Abstract
Biological treatment using sulfate-reducing bacteria (SRB) is a promising approach to remediate acid rock drainage (ARD). Our purpose was to assess the performance of a sequential system consisting of a limestone bed filter followed by a sulfate-reducing bioreactor treating synthetic ARD for 375 days and to evaluate changes in microbial composition. The treatment system was effective in increasing the pH of the ARD from 2.7 to 7.5 and removed total Cu(II) and Zn(II) concentrations by up to 99.8% and 99.9%, respectively. The presence of sulfate in ARD promoted sulfidogenesis and changed the diversity and structure of the microbial communities. Methansarcina spp. was the most abundant amplicon sequence variant (ASV); however, methane production was not detected. Biodiversity indexes decreased over time with the bioreactor operation, whereas SRB abundance remained stable. Desulfobacteraceae, Desulfocurvus, Desulfobulbaceae and Desulfovibrio became more abundant, while Desulfuromonadales, Desulfotomaculum and Desulfobacca decreased. Geobacter and Syntrophobacter were enriched with bioreactor operation time. At the beginning, ASVs with relative abundance <2% represented 65% of the microbial community and 21% at the end of the study period. Thus, the results show that the microbial community gradually lost diversity while the treatment system was highly efficient in remediating ARD.
Keywords:
sulfate-reducing; bioreactor; anaerobic; acid rock drainage; metagenomics; microbial; community; dynamics; diversity 1. Introduction
Massive generation of acid rock drainage (ARD) is one of the most important environmental impacts of mining activities. ARD contains high concentrations of heavy metals, metalloids and sulfate, and low pH values [1,2,3].
Bioremediation of ARD through metal bioprecipitation is mediated by sulfate reducing bacteria (SRB), a diverse group of microorganisms which use sulfate as a terminal electron acceptor [4,5,6,7,8]. In the presence of a suitable electron-donating substrate, either an organic compound or hydrogen, SBRs catalyze the reduction of sulfate into sulfide. Sulfide anions react with many metal cations (e.g., Cu(II), Zn(II), Ni(II), Hg(II), Cd(II), etc.) forming sparingly soluble metal sulfides that can be easily removed from solution by gravity settling sulfides [5,9,10]. In addition, the microbial oxidation of the electron-donating substrate by SRB generates alkalinity and increases pH [6,9,11,12].
SRBs constitute a heterogeneous bacterial group of ubiquitous, metabolically flexible, and free-living microorganisms [7,13]. Both in natural and in engineered environments, SRBs are part of consortia with other anaerobic microorganisms, such as methanogenic archaea and syntrophic bacteria [5,14,15,16]. The assessment of the synergistic or antagonistic effects between members of a microbial consortium and the multiple interactions influenced by contaminants, metabolites, and inhibitory substances, is crucial in the development and improvement of innovative bioremediation [17,18,19].
The effects of heavy metals, pH and different electron donors and carbon sources on sulfate reduction and methanogenesis in anaerobic bioreactors are well-documented [12,20,21,22,23,24]. However, less information is available on the temporal changes in the structure of the microbial communities that drive the sulfate reduction process in bioreactors treating ARD [15,25,26].
Microbial community dynamics are linked to the ecological functionality of community members and adaptation to external disturbances [24,27,28,29]. Therefore, we hypothesize that metal-bioprecipitation of the influent may influence the microbial communities and may result in the assembly of a robust and long-lasting consortium for ARD bioremediation.
The objective of this study was to evaluate the performance of a sequential system, consisting of a limestone bed filter followed by a sulfate-reducing bioreactor, treating synthetic ARD composed of copper (II), zinc (II), sulfate and acetate as an organic carbon source, and to characterize the diversity of the microbial communities responsible for the biological treatment of ARD during 375 days of operation. We used 16S ribosomal RNA (rRNA) gene sequencing to assess microbial community structure, abundance, and diversity.
2. Materials and Methods
2.1. Configuration and Operation of the Treatment System
The pilot-scale sulfate-reducing (SR) bioreactor with a limestone pre-column (Figure 1) was described in a previous study [30]. The volume of the limestone precolumn was 0.40-L and the volume of the biological reactor was 0.49-L. Before starting our research, the system pre-column was packed with fresh and pre-sieved limestone, as recommended by Mendez and coworkers [30].
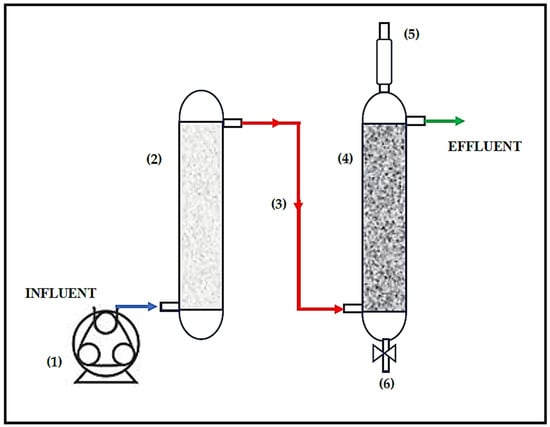
Figure 1.
Scheme of the treatment system. (1) Peristaltic pump, (2) limestone pre-column (height: 25 cm, internal diameter (i.d.): 5.5 cm), (3) limestone pre-column effluent and/or bioreactor influent, (4) biological reactor (height: 43.2 cm, i.d.: 5.5 cm), (5) biogas, (6) sampling port of sludge.
