Genetic Diversity of Antimicrobial Resistance and Key Virulence Features in Two Extensively Drug-Resistant Acinetobacter baumannii Isolates
Abstract
1. Introduction
2. Results and Discussion
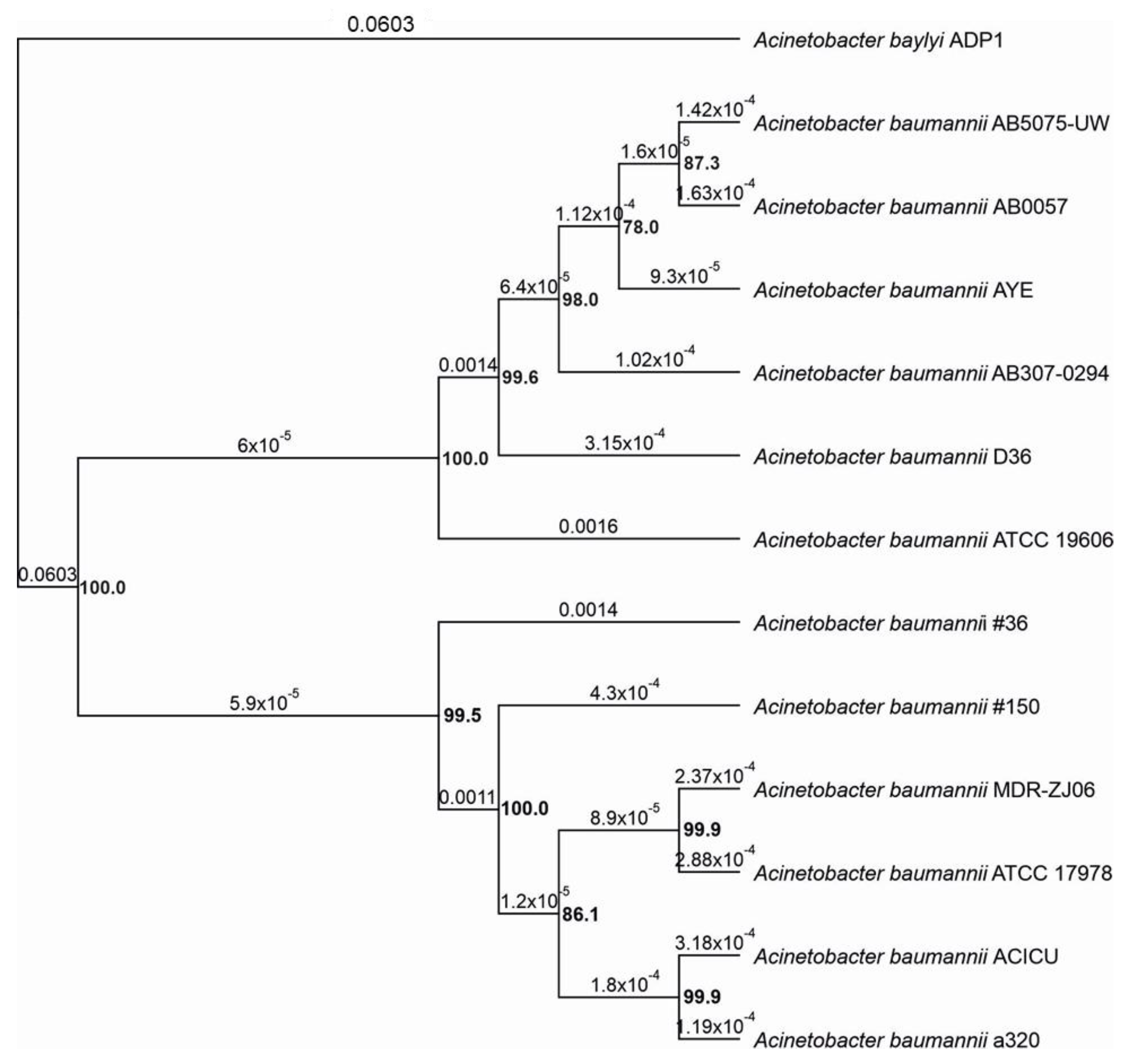
2.1. Genome Sequncing of A. baumannii Isolates and Phylogenetic Analysis of Selected Strains
2.2. Insertion Sequences (ISs) and Transposons
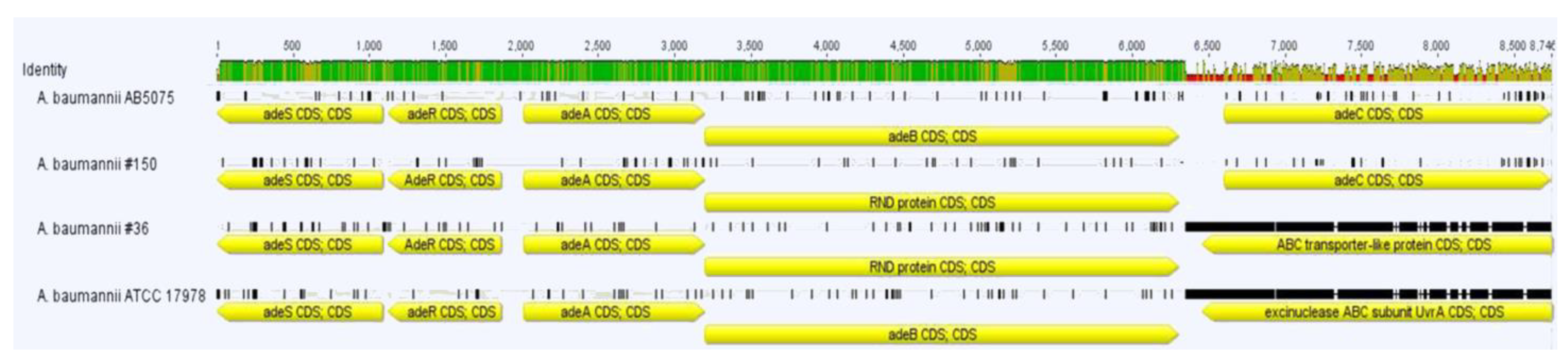
2.3. Resistance Genes
2.4. Secretion Systems
2.5. Outer Membrane Proteins, LPS and Capsule
2.6. Iron Scavengers
3. Materials and Methods
3.1. Bacterial Strains and Antimicrobial Susceptibility Testing (AST)
3.2. Genome Sequencing
3.3. Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bassetti, M.; Labate, L.; Russo, C.; Vena, A.; Giacobbe, D.R. Therapeutic options for difficult-to-treat Acinetobacter baumannii infections: A 2020 perspective. Expert Opin. Pharmacother. 2021, 22, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Sarshar, M.; Behzadi, P.; Scribano, D.; Palamara, A.T.; Ambrosi, C. Acinetobacter baumannii: An Ancient Commensal with Weapons of a Pathogen. Pathogens 2021, 10, 387. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef]
- Pompilio, A.; Scribano, D.; Sarshar, M.; Di Bonaventura, G.; Palamara, A.T.; Ambrosi, C. Gram-Negative Bacteria Holding Together in a Biofilm: The Acinetobacter baumannii Way. Microorganisms 2021, 9, 1353. [Google Scholar] [CrossRef] [PubMed]
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Bonomo, R.A.; Tolmasky, M.E. Carbapenemases: Transforming Acinetobacter baumannii into a Yet More Dangerous Menace. Biomolecules 2020, 10, 720. [Google Scholar] [CrossRef] [PubMed]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.; Wisplinghoff, H.; Rodríguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef] [PubMed]
- Scribano, D.; Marzano, V.; Levi Mortera, S.; Sarshar, M.; Vernocchi, P.; Zagaglia, C.; Putignani, L.; Palamara, A.T.; Ambrosi, C. Insights into the Periplasmic Proteins of Acinetobacter baumannii AB5075 and the Impact of Imipenem Exposure: A Proteomic Approach. Int. J. Mol. Sci. 2019, 20, 3451. [Google Scholar] [CrossRef]
- Ambrosi, C.; Scribano, D.; Sarshar, M.; Zagaglia, C.; Singer, B.B.; Palamara, A.T. Acinetobacter baumannii Targets Human Carcinoembryonic Antigen-Related Cell Adhesion Molecules (CEACAMs) for Invasion of Pneumocytes. mSystems 2020, 5. [Google Scholar] [CrossRef]
- Piran, A.; Fereshteh, S.; Badmasti, F. Lessons from Comparative Genome Analysis of Acinetobacter baumannii Strains. Health Biotechnol. Biopharma 2021, 5, 58–72. [Google Scholar]
- Antunes, L.C.; Imperi, F.; Carattoli, A.; Visca, P. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS ONE 2011, 6, e22674. [Google Scholar] [CrossRef]
