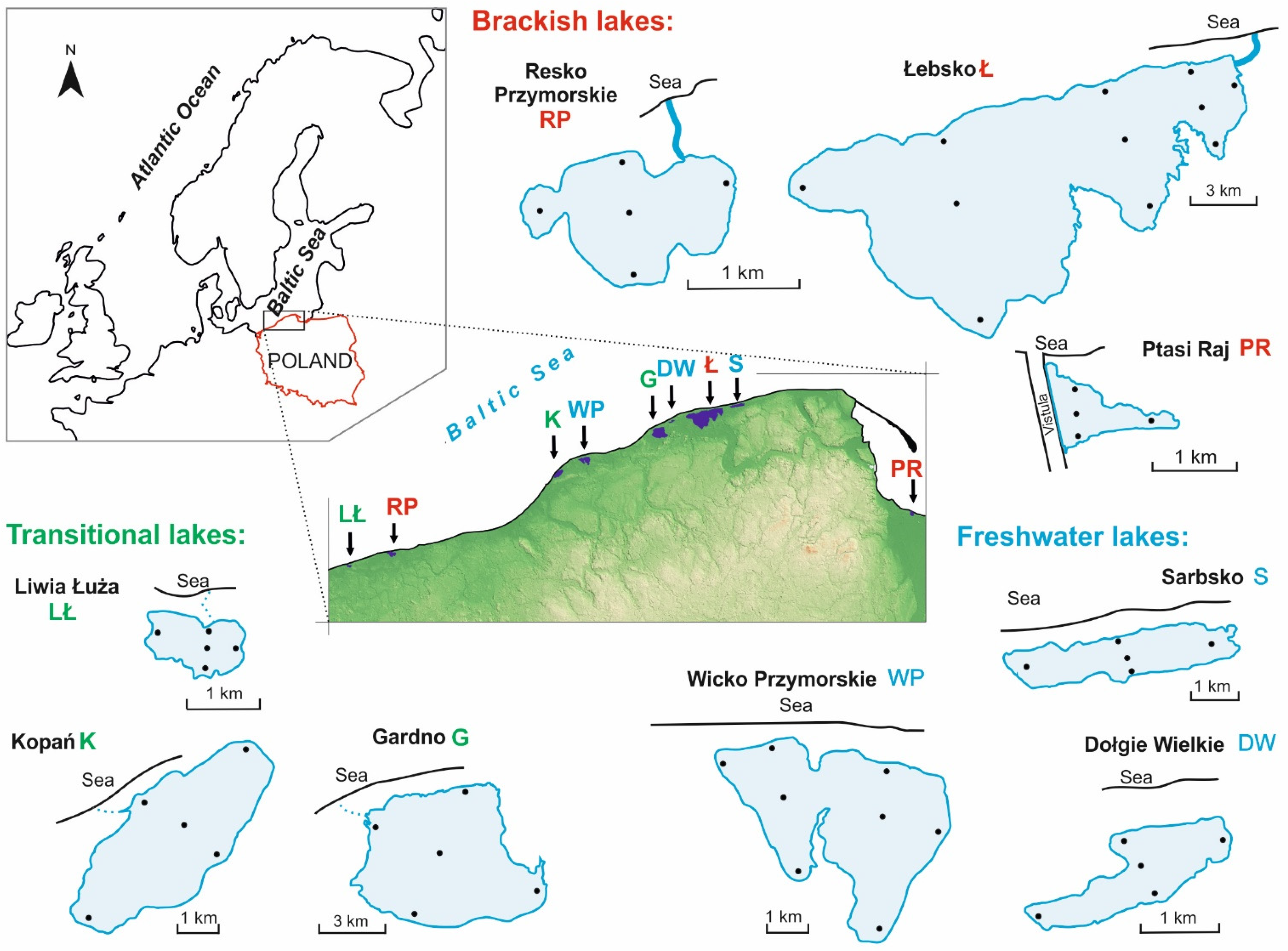
| Lake Name | Geographic Coordinates | Area (ha) | Mean Depth (m) | Type of Lake According to Venetian Classification | Habitat |
|---|---|---|---|---|---|
| Liwia Łuża | 54°05′ N, 15°06′ E | 172 | 1.0 | Limnetic/oligohaline | Transitional |
| Resko Przymorskie | 54°09′ N, 15°21′ E | 554 | 1.6 | Mesohaline | Brackish |
| Wicko Przymorskie | 54°33′ N, 16°38′ E | 977 | 2.4 | Limnetic | Freshwater |
| Kopań | 54°29′ N, 16°27′ E | 753 | 1.5 | Limnetic/Oligohaline | Transitional |
| Gardno | 54°39′ N, 17°07′ E | 2261 | 1.4 | Oligohaline /Limnetic | Transitional |
| Łebsko | 54°43′ N, 17°25′ E | 7020 | 1.6 | Oligohaline/Mesohaline | Brackish |
| Dołgie Wielkie | 54°42′ N, 17°12′ E | 136 | 1.4 | Limnetic | Freshwater |
| Sarbsko | 54°46′ N, 17°38′ E | 610 | 1.2 | Limnetic | Freshwater |
| Ptasi Raj | 54°22′ N, 18°48′ E | 53 | 1.2 | Mesohaline | Brackish |
| Probe | Sequence | Target rRNA | Specificity | FA ** |
|---|---|---|---|---|
| [%] | ||||
| EUB338 | 5’-GCT GCC TCC CGT AGG AGT-3’ | 16S | Most bacteria | 35 |
| EUB338II | 5’-GCA GCC ACC CGT AGG TGT-3’ | 16S | Planctomycetales | 35 |
| EUB338III | 5’-GCT GCC ACC CGT AGG TGT-3’ | 16S | Verrucomicrobiales | 35 |
| NON338 | 5’-ACT CCT ACG GGA GGC AGC-3’ | 16S | Control probe complementary to EUB338 | 35 |
| ALF968 | 5’-GGT AAG GTT CTG CGC GTT-3’ | 16S | Alphaproteobacteria, except of Rickettsiales | 20 |
| BET42a | 5’-GCC TTC CCA CTT CGT TT-3’ | 23S | Betaproteobacteria | 35 |
| 5’-GCC TTC CCA CAT CGT TT-3’ * | ||||
| GAM42a | 5’-GCC TTC CCA CAT CGT TT-3’ | 23S | Gammaproteobacteria | 35 |
| 5’-GCC TTC CCA CTT CGT TT-3’ * | ||||
| DELTA495a | 5’-AGT TAG CCG GTG CTT CCT-3’ | 16S | Most Deltaproteobacteria | 35 |
| 5’-AGT TAG CCG GTG CTT CTT-3’ * | and most Gemmatimonadetes | |||
| DELTA495b | 5’-AGT TAG CCG GCG CTT CCT-3’ | 16S | Some Deltaproteobacteria | 35 |
| 5’-AGT TAG CCG GCG CTT CKT-3’ * | ||||
| DELTA495c | 5’-AAT TAG CCG GTG CTT CCT-3’ | 16S | Some Deltaproteobacteria | 35 |
| 5’-AAT TAG CCG GTG CTT CTT-3’ * | ||||
| CF319a | 5’-TGG TCC GTG TCT CAG TAC-3’ | 16S | Most Flavobacteria, some Bacteroidetes, some Sphingobacteria | 35 |