The treatment system was fed with a synthetic ARD composed of a basal mineral medium [30] supplemented with sulfate (2000 mg L−1), acetate as an electron donor and an organic carbon source with a chemical oxygen demand (COD) of 2500 mg COD L−1, Cu(II) (15 mg L−1) and Zn(II) (15 mg L−1). The medium pH was adjusted to 2.7 by adding hydrochloric acid (HCl). The reactor was operated at 30 ± 2 °C under darkness conditions to avoid anoxygenic photosynthesis. The inoculum (sludge) contained 7.18% volatile suspended solids (VSS) in wet-weight, with a maximum sulfate reducing activity of 4.25 mg S2− L−1 d−1.
During period I (day 0–24), the bioreactor was operated as a stand-alone reactor without metal addition or acidification at a volumetric loading rate of 2.8 g acetate-COD L−1 d−1 and a hydraulic retention time (HRT) of 2.0 ± 0.37 d. In periods II and III, the bioreactor was operated for 201 and 150 days, respectively, with the lime-stone bed reactor. Due to the toxicity of the acidic conditions and high metal concentrations of the influent, the purpose of the limestone reactor was to increase pH by alkalinity generation and to reduce heavy metal levels by precipitation as hydroxides or carbonates [11,31]. In period II, 15 mg L−1 of Cu(II) was added (as CuCl2·2H2O), and, in period III, in addition to Cu(II), 15 mg L−1 of Zn(II) was dosed (as ZnCl2). The volumetric loading rate in periods II and III averaged 2.8 g acetate-COD L−1 d−1 with an HRT of 2.0 ± 0.37 d. For preparation of synthetic ARD, concentrations of Cu(II) and Zn(II) were selected to simulate the metal concentrations found in real ARD collected in mining zones in Ecuador [30]. Influent and effluent samples were sampled and analyzed weekly. During period I, the pH of the influent was not modified, while in periods II and III, the influent pH was acidified to a value of 2.7.
2.2. Analytical Methods
Sulfate was determined using the turbidimetric method 4500-SO42− [32]. Sulfide was analyzed immediately after sample collection using the methylene blue method [33]. Chemical oxygen demand was determined by the colorimetric method 5220D according to standard methods [32]. Before sulfate, COD and metals analyses, samples were filtered using a 0.45-µm filter. Sulfate reduction and COD removal were determined as the difference between the influent and the effluent sulfate and COD concentrations, respectively.
pH was measured using a portable multi-parameter Thermo Scientific Orion 5-Star meter (Thermo Scientific, Beverly, MA, USA) according to the methods 2510A and 4500A, respectively [32].
Soluble metal concentrations of Cu(II) and Zn(II) in liquid samples were analyzed by inductively coupled plasma optical emission spectrometry (ICP-OES, Thermo Scientific ICAP 7400, Thermo Scientific, Beverly, MA, USA). Calibration curves were conducted for each metal in 2% HNO3 trace metals. Samples were constructed according to method 3120B [32].
Methane (CH4) produced in the sulfate-reducing bioreactor was estimated by the liquid displacement method through a sodium hydroxide (NaOH) solution (2%) to remove carbon dioxide (CO2) and hydrogen sulfide (H2S) [34]. The H2S concentration in the biogas was calculated from the H2S concentration in the liquid phase, assuming equilibrium between phases and a dimensionless Henry’s factor of 0.36 [20,35]. The electron equivalents of reducing power, as percentage, fed to the reactor (CODin, as g COD L−1 reactor d−1) used for methane (% CH4-COD) and sulfide (% H2S-COD) generation, were calculated as described in our previous publication [20].
Standard deviations and analysis of variance (ANOVA), were calculated using SPSS Statistics software (IBM Corp., New York, NY, USA). Thus, the media values of analytical parameters in the different operational periods were compared by ANOVA, with 95% confidence intervals.
2.3. Sludge Sample Collection
Samples were collected for 375 days in sterile plastic recipients. Two samples of sludge were collected during period I (adaptation phase), day 0 and day 24, two during period II (15 mg Cu(II) L−1), day 181 and day 217, and two during period III (15 mg Cu(II) L−1 and 15 mg Zn(II) L−1), day 356 and day 372, six-bioreactor sludge samples in total [35]. Approximately 15–20 g of material was collected per sample. Genomic DNA was immediately extracted and maintained at −80 °C prior to molecular analyses.
2.4. DNA Extraction
DNA extraction was performed as previously described (Ben-Dov et al., 2007), using DNeasy® PowerSoil® Kit (QIAGEN GmbH., Hilden, Germany). The concentration of the resulting DNA preparation was determined by a Qubit® fluorometer (Thermo Fisher Scientific Inc, Waltham, USA). DNA integrity was evidenced by electrophoresis and DNA samples were stored at −80 °C until use in molecular analysis [35]. For 16S rRNA analysis, DNA samples were lyophilized in a freeze dryer (ilShin BioBase, Dongducheon, Korea), prior to submission for next generation sequencing.
2.5. rRNA Analysis
V3 and V4 regions of 16S rRNA genes were sequenced (Primers: Bakt_341F: CCTACGGGNGGCWGCAG and Bakt_805R: GACTACHVGGGTATCTAATCC) in Macrogen (Seoul, Korea) using an Illumina MiSeq platform (Albany, NY, USA) and MCS Sequencing Control Software, yielding a total of ~120.3 Mbp of sequencing reads. Only reads with 100% correct primer sequences were used. After primer sequences were removed, the resulting sequences were collapsed into amplicon sequence variants (ASVs) using the R package DADA2 version 1.8.1 [36,37]. A total of 766,178 sequences were merged in 703 ASVs. Taxonomic assignment of each ASV was performed using the naïve Bayes k-mer method implemented in the MOTHUR package [38] using the Silva 132 database as the training reference [39]. All samples were rarefied to 69,000 reads per sample before analysis.