- Jacobs, A.C.; Thompson, M.G.; Black, C.C.; Kessler, J.L.; Clark, L.P.; McQueary, C.N.; Gancz, H.Y.; Corey, B.W.; Moon, J.K.; Si, Y. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 2014, 5, e01076-14. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, C.; Scribano, D.; Aleandri, M.; Zagaglia, C.; Di Francesco, L.; Putignani, L.; Palamara, A.T. Acinetobacter baumannii Virulence Traits: A Comparative Study of a Novel Sequence Type with Other Italian Endemic International Clones. Front. Microbiol. 2017, 8, 1977. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, C.; Aleandri, M.; Giordano, A.; Scribano, D.; Marazzato, M.; Zagaglia, C.; Conte, M.P.; Palamara, A.T. Molecular characterisation of extensively drug-resistant Acinetobacter baumannii: First report of a new sequence type in Italy. J. Glob. Antimicrob. Resist. 2016, 7, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA-an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Bryant, D.; Moulton, V. Neighbor-net: An agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 2004, 21, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Ayoub Moubareck, C.; Hammoudi Halat, D. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef]
- Turton, J.F.; Ward, M.E.; Woodford, N.; Kaufmann, M.E.; Pike, R.; Livermore, D.M.; Pitt, T.L. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 2006, 258, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef]
- Snitkin, E.S.; Zelazny, A.M.; Montero, C.I.; Stock, F.; Mijares, L.; Murray, P.R.; Segre, J.A. Genome-wide recombination drives diversification of epidemic strains of Acinetobacter baumannii. Proc. Natl. Acad. Sci. USA 2011, 108, 13758–13763. [Google Scholar] [CrossRef] [PubMed]
- Fouts, D.E. Phage_Finder: Automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 2006, 34, 5839–5851. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Liu, Z.; Ling, B.; Zhou, L. Prevalence of 16S rRNA methylase, modifying enzyme, and extended-spectrum beta-lactamase genes among Acinetobacter baumannii isolates. J. Chemother. 2015, 27, 207–212. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Wang, R.; Liu, D.; Walsh, T.R.; Zhang, R.; Lv, Y.; Ke, Y.; Ji, Q.; Wei, R.; Liu, Z. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat. Microbiol. 2019, 4, 1450–1456. [Google Scholar] [CrossRef]
- Hujer, K.M.; Hamza, N.S.; Hujer, A.M.; Perez, F.; Helfand, M.S.; Bethel, C.R.; Thomson, J.M.; Anderson, V.E.; Barlow, M.; Rice, L.B.; et al. Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 beta-lactamase: Defining a unique family of class C enzymes. Antimicrob. Agents Chemother. 2005, 49, 2941–2948. [Google Scholar] [CrossRef] [PubMed]
- Sheikhalizadeh, V.; Hasani, A.; Ahangarzadeh Rezaee, M.; Rahmati-Yamchi, M.; Hasani, A.; Ghotaslou, R.; Goli, H.R. Comprehensive study to investigate the role of various aminoglycoside resistance mechanisms in clinical isolates of Acinetobacter baumannii. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2017, 23, 74–79. [Google Scholar] [CrossRef]