| HGC69a | 5’-TAT AGT TAC CAC CGC CGT-3’ | 23S | Actinobacteria (high G+C Gram-positive bacteria) | 25 |
| 5’-TAT AGT TAC GGC CGC CGT-3’ * |
| Brackish (N = 90) | Transitional (N = 90) | Freshwater (N = 102) | |||||
|---|---|---|---|---|---|---|---|
| Mean | ±SD | Mean | ±SD | Mean | ±SD | ||
| EC | µS/cm | 6731 c | 3567 | 1802 b | 1150 | 342 a | 254 |
| pH * | - | 8.43 a | - | 8.80 c | - | 8.65 b | - |
| DO | % | 92.05 a | 27.37 | 113.68 c | 24.05 | 102.18 b | 22.34 |
| Chl-a | µg/L | 23.26 a | 32.84 | 30.601 a | 46.56 | 71.31 b | 132.97 |
| Salinity | PSU | 5.02 c | 2.83 | 1.17 b | 0.86 | 0.24 a | 0.15 |
| TOC | mg/L | 12.74 a | 5.05 | 20.02 b | 13.87 | 21.623 b | 13.97 |
| DOC | mg/L | 7.18 a | 3.23 | 11.55 b | 6.91 | 12.11 b | 5.29 |
| N-NO2− | mg/L | 0.006 b | 0.001 | 0.006 b | 0.001 | 0.005 a | 0.001 |
| N-NO3− | mg/L | 0.89 b | 0.73 | 0.76 b | 0.58 | 0.55 a | 0.42 |
| N-NH4+ | mg/L | 0.37 b | 0.32 | 0.378 b | 0.37 | 0.19 a | 0.13 |
| TIN | mg/L | 1.25 b | 0.72 | 1.14 b | 0.61 | 0.75 a | 0.40 |
| TP | mg/L | 0.40 | 0.24 | 0.34 | 0.28 | 0.38 | 0.25 |
| P-PO43− | mg/L | 0.14 b | 0.139 | 0.10 a | 0.080 | 0.13 b | 0.094 |
| Freshwater (N = 102) | Transitional (N = 90) | Brackish (N = 90) | ||||
|---|---|---|---|---|---|---|
| Mean | ±SD | Mean | ±SD | Mean | ±SD | |
| Alphaproteobacteria | 4.37 c | 1.12 | 5.27 b | 1.94 | 6.70 a | 1.47 |
| Betaproteobacteria | 17.25 a | 4.70 | 16.13 a | 6.33 | 12.22 b | 3.74 |
| Gammaproteobacteria | 9.51 b | 3.80 | 11.42 a | 3.11 | 12.23 a | 3.03 |
| Deltaproteobacteria | 10.27 a | 3.20 | 12.33 b | 5.09 | 10.10 a | 3.74 |
| Cytophaga-Flavobacterium | 15.21 a | 4.67 | 14.06 a | 4.81 | 14.09 a | 4.54 |
| Actinobacteria | 30.07 a | 6.83 | 31.79 a | 5.74 | 26.82 b | 10.05 |
| Total (TBN × 106 cells/mL) | 6.93 b | 3.58 | 5.48 a | 2.91 | 4.79 a | 2.26 |
| Groups of Bacteria | |||||||
|---|---|---|---|---|---|---|---|
| Actino | Beta- | C-F | Delta- | Gamma- | Alpha- | ||
| Average dissimilarity | 5.0 | 3.5 | 3.14 | 2.85 | 2.38 | 1.3 | |
| Contribution % | 27.6 | 19.3 | 17.23 | 15.65 | 13.04 | 7.2 | |