2.6. Microbial Diversity and Statistical Analysis
Alpha diversity metrics and the Bray–Curtis dissimilarity index were calculated using the vegan package version 2.5–3 [40]. Five measures for the sludge diversity and richness were used: Shannon index (diversity index), inverse Simpson index (to quantify average proportional abundance of species in a sample), richness (count of unique species), Chao1 (abundance of species-based index), and evenness (how equally abundant species are) [41]. Beta-diversity analyses (principal coordinate analysis) were based on Bray–Curtis dissimilarity calculated from the rarefied abundance tables [42,43]. To visualize the composition of the communities, a stacked bar chart was built by grouping all ASVs assigned to known methanogenic archaea [44], non-syntrophic SRB [45] and syntrophic SRB [46]; then, all the remaining ASVs with an abundance higher than 2% were collapsed to genus level. A heatmap plotting log10 of the counts for methanogenic archaea, SRB and syntrophic bacteria ASVs was created using the Pheatmap package [47]. Finally, a phylogenetic tree was assembled using the ASV sequences of the methanogenic archaea, non-syntrophic SRB and syntrophic SRB using MEGA-X software [48]. The sequences were aligned using the default parameters of the Clustal-W algorithm [49,50] and a maximum likelihood tree was constructed using the GTR+G+I model with 500 bootstrap replications [48]. The final consensus tree (70% cut-off value) was visualized and annotated with the online tool ‘Interactive Tree of Life’ (https://itol.embl.de (accessed on 27 August 2021)) [51].
3. Results
3.1. Performance of the Treatment System
The sulfate-reducing bioreactor with a limestone pre-column (Figure 1) was operated for 375 days. In period I, or the phase of adaptation, the treatment system was fed with non-acidified synthetic ARD (pH: 8.04 ± 0.36). In period II, 15 mg L−1 of Cu(II) was added, and the pH of synthetic ARD was 2.7. In period III, 15 mg L−1 of Cu(II) and 15 mg L−1 of Zn(II) were simultaneously added to the pH-2.7 synthetic ARD (Table 1).

Table 1.
Set up of the experiment, influent composition, and conditions of operation of the treatment system during the three periods of operation.
Treatment of pH of the acidic ARD in the limestone pre-column during periods II and II increased the pH from 2.7 to neutral pH (Figure 2A). Following treatment in the SR bioreactor, the effluent pH values were 8.04, 7.45 and 7.43 in the periods I, II and III, respectively.
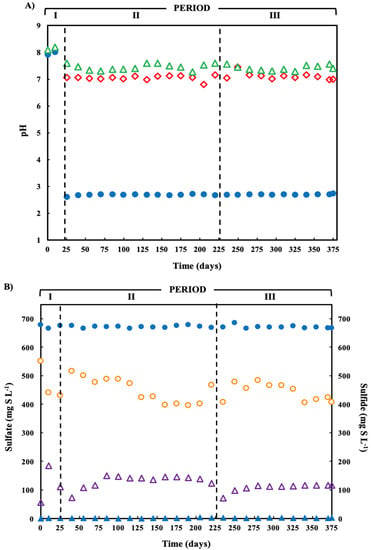
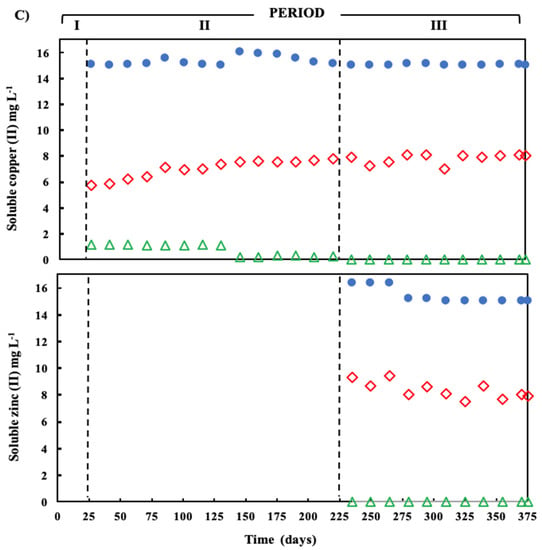
Figure 2.
(A) pH variation in the operation time of the treatment system fed with a pH-2.7 synthetic ARD containing sulfate (2000 mg L−1), acetate as electron donor (2.5 g COD L−1), Cu(II) (15 mg L−1 during periods II and III) and Zn(II) (15 mg L−1 during period III): limestone pre-column influent (●), limestone pre-column effluent/bioreactor influent (  ), and bioreactor effluent (
), and bioreactor effluent (  ). (B) Sulfate reduction (primary axis-left) and sulfide production (secondary axis-right) in the treatment system fed with a synthetic ARD containing sulfate (2000 mg L−1), acetate as electron donor (2.5 g COD L−1), Cu(II) (15 mg L−1 during periods II and III) and Zn(II) (15 mg L−1 during period III): sulfate (●) and sulfide (▲) in the influent and sulfate (
). (B) Sulfate reduction (primary axis-left) and sulfide production (secondary axis-right) in the treatment system fed with a synthetic ARD containing sulfate (2000 mg L−1), acetate as electron donor (2.5 g COD L−1), Cu(II) (15 mg L−1 during periods II and III) and Zn(II) (15 mg L−1 during period III): sulfate (●) and sulfide (▲) in the influent and sulfate (  ) and sulfide (
) and sulfide (  ) in the effluent. (C) Concentration of soluble Cu(II) and soluble Zn(II) during the operation of the treatment system fed with a synthetic ARD containing sulfate (2000 mg L−1), acetate as electron donor (2.5 g COD L−1), Cu (II) (15 mg L−1 during periods II and III) and Zn(II) (15 mg L−1 during period III): limestone pre-column influent (●), limestone pre-column effluent/bioreactor influent (
) in the effluent. (C) Concentration of soluble Cu(II) and soluble Zn(II) during the operation of the treatment system fed with a synthetic ARD containing sulfate (2000 mg L−1), acetate as electron donor (2.5 g COD L−1), Cu (II) (15 mg L−1 during periods II and III) and Zn(II) (15 mg L−1 during period III): limestone pre-column influent (●), limestone pre-column effluent/bioreactor influent (  ), and bioreactor effluent (
), and bioreactor effluent (  ).