- Ostadi, Y.; Rezai, A.A.; Moghadampour, M.; Faghri, J. The involvement of drug efflux system in amikacin resistance of multiple drug resistant Acintobacter baumannii isolates in Isfahan, Iran. J. Med. Bacteriol. 2019, 8, 13–20. [Google Scholar]
- Abdi, S.N.; Ghotaslou, R.; Ganbarov, K.; Mobed, A.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Kafil, H.S. Acinetobacter baumannii Efflux Pumps and Antibiotic Resistance. Infect. Drug Resist. 2020, 13, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.-J.; Courvalin, P.; Grillot-Courvalin, C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: Major role for AdeABC overexpression and AdeRS mutations. Antimicrob. Agents Chemother. 2013, 57, 2989–2995. [Google Scholar] [CrossRef]
- Adams, M.D.; Goglin, K.; Molyneaux, N.; Hujer, K.M.; Lavender, H.; Jamison, J.J.; MacDonald, I.J.; Martin, K.M.; Russo, T.; Campagnari, A.A. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 2008, 190, 8053–8064. [Google Scholar] [CrossRef] [PubMed]
- Pérez, A.; Pérez-Llarena, F.J.; García, P.; Kerff, F.; Beceiro, A.; Galleni, M.; Bou, G. New mutations in ADC-type β-lactamases from Acinetobacter spp. affect cefoxitin and ceftazidime hydrolysis. J. Antimicrob. Chemother. 2014, 69, 2407–2411. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tian, G.-B.; Adams-Haduch, J.M.; Taracila, M.; Bonomo, R.A.; Wang, H.-N.; Doi, Y. Extended-spectrum AmpC cephalosporinase in Acinetobacter baumannii: ADC-56 confers resistance to cefepime. Antimicrob. Agents Chemother. 2011, 55, 4922–4925. [Google Scholar] [CrossRef]
- Rodríguez-Martínez, J.-M.; Nordmann, P.; Ronco, E.; Poirel, L. Extended-spectrum cephalosporinase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 3484–3488. [Google Scholar] [CrossRef] [PubMed]
- Wareth, G.; Linde, J.; Nguyen, N.H.; Nguyen, T.N.; Sprague, L.D.; Pletz, M.W.; Neubauer, H. WGS-Based Analysis of Carbapenem-Resistant Acinetobacter baumannii in Vietnam and Molecular Characterization of Antimicrobial Determinants and MLST in Southeast Asia. Antibiotics 2021, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Chapartegui-González, I.; Lázaro-Díez, M.; Redondo-Salvo, S.; Navas, J.; Ramos-Vivas, J. Antimicrobial Resistance Determinants in Genomes and Plasmids from Acinetobacter baumannii Clinical Isolates. Antibiotics 2021, 10, 753. [Google Scholar] [CrossRef] [PubMed]
- Hujer, A.M.; Hujer, K.M.; Leonard, D.A.; Powers, R.A.; Wallar, B.J.; Mack, A.R.; Taracila, M.A.; Rather, P.N.; Higgins, P.G.; Prati, F. A comprehensive and contemporary “snapshot” of β-lactamases in carbapenem resistant Acinetobacter baumannii. Diagn. Microbiol. Infect. Dis. 2021, 99, 115242. [Google Scholar] [CrossRef]
- Dortet, L.; Bonnin, R.A.; Bernabeu, S.; Escaut, L.; Vittecoq, D.; Girlich, D.; Imanci, D.; Fortineau, N.; Naas, T. First occurrence of OXA-72-producing Acinetobacter baumannii in Serbia. Antimicrob. Agents Chemother. 2016, 60, 5724–5730. [Google Scholar] [CrossRef] [PubMed]