| Cumulative % | 27.6 | 46.9 | 64.2 | 79.8 | 92.9 | 100.0 | |
| Mean | Freshwater | 30.1 | 17.3 | 15.2 | 10.3 | 9.51 | 4.4 |
| Transitional | 31.8 | 16.1 | 14.1 | 12.3 | 11.4 | 5.3 | |
| Brackish | 26.8 | 12.2 | 14.1 | 10.1 | 12.2 | 6.7 | |
| Bacteria Group | Group Identifier for Group with Maximum Observed | Observed Indicator Value (IndVal) | IndVal from Randomised Groups | ||
|---|---|---|---|---|---|
| Mean | ±SD | p | |||
| Alphaproteobacteria | brackish | 41.0 | 34.3 | 0.72 | 0.0002 |
| Betaproteobacteria | freshwater | 37.8 | 34.4 | 0.76 | 0.0004 |
| Deltaproteobacteria | transitional | 37.7 | 34.5 | 0.80 | 0.0004 |
| Gammaproteobacteria | brackish | 36.9 | 34.3 | 0.72 | 0.0002 |
| Actinobacteria | transitional | 35.8 | 34.1 | 0.65 | <0.002 |
| Cytophaga-Flavobacteria | freshwater | 35.1 | 34.3 | 0.72 | 0.08 |
| Mean | 37.4 | 34.3 | 0.73 | 0.014 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lew, S.; Glińska-Lewczuk, K.; Burandt, P.; Kulesza, K.; Kobus, S.; Obolewski, K. Salinity as a Determinant Structuring Microbial Communities in Coastal Lakes. Int. J. Environ. Res. Public Health 2022, 19, 4592. https://doi.org/10.3390/ijerph19084592
Lew S, Glińska-Lewczuk K, Burandt P, Kulesza K, Kobus S, Obolewski K. Salinity as a Determinant Structuring Microbial Communities in Coastal Lakes. International Journal of Environmental Research and Public Health. 2022; 19(8):4592. https://doi.org/10.3390/ijerph19084592
Chicago/Turabian StyleLew, Sylwia, Katarzyna Glińska-Lewczuk, Paweł Burandt, Klaudia Kulesza, Szymon Kobus, and Krystian Obolewski. 2022. "Salinity as a Determinant Structuring Microbial Communities in Coastal Lakes" International Journal of Environmental Research and Public Health 19, no. 8: 4592. https://doi.org/10.3390/ijerph19084592
APA StyleLew, S., Glińska-Lewczuk, K., Burandt, P., Kulesza, K., Kobus, S., & Obolewski, K. (2022). Salinity as a Determinant Structuring Microbial Communities in Coastal Lakes. International Journal of Environmental Research and Public Health, 19(8), 4592. https://doi.org/10.3390/ijerph19084592