).
 ), and bioreactor effluent (
), and bioreactor effluent (  ). (B) Sulfate reduction (primary axis-left) and sulfide production (secondary axis-right) in the treatment system fed with a synthetic ARD containing sulfate (2000 mg L−1), acetate as electron donor (2.5 g COD L−1), Cu(II) (15 mg L−1 during periods II and III) and Zn(II) (15 mg L−1 during period III): sulfate (●) and sulfide (▲) in the influent and sulfate (
). (B) Sulfate reduction (primary axis-left) and sulfide production (secondary axis-right) in the treatment system fed with a synthetic ARD containing sulfate (2000 mg L−1), acetate as electron donor (2.5 g COD L−1), Cu(II) (15 mg L−1 during periods II and III) and Zn(II) (15 mg L−1 during period III): sulfate (●) and sulfide (▲) in the influent and sulfate (  ) and sulfide (
) and sulfide (  ) in the effluent. (C) Concentration of soluble Cu(II) and soluble Zn(II) during the operation of the treatment system fed with a synthetic ARD containing sulfate (2000 mg L−1), acetate as electron donor (2.5 g COD L−1), Cu (II) (15 mg L−1 during periods II and III) and Zn(II) (15 mg L−1 during period III): limestone pre-column influent (●), limestone pre-column effluent/bioreactor influent (
) in the effluent. (C) Concentration of soluble Cu(II) and soluble Zn(II) during the operation of the treatment system fed with a synthetic ARD containing sulfate (2000 mg L−1), acetate as electron donor (2.5 g COD L−1), Cu (II) (15 mg L−1 during periods II and III) and Zn(II) (15 mg L−1 during period III): limestone pre-column influent (●), limestone pre-column effluent/bioreactor influent (  ), and bioreactor effluent (
), and bioreactor effluent (  ).
).
The average sulfate removal was approximately 38% in periods I and II (Table 1), reaching a sulfate removal of 48.8%. The average concentration of biogenic sulfide formed during the three operation periods was 160, 191 and 158 mg H2S-S L−1, respectively.
The organic COD removal efficiency increased by approximately 70% in relation to the initial COD concentration in the influent (Table 1). In the same manner, Table 1 summarizes the fraction of acetate (organic substrate), as %COD, used primarily by SRBs for H2S production; there was no methane production. The percentages of organic COD removed by SRB were 49.4, 51.3 and 42.4% during periods I, II and III, respectively. No significant statistical differences were observed. The time course of sulfate reduction showed reduction in sulfate concentration in all three periods, while sulfide concentration increased until the values stabilized (Figure 3B).
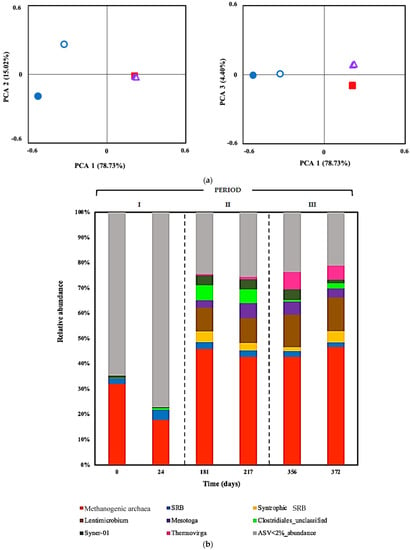
Figure 3.
(a) Analysis performed on Bray–Curtis distances or dissimilarities for six sludge samples during the three different operation periods of the sulfate-reducing bioreactor with the limestone pre-column system. Two samples or biological replicates were collected in each period. Period I (adaptation phase): day 0 (●) and day 24 (○). Period II (15 mg Cu(II) L−1): day 181 (■) and day 217 (□). Period II (15 mg Cu(II) L−1 and 15 mg Zn(II) L−1: day 356 (▲) and day 372 (  ). (b) Relative abundances of most abundant amplicon sequence variants (ASVs) at genus level in six sludge samples during the three different operation periods of the sulfate-reducing bioreactor with the limestone pre-column system. Two samples or biological replicates were collected in each period.
). (b) Relative abundances of most abundant amplicon sequence variants (ASVs) at genus level in six sludge samples during the three different operation periods of the sulfate-reducing bioreactor with the limestone pre-column system. Two samples or biological replicates were collected in each period.
 ). (b) Relative abundances of most abundant amplicon sequence variants (ASVs) at genus level in six sludge samples during the three different operation periods of the sulfate-reducing bioreactor with the limestone pre-column system. Two samples or biological replicates were collected in each period.
). (b) Relative abundances of most abundant amplicon sequence variants (ASVs) at genus level in six sludge samples during the three different operation periods of the sulfate-reducing bioreactor with the limestone pre-column system. Two samples or biological replicates were collected in each period.