- Rossi, I.; Royer, S.; Ferreira, M.; Braga, I.A.; Campos, P.; Batistão, D.; Fuga, B.; Cerdeira, L.; Lincopan, N.; Gontijo-Filho, P.P. Novel ST1465/CC216 Nosocomial Lineage of Carbapenem-Resistant Acinetobacter baumannii Harboring an Unusual Plasmid Carrying bla NDM-1 Gene. Microb. Drug Resist. 2021, 27, 471–475. [Google Scholar] [CrossRef]
- Elhosseiny, N.M.; Attia, A.S. Acinetobacter: An emerging pathogen with a versatile secretome. Emerg. Microbes Infect. 2018, 7, 1–15. [Google Scholar] [CrossRef]
- Pena, R.T.; Blasco, L.; Ambroa, A.; González-Pedrajo, B.; Fernández-García, L.; López, M.; Bleriot, I.; Bou, G.; García-Contreras, R.; Wood, T.K. Relationship between quorum sensing and secretion systems. Front. Microbiol. 2019, 10, 1100. [Google Scholar] [CrossRef]
- Korotkov, K.V.; Sandkvist, M.; Hol, W.G. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012, 10, 336–351. [Google Scholar] [CrossRef] [PubMed]
- Koskiniemi, S.; Lamoureux, J.G.; Nikolakakis, K.C.; de Roodenbeke, C.t.K.; Kaplan, M.D.; Low, D.A.; Hayes, C.S. Rhs proteins from diverse bacteria mediate intercellular competition. Proc. Natl. Acad. Sci. USA 2013, 110, 7032–7037. [Google Scholar] [CrossRef] [PubMed]
- Zusman, T.; Feldman, M.; Halperin, E.; Segal, G. Characterization of the icmH and icmF genes required for Legionella pneumophila intracellular growth, genes that are present in many bacteria associated with eukaryotic cells. Infect. Immun. 2004, 72, 3398–3409. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.G.; Gianoulis, T.A.; Pukatzki, S.; Mekalanos, J.J.; Ornston, L.N.; Gerstein, M.; Snyder, M. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007, 21, 601–614. [Google Scholar] [CrossRef]
- Liu, C.-C.; Kuo, H.-Y.; Tang, C.Y.; Chang, K.-C.; Liou, M.-L. Prevalence and mapping of a plasmid encoding a type IV secretion system in Acinetobacter baumannii. Genomics 2014, 104, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Pérez, A.; Merino, M.; Rumbo-Feal, S.; Álvarez-Fraga, L.; Vallejo, J.A.; Beceiro, A.; Ohneck, E.J.; Mateos, J.; Fernández-Puente, P.; Actis, L.A.; et al. The FhaB/FhaC two-partner secretion system is involved in adhesion of Acinetobacter baumannii AbH12O-A2 strain. Virulence 2017, 8, 959–974. [Google Scholar] [CrossRef]
- Roussin, M.; Rabarioelina, S.; Cluzeau, L.; Cayron, J.; Lesterlin, C.; Salcedo, S.P.; Bigot, S. Identification of a Contact-Dependent Growth Inhibition (CDI) System That Reduces Biofilm Formation and Host Cell Adhesion of Acinetobacter baumannii DSM30011 Strain. Front. Microbiol. 2019, 10, 2450. [Google Scholar] [CrossRef]
- Weidensdorfer, M.; Ishikawa, M.; Hori, K.; Linke, D.; Djahanschiri, B.; Iruegas, R.; Ebersberger, I.; Riedel-Christ, S.; Enders, G.; Leukert, L. The Acinetobacter trimeric autotransporter adhesin Ata controls key virulence traits of Acinetobacter baumannii. Virulence 2019, 10, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.S.; Kinsella, R.L.; Harding, C.M.; Feldman, M.F. The secrets of Acinetobacter secretion. Trends Microbiol. 2017, 25, 532–545. [Google Scholar] [CrossRef]
- Ruiz, F.M.; Lopez, J.; Ferrara, C.G.; Santillana, E.; Espinosa, Y.R.; Feldman, M.F.; Romero, A. Structural characterization of TssL from Acinetobacter baumannii: A key component of the type VI secretion system. J. Bacteriol. 2020, 202, e00210-20. [Google Scholar] [CrossRef]
- Wright, M.S.; Haft, D.H.; Harkins, D.M.; Perez, F.; Hujer, K.M.; Bajaksouzian, S.; Benard, M.F.; Jacobs, M.R.; Bonomo, R.A.; Adams, M.D. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. mBio 2014, 5, e00963-13. [Google Scholar] [CrossRef] [PubMed]
- Russell, A.B.; Hood, R.D.; Bui, N.K.; LeRoux, M.; Vollmer, W.; Mougous, J.D. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 2011, 475, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.S.; Miyata, S.T.; Iwashkiw, J.A.; Mortensen, B.L.; Skaar, E.P.; Pukatzki, S.; Feldman, M.F. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS ONE 2013, 8, e55142. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, J.-Y.; Lee, H.; Choi, J.Y.; Kim, D.H.; Wi, Y.M.; Peck, K.R.; Ko, K.S. Microbiological features and clinical impact of the type VI secretion system (T6SS) in Acinetobacter baumannii isolates causing bacteremia. Virulence 2017, 8, 1378–1389. [Google Scholar] [CrossRef]
- Meumann, E.M.; Anstey, N.M.; Currie, B.J.; Piera, K.A.; Kenyon, J.J.; Hall, R.M.; Davis, J.S.; Sarovich, D.S. Genomic epidemiology of severe community-onset Acinetobacter baumannii infection. Microb. Genom. 2019, 5, e000258. [Google Scholar] [CrossRef]
- Traglia, G.; Chiem, K.; Quinn, B.; Fernandez, J.S.; Montaña, S.; Almuzara, M.; Mussi, M.A.; Tolmasky, M.E.; Iriarte, A.; Centrón, D. Genome sequence analysis of an extensively drug-resistant Acinetobacter baumannii indigo-pigmented strain depicts evidence of increase genome plasticity. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Park, J.S.; Lee, W.C.; Yeo, K.J.; Ryu, K.S.; Kumarasiri, M.; Hesek, D.; Lee, M.; Mobashery, S.; Song, J.H.; Kim, S.I.; et al. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2012, 26, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.; Hu, Y.; Chen, Z.; Li, M.; Hou, Z.; Luo, X.; Mao, X.; Xue, X. Outer membrane protein A (OmpA) as a potential therapeutic target for Acinetobacter baumannii infection. J. Biomed. Sci. 2020, 27, 26. [Google Scholar] [CrossRef]
- Ambrosi, C.; Pompili, M.; Scribano, D.; Zagaglia, C.; Ripa, S.; Nicoletti, M. Outer membrane protein A (OmpA): A new player in shigella flexneri protrusion formation and inter-cellular spreading. PLoS ONE 2012, 7, e49625. [Google Scholar] [CrossRef]
- Singh, J.K.; Adams, F.G.; Brown, M.H. Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front. Microbiol. 2019, 9, 3301. [Google Scholar] [CrossRef]
- Kenyon, J.J.; Nigro, S.J.; Hall, R.M. Variation in the OC locus of Acinetobacter baumannii genomes predicts extensive structural diversity in the lipooligosaccharide. PLoS ONE 2014, 9, e107833. [Google Scholar] [CrossRef] [PubMed]