The sequential treatment system was highly effective for removing heavy metals. About a half of the initial Cu(II) and Zn(II) concentration was removed in the limestone pre-column and the remaining fraction in the bioreactor (Figure 4a). Cu(II) removal efficiency by the complete treatment system was 98.5 and 99.9%, in periods II and III, respectively, whereas Zn(II) removal during period III was close to 100%. In the limestone pre-column, copper removal efficiency was about 50% in periods II and III, and in period III Zn removal efficiency was 47.1% (Table 2). In the SR bioreactor, the average concentration of soluble Cu(II) was reduced from 6.98 to 0.22 mg L−1 in period II, and from 7.54 to 0.01 mg L−1 in period III. Zn(II) removal in the bioreactor was 51.8%.
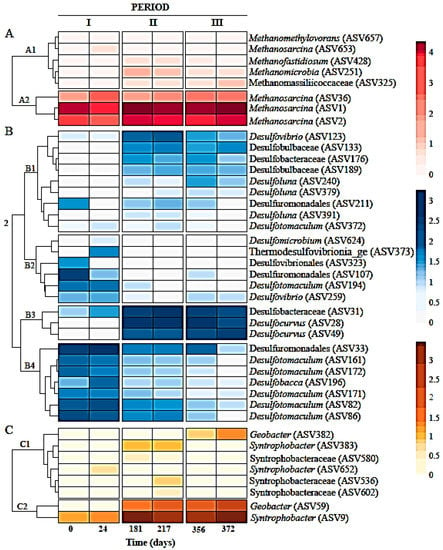
Figure 4.
Clusters and heat maps plotting log10 of the counts for methanogenic archaea, SRB and syntrophic bacteria amplicon sequence variants (ASVs).

Table 2.
Average removal of soluble Cu(II) and Zn(II) attained by the treatment system during the various periods of operation.
3.2. Microbial Community Analysis
Genomic DNA from sludge samples collected in three operation periods of a sulfate-reducing bioreactor were used for 16S rRNA gene sequencing. A total of 69,000 reads per sample were obtained after the exclusion of singletons and rarefaction. All indices of alpha (α) diversity (Table 3) showed differences in reference to lesser values along the operation of the bioreactor. The Inverse Simpson Index, a measure of the richness in a community with a uniform evenness that would have the same level of diversity, decreased by approximately a half at period III compared to the adaptation stage.

Table 3.
Alpha diversity indices of the mixed microbial culture during the various periods of operation of the sulfate-reducing bioreactor.
Figure 3a shows beta (β) diversity statistics using a principal components plot (PCA) plotting the Bray–Curtis distance. These graphics show dissimilarities in the microbial composition of two sludge samples obtained during period I or adaptation phase compared with periods II and III, and that >93% of the variance was explained by changes in pH and the addition of metals to ARD fed to the treatment system. Conversely, there were no significant differences in microbial communities between samples of periods II and III, showing the same cluster patterns; the second PCA plot shows a better differentiation among assemblages at periods II and III explained by the addition of Zn(II) during period III.
To analyze community structure, the relative prokaryotic abundances were organized at genus level in six sludge samples obtained during the three different operation periods of the treatment system (Figure 3b). The archaeal community was represented by methanogens, with relative abundance ranging from 32.1% to 46.7% in the sludge samples. The non-syntrophic SRB population remained stable, around a relative abundance of 2.0–1.8%. SRB syntrophic bacteria became significantly more abundant, 0.02% to 4.4%. Members of the genus Lentimicrobium were present from 0.1% at the beginning, and achieving 13.4% of total encountered ASVs at the end of the study. In period I, microbial populations belonging to the genus Mesotoga presented a very low abundance proportion (0.1%) and increased their relative abundance to 3.0 to 6% in period II, but they decreased in period III by approximately ~3.5 to 1.5%. Unclassified Clostidriales had low abundance (0.1%) in sludge samples during period I or the adaptation phase and increased their relative abundance to 3.0 to 6% in period II, but they decreased in period III by 0.7 to 2.0%. ASVs corresponding to Syner-01 increased from a relative abundance of 1% at the beginning of the period I to around 4% in period II and finally decreased to 1.5%. Thermovirga was not detected during period I and then increased to 5.6%. Other ASVs with relative abundance less than 2% reduced from 64.6 to 21.1% in the time course of the bioreactor operation with the limestone pre-column system.
Figure 4 shows heatmaps clustered as methanogenic archaea, non-syntrophic and syntrophic sulfate-reducing bacteria ASVs. Methanogens included the genera Methanosarcina (ASV1, ASV2, ASV36 and ASV653), Methanomicrobia unclassified (ASV251), Candidatus Methanofastidiosum (ASV428), Methanomethylovorans (ASV657), and Methanomassiliicoccaceae (ASV325). The most abundant methanogens in all sludge samples were Methanosarcina and those populations increased with bioreactor operation, except ASV653 that was relatively stable, as well as the remainder of the identified methanogens. Sulfate-reducing bacteria were represented by members of Desulfocurvus (ASV28 and ASV49), Desulfobacteraceae (ASV31 and ASV176), Desulfuromonadales (ASV33, ASV107 and ASV211), Desulfotomaculum (ASV82, ASV86, ASV161, ASV171, ASV172, ASV194 and ASV372), Desulfovibrio (ASV123 and ASV259), Desulfobulbaceae (ASV133 and ASV189), Desulfobacca (ASV196), Desulfoluna (ASV240, ASV379 and ASV391), Desulfovibrionales (ASV323), Thermodesulfovibrionia_ge (ASV373) and Desulfomicrobium (ASV624). Sulfate-reducing populations belonging to genera Desulfobacteraceae, Desulfocurvus, Desulfobulbaceae and Desulfovibrio became more abundant, while Desulfuromonadales, Desulfotomaculum and Desulfobacca decreased. Syntrophic SRB corresponded to genera Geobacter (ASV59 and ASV3829), Syntrophobacter (ASV9, ASV383 and ASV652), and members of Syntrophobacteraceae (ASV536, ASV580 and ASV602). Syntrophobacter (ASV9) and ASVs belonging to the Geobacter genus were enriched with bioreactor operation. Other ASVs affiliated to SRB syntrophic bacteria remained stable.