- Runci, F.; Gentile, V.; Frangipani, E.; Rampioni, G.; Leoni, L.; Lucidi, M.; Visaggio, D.; Harris, G.; Chen, W.; Stahl, J. Contribution of active iron uptake to Acinetobacter baumannii pathogenicity. Infect. Immun. 2019, 87, e00755-18. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, J.R.; Skaar, E.P. Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog. 2020, 16, e1008995. [Google Scholar] [CrossRef]
- Shapiro, J.A.; Wencewicz, T.A. Acinetobactin isomerization enables adaptive iron acquisition in Acinetobacter baumannii through pH-triggered siderophore swapping. ACS Infect. Dis. 2016, 2, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Moynié, L.; Serra, I.; Scorciapino, M.A.; Oueis, E.; Page, M.G.; Ceccarelli, M.; Naismith, J.H. Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogen Acinetobacter baumannii. eLife 2018, 7, e42270. [Google Scholar] [CrossRef] [PubMed]
- Klebba, P.E.; Newton, S.M.; Six, D.A.; Kumar, A.; Yang, T.; Nairn, B.L.; Munger, C.; Chakravorty, S. Iron Acquisition Systems of Gram-negative Bacterial Pathogens Define TonB-Dependent Pathways to Novel Antibiotics. Chem. Rev. 2021, 121, 5193–5239. [Google Scholar] [CrossRef]
- Andrews, S.; FASTQC. A quality control tool for high throughput sequence data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 4 January 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Hasan, T.; Choi, C.H.; Oh, M.H. Genes Involved in the Biosynthesis and Transport of Acinetobactin in Acinetobacter baumannii. Genom. Inform. 2015, 13, 2. [Google Scholar] [CrossRef] [PubMed]
- Penwell, W.F.; Arivett, B.A.; Actis, L.A. The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS ONE 2012, 7, e36493. [Google Scholar] [CrossRef]
- Gallagher, L.A.; Ramage, E.; Weiss, E.J.; Radey, M.; Hayden, H.S.; Held, K.G.; Huse, H.K.; Zurawski, D.V.; Brittnacher, M.J.; Manoil, C. Resources for Genetic and Genomic Analysis of Emerging Pathogen Acinetobacter baumannii. J. Bacteriol. 2015, 197, 2027–2035. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Tang, H. ISEScan: Automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics 2017, 33, 3340–3347. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Antunes, L.C.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a global pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef]
- Imperi, F.; Antunes, L.C.; Blom, J.; Villa, L.; Iacono, M.; Visca, P.; Carattoli, A. The genomics of Acinetobacter baumannii: Insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 2011, 63, 1068–1074. [Google Scholar] [CrossRef]
- Yakkala, H.; Samantarrai, D.; Gribskov, M.; Siddavattam, D. Comparative genome analysis reveals niche-specific genome expansion in Acinetobacter baumannii strains. PLoS ONE 2019, 14, e0218204. [Google Scholar] [CrossRef]
- Badmasti, F.; Siadat, S.D.; Bouzari, S.; Ajdary, S.; Shahcheraghi, F. Molecular detection of genes related to biofilm formation in multidrug-resistant Acinetobacter baumannii isolated from clinical settings. J. Med. Microbiol. 2015, 64, 559–564. [Google Scholar] [CrossRef]
- Jalal, D.; Elzayat, M.G.; Diab, A.A.; El-Shqanqery, H.E.; Samir, O.; Bakry, U.; Hassan, R.; Elanany, M.; Shalaby, L.; Sayed, A.A. Deciphering Multidrug-Resistant Acinetobacter baumannii from a Pediatric Cancer Hospital in Egypt. Msphere 2021, 6, e00725-21. [Google Scholar] [CrossRef]
- Ribeiro, M.; Simões, M. Advances in the antimicrobial and therapeutic potential of siderophores. Environ. Chem. Lett. 2019, 17, 1485–1494. [Google Scholar] [CrossRef]


| Insertion Sequences | Family | No. in #36 | No. in #150 |
|---|---|---|---|
| ISAba1 | IS4 | 2 | 21 |
| ISAba12 | IS5 | 1 | 0 |
| ISAba13 | IS5 | 0 | 1 |
| ISAba14 | IS3 | 1 | 0 |
| ISAba17 | IS66 | 0 | 2 |
| ISAba19 | IS3 | 0 | 1 |
| ISAba25 | IS66 | 2 | 0 |
| ISAba27 | IS5 | 0 | 2 |
| ISAba125 | IS30 | 0 | 3 |
| ISAlw27 | IS3 | 1 | 0 |
| IS1006 | IS6 | 2 | 0 |
| IS26 | IS6 | 0 | 1 |
| ISVsa3 | IS91 | 2 | 0 |
| Unknown | IS4/IS481 | 0 | 3 |
| Antibiotics | Strain | |
|---|---|---|
| #36 | #150 | |
| Amikacin | 16 S | ≥64 R |
| Amoxycillin/clavulanic acid | ≥32 R | ≥32 R |
| Ampicillin | ≥32 R | ≥32 R |
| Cefepime | 16 IE | ≥64 R |
| Cefotaxime | ≥64 R | ≥64 R |
| Ceftazidime | ≥64 R | ≥64 R |
| Ciprofloxacin | ≥4 R | ≥4 R |
| Colistin | ≤0.5 S | ≤0.5 S |
| Gentamicin | ≥16 R | ≥16 R |
| Imipenem | ≥8 R | ≥8 R |
| Piperacillin/Tazobactam | ≥128 R | ≥128 R |
| Tigecycline | ≤0.5 S | ≥8 R |
| Trimethoprim/Sulfamethoxazole | ≥160 R | ≥160 R |
| Type of Secretion System (TSS) | Components | Translocation System | Genes Found in Both Strains #36 and #150 | Reference |
|---|---|---|---|---|
| Type I secretion system (T1SS) | IM ABC transporter protein, a membrane fusion protein (MFP) and an OM protein | Single procedure (directly from cytoplasm to outside cell) | Type I SS permease/ATPase Type I secretion C-terminal target domain-containing protein HlyD family secretion protein 1 HlyD family secretion protein 2 HlyD family secretion protein 3 HlyD family secretion protein 4 HlyD family secretion protein 5 HlyD family type I secretion periplasmic adaptor subunit | [40] |
| Type II secretion system (T2SS) | IM SecYEG/Tat pathways, 15 general secretion pathway proteins (Gsp) | Double step procedure (Sec/Tat transfer the substrates of T2SS and T5SS across the inner membrane) | Type II secretion system F family protein gspD gspE gspF gspG gspH gspI gspJ gspK gspL gspM gspN | [49] |
| Type IV secretion system (T4SS) | Three type IVa ATPases, three IM proteins, a PP protein, two OM proteins, three surface/pilus proteins (tra and vir genes), eight genes homologous to the Legionella/Coxiella type IV virulence/secretion apparatus Dot/Icm | Single procedure (directly from cytoplasm into the outside of the cell) | icmH traC (#150) Type IV secretion system DNA-binding domain-containing protein (#150) Type IV secretion protein Rhs (#36) | [46,47,50,51] |