Forty-one ASVs were considered to build a consensus tree using the V3 and V4 regions of 16SrRNA sequences datasets (Figure 5). A phylogenetic tree that represented methanogenic, non-syntrophic and syntrophic sulfate reducing populations, that grouped 8, 25 and 8 ASVs, respectively, was constructed. In addition, the analysis included reference nomenclature of the clusters obtained from heatmaps represented in Figure 4. Methanosarcina ASVs belonging to cluster A2, the most abundant methanogens in this study, were as closely related to Methanosarcina (ASV 653) as Methanomethylovorans, both with low abundance from cluster A1. Therefore, according to the consensus tree, the bacterial community was represented as phylogenetically coherent groups, including sequences of non-syntrophic SRBs and syntrophic SRBs.
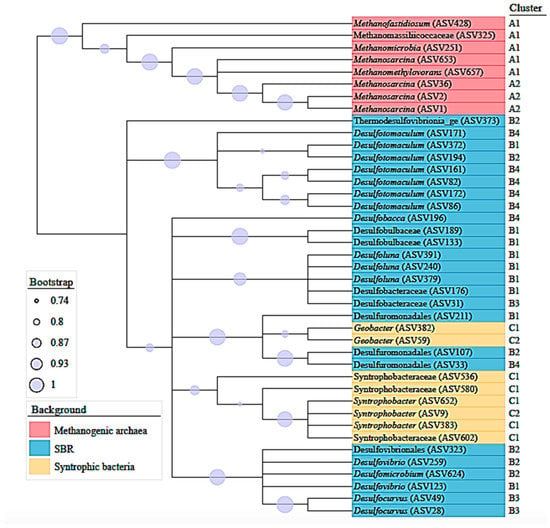
Figure 5.
Phylogenetic tree based on V3 and V4 regions of 16SrRNA sequences datasets, showing communities of methanogenic archaea (red), SRB (blue) and syntrophic bacteria (yellow). The branches correspond to amplicon sequence variants (ASVs) reported in this study. Bootstrap values are shown as circles.
4. Discussion
4.1. Bioreactor Performance: Sulfate Removal, Sulfide Production and Metals Removal
Sulfate removal is highly related to the ability of the treatment system to promote bioprecipitation of metals [52]. In the same bioreactor, Mendez et al. reported continuous increase in sulfate removal, reaching a maximum value of 35.7% when synthetic ARD was supplemented with Cu(II) concentrations of 10, 20, 30 and 40 mg L−1 [30]. While, our results showed better sulfate reducing activity, suggesting that growing of SRBs and sulfidogenesis were not affected by the presence of Cu(II) and Zn(II). This could be due to a better adaptation by SRBs in sludge, or to the combination of two or more metals in higher concentrations [53,54,55,56].
Previous research evaluated the inhibitory concentrations of Cu(II) and Zn(II), alone or in a mixture, using sludge inoculated in an SR bioreactor with a limestone pre-column system [57]. Those results showed that 15 mg L−1 of Cu(II) caused approximately 30% inhibition of the sulfate reducing activity. On the other hand, 15 mg Cu(II) L−1 and 15 mg Zn(II) L−1 caused less than 20% inhibition. In addition, depending on the nature of the metals in the mixture, their influence over anaerobic digestion can generate synergistic or antagonistic effects [19,52]. Despite this, no inhibitory effect due to Cu(II) and/or Zn(II) presence was observed during periods II and III of operation of the SR bioreactor in the current study. This could be attributed, mainly, to the low concentrations of these metals in the bioreactor, since in the limestone pre-column about 50% metal precipitation occurs as hydroxides and carbonates [53]. Moreover, the chemisorption in the sand (solid phase) into the bioreactor, could provide support and protection to anaerobes of the inoculated sludge through formation of biofilm [58].
The absence of methane in the SR bioreactor is not surprising since SRB can compete better than methanogens for common organic and inorganic substrates [52]. Typically, the organic substrate affinity of SRB for acetate is ten-fold higher than that of methanogens; consequently, the result of this competition is methanogenic inhibition, and methanogenesis is the predominant process [5,14,15]. On the other hand, an additional inhibition effect could be attributed to the toxicity of sulfide, the product of sulfate reduction, for other microbial groups [52]. For example, Moosa and Harrison [59] reported microbial inhibition in terms of a reduction in sulfate reduction activity and an increase when sulfide concentration increased in a chemo-stat culture at pH 7.0 ± 0.2 during the treatment of acid mine drainage. In addition, an excess of sulfate (e.g., 0.6 g COD/g SO42−), could promote the dominance of SRBs over methanogens at pH levels ranging from 7.4 to 8.0 [20]. Moreover, the presence of Cu(II) and/or Zn(II) can be toxic to methanogens and other microorganisms, through mechanisms such as the generation of reactive species of oxygen, or transmembrane interference in nutrient and energy transport, among others; consequently, methanogenesis could be affected [53,54,60]. Toxicity threshold concentrations of Cu(II) and Zn(II) to methanogens have been reported in several studies in the literature, which vary widely depending on the conditions under which anaerobic degradation was evaluated [61,62,63].