| Type V secretion system (T5SS) | An N-terminal Sec-dependent signal peptide, a central passenger domain and C-terminal β barrel | Two-partner and autotransporter | abfhaB (#36) cdiB1 (#36) cdiB2 (#36) | [2] |
| Type VI secretion system (T6SS) | Thirteen core components: membrane-spanning complex, baseplate components and priming protein TssA. VgrG-tipped Hcp tubule wrapped in the TssB/C sheath. No TssJ | Single procedure (directly from cytoplasm into the outside of the cell) | tssA tssB tssC tssE tssF tssG tssH tssK tssL tssM Type VI secretion system tip protein vgrg1 Type VI secretion system tip protein vgrg2 Type VI secretion system tip protein vgrg3 Type VI secretion system tube protein Hcp tagF | [49] |
| Strain | Bioproject | Biosample | Accession No. | Reference |
|---|---|---|---|---|
| #36 | PRJNA803948 | SAMN25691074 | - | This study |
| #150 | PRJNA803948 | SAMN25691075 | - | This study |
| AB5075-UW | PRJNA224116 | SAMN02894434 | NZ_CP008706.1 | [71] |
| ATCC 17978 | PRJNA17477 | SAMN02604331 | NZ_CP053098.1 | ATCC * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marazzato, M.; Scribano, D.; Sarshar, M.; Brunetti, F.; Fillo, S.; Fortunato, A.; Lista, F.; Palamara, A.T.; Zagaglia, C.; Ambrosi, C. Genetic Diversity of Antimicrobial Resistance and Key Virulence Features in Two Extensively Drug-Resistant Acinetobacter baumannii Isolates. Int. J. Environ. Res. Public Health 2022, 19, 2870. https://doi.org/10.3390/ijerph19052870
Marazzato M, Scribano D, Sarshar M, Brunetti F, Fillo S, Fortunato A, Lista F, Palamara AT, Zagaglia C, Ambrosi C. Genetic Diversity of Antimicrobial Resistance and Key Virulence Features in Two Extensively Drug-Resistant Acinetobacter baumannii Isolates. International Journal of Environmental Research and Public Health. 2022; 19(5):2870. https://doi.org/10.3390/ijerph19052870
Chicago/Turabian StyleMarazzato, Massimiliano, Daniela Scribano, Meysam Sarshar, Francesca Brunetti, Silvia Fillo, Antonella Fortunato, Florigio Lista, Anna Teresa Palamara, Carlo Zagaglia, and Cecilia Ambrosi. 2022. "Genetic Diversity of Antimicrobial Resistance and Key Virulence Features in Two Extensively Drug-Resistant Acinetobacter baumannii Isolates" International Journal of Environmental Research and Public Health 19, no. 5: 2870. https://doi.org/10.3390/ijerph19052870
APA StyleMarazzato, M., Scribano, D., Sarshar, M., Brunetti, F., Fillo, S., Fortunato, A., Lista, F., Palamara, A. T., Zagaglia, C., & Ambrosi, C. (2022). Genetic Diversity of Antimicrobial Resistance and Key Virulence Features in Two Extensively Drug-Resistant Acinetobacter baumannii Isolates. International Journal of Environmental Research and Public Health, 19(5), 2870. https://doi.org/10.3390/ijerph19052870