The complete treatment system composed of the limestone pre-column and the bioreactor efficiently removed Cu(II) and Zn(II). The limestone pre-column drove ARD neutralization and promoted metal precipitation as carbonates and hydroxides [53,64]. On the other hand, the bioreactor demonstrated strong Cu(II) and Zn(II) removal efficiencies through bioprecipitation. Sierra-Alvarez et al. [20] reported Cu(II) removal efficiencies higher than 99% in a system integrating a SR bioreactor with a fluidized bed crystallization reactor for semiconductor manufacturing wastewater treatment. In a study evaluating the bioremediation of ARD in flow-through columns testing zero-valent iron (ZVI), Ayala-Parra et al. obtained high removal efficiencies of Cu(II), Cd(II) and Pb(II) [53]. In bioreactors for treatment of ARD with very high concentrations of metals, efficiencies of the order of 90% and higher were achieved during the bioprecipitation of Cu (II), Zn (II), Cd (II), Pb (II), Ag (II), and Fe (II) [65].
Our data indicate that biogenic sulfide production was a confirmation of Cu(II) and Zn(II) removal by bioprecipitation. Nevertheless, pH neutralization and abiotic metal precipitation that occurred in the limestone pre-column, significantly contributed to heavy metal immobilization [30]. For that reason, the limestone in the bed reactor should be replaced when exhausted [66].
4.2. Microbial Diversity and Community Structure
Alpha diversity refers to which habitat contains the most taxa within each sample, while beta diversity refers to the habitat within samples [67,68,69]. This study showed a reduction in alpha and beta diversity with bioreactor operation time. This was supported by concomitant reduction of the richness, evenness, and dissimilarity metrics [69,70,71,72]. In addition, our findings indicated a higher dynamic alteration of low abundance ASVs and a stable persistence of a few dominant ASVs that contributed to the sulfate reduction process.
In bioreactors treating ARD, methanogens, such as Methanobacterium, Methanosaeta and Methanosarcina, among others, have been detected [14,30,73]. In the present research, the archaeal community was mainly constituted by ASVs belonging to Methanosarcina, which is the most abundant group of ASVs in the prokaryotes of all sludge samples assessed. These archaea are mainly acetoclastic methanogens, a robust and tolerant group against different stressors compared to other methanogens [14,74]. However, the Methanosarcina genus also includes hydrogenotrophic and methylotrophic methanogens [75]. Methanosarcina dominated the archaeal community, possibly displacing other methanogens. De Vrieze et al. [74] reported that the growth rate of Methanosarcina spp. is higher than those of Methanosaeta species in mesophilic anaerobic digestion with acetate concentrations up to 100 mg L−1. Differences in growth rates could generate the dominance of Methanosarcina populations over other methanogen species in the archaeal community.
Even though methanogens were the most abundant microorganisms, no methane production was detected. In addition to inhibition due to the sulfate reduction process, the absence of methane could be hypothesized to occur in the following two possible scenarios: (1) biological oxidation of methane or methanotrophy, that could occur by a combination of biotic and abiotic factors. Methanotrophy is driven by bacteria affiliated with Gamma-proteobacteria and Alpha-proteobacteria classes and anaerobic methanotrophic archaea (ANME) [76]. (2) reverse methanogenesis—this chemical mechanism occurs when SRBs consume hydrogen entirely, then methane concentration increases and the reverse reaction is thermodynamically possible [76,77]. In addition, due to the versatile and complex metabolisms of members of Methanosarcina, literature studies report that they are capable of growing and competing under anaerobic conditions without producing methane [78]. Finally, methane analysis is susceptible to being optimized throughout the employment of gas chromatography for detection and quantification [79].
In the current investigation, all non-syntrophic SRB populations increased during bioreactor operation and were affiliated to the Deltaproteobacteria class. While those which decreased (i.e., Desulfuromonadales, Desulfotomaculum and Desulfobacca), were affiliated to the Deltaproteobacteria and Clostridia classes [5]. The class Deltaproteobacteria is recognized to be a monophyletic bacteria group which are Gram-negative staining, mesophilic, sulfur and sulfate reducing, and able to reduce elemental metals [80]. On the other hand, in the phylum Firmicutes, Clostridia is a polyphyletic class that groups mainly Gram-positive bacteria and includes species of the Desulfotomaculum genus [5,81]. According to the results, the metal concentrations tested in the current study did not cause upset by changes in function and structure of enzymes that intervene in the sulfate reducing process [30]. This is due to the presence of metals that could generate endogenous dynamics in the composition of SBRs and select resistant populations. Mendez-Garcia et al. [25] reported that the presence of these microorganisms at acid mine drainage habitats is restricted to Deltaproteobacteria and Firmicutes members. A deeper biological knowledge of microbial resistance strategies to heavy metals requires an evolutionary genetics, proteomics and metabolomics approach [82,83].
Species of Geobacter and Syntrophobacter are acetate-oxidizing bacteria, and capable of metal reduction and sulfate reduction [25,46]. Both are affiliated with the Deltaproteobacteria class and became markedly more abundant during operation of the SR bioreactor [5,81]. A potential syntrophic partnership between bacteria, including Geobacter and Syntrophobacter, and hydrogenotrophic methanogens has been reported when acetoclastic methanogens, such as Metanosarcina spp., become more abundant in a sulfidogenic dominant environment [15,16,84].
As expected, the consensus tree provides a robust phylogenetic framework for the current study. At class level, we obtained clusters following the taxonomy, traits, and dynamics of microorganisms during the bioreactor operation [29,85].
Further studies are needed to determine how some factors, such as utilization of sustainable sources of electron donors (e.g., lignocellulosic or non-lignocellulosic materials) may play a significant role in microbiological interactions during sulfate reducing processes. In addition, it is relevant to study other microbial populations with syntrophic and fermentative lifestyles, and their metabolic networks.
5. Conclusions
This study confirmed that the treatment system, integrating a limestone pre-column and an SR bioreactor, is a successful technology for the removal of Cu(II) and Zn(II) from synthetic ARD using acetate as an organic carbon source and sulfate. Both metals were removed in the limestone pre-column and in the sulfate-reducing bioreactor.
The microbial community was comprised of archaeal and bacterial populations. However, sulfidogenesis was the dominant process and methanogenesis was not observed. An analysis of microbial diversity demonstrated changes throughout stabilization of the sulfate-reducing process and before the addition of metals. Gradually, the microbial community in the bioreactor became less diverse and interactions resulted in syntrophic associations. Thereby, the bioprecipitation process was not affected.
These findings provide new insights for understanding the metabolism and the functionality of microbial populations involved in sulfate reducing processes.
Author Contributions
Conceptualization, V.O.-H.; methodology, V.O.-H., R.S.-A., A.L.-R. and G.T.; software, A.Z.-R., D.X.R.-V. and P.C.; validation, V.O.-H. and A.Z.-R.; formal analysis, V.O.-H., R.S.-A., A.L.-R. and G.T.; investigation, V.O.-H. and A.Z.-R.; resources, V.O.-H.; data curation, V.O.-H., R.S.-A., G.T. and. P.C.; writing—original draft preparation, A.Z.-R.; writing—review and editing, V.O.-H., R.S.-A., A.L.-R., G.T., D.X.R.-V. and P.C.; visualization, D.X.R.-V. and A.Z.-R.; supervision, V.O.-H.; project administration, V.O.-H.; funding acquisition, V.O.-H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Universidad San Francisco de Quito’s Chancellor Grant 2016 and MiniGrant 2017 of the Institute of Microbiology.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Amplicon sequencing data, count tables and relevant data files can be found at GitHub (https://github.com/darioxr/SR_bioreactor_Cu_Zn (accessed on 8 January 2022)). This research was carried out with permit number MAE-DNB-CM-2018-0085 of the Ecuadorian Ministry of the Environment.
Acknowledgments
The authors thank Laboratorio de Ingeniería Ambiental (LIA-USFQ) for performing Cu(II) and Zn(II) analyses, and the Laboratorio de Agrobiotecnología USFQ for support in molecular analyses.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Simate, G.S.; Ndlovu, S. Acid Mine Drainage: Challenges and Opportunities. J. Environ. Chem. Eng. 2014, 2, 1785–1803. [Google Scholar] [CrossRef]
- Nieto, J.M.; Sarmiento, A.M.; Olías, M.; Canovas, C.R.; Riba, I.; Kalman, J.; Delvalls, T.A. Acid Mine Drainage Pollution in the Tinto and Odiel Rivers (Iberian Pyrite Belt, SW Spain) and Bioavailability of the Transported Metals to the Huelva Estuary. Environ. Int. 2007, 33, 445–455. [Google Scholar] [CrossRef]
- Zipper, C.; Skounsen, J. Acid Mine Drainage, Rock Drainage, and Acid Sulfate Soils; Jacobs, J.A., Lehr, J.H., Testa, S.M., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2014; ISBN 978-1-118-74919-7. [Google Scholar]
- Dar, S.A.; Yao, L.; van Dongen, U.; Kuenen, J.G.; Muyzer, G. Analysis of Diversity and Activity of Sulfate-Reducing Bacterial Communities in Sulfidogenic Bioreactors Using 16S RRNA and DsrB Genes as Molecular Markers. Appl. Environ. Microbiol. 2007, 73, 594–604. [Google Scholar] [CrossRef] [Green Version]
- Muyzer, G.; Stams, A.J.M. The Ecology and Biotechnology of Sulphate-Reducing Bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef]
- Kaksonen, A.H.; Puhakka, J.A. Sulfate Reduction Based Bioprocesses for the Treatment of Acid Mine Drainage and the Recovery of Metals. Eng. Life Sci. 2007, 7, 541–564. [Google Scholar] [CrossRef]
- Freeman, S.A.; Sierra-Alvarez, R.; Altinbas, M.; Hollingsworth, J.; Stams, A.J.M.; Smidt, H. Molecular Characterization of Mesophilic and Thermophilic Sulfate Reducing Microbial Communities in Expanded Granular Sludge Bed (EGSB) Reactors. Biodegradation 2008, 19, 161–177. [Google Scholar] [CrossRef]
- Hulshoff, L.W.; Lens, P.N.L.; Weijma, J.; Stams, A.J.M. New Developments in Reactor and Process Technology for Sulfate Reduction. Water Sci. Technol. 2001, 44, 67–76. [Google Scholar] [CrossRef]
- Xu, Y.-N.; Chen, Y. Advances in Heavy Metal Removal by Sulfate-Reducing Bacteria. Water Sci. Technol. 2020, 81, 1797–1827. [Google Scholar] [CrossRef]
- Hui, C.; Guo, Y.; Liu, L.; Yi, J. Recent Advances in Bacterial Biosensing and Bioremediation of Cadmium Pollution: A Mini-Review. World J. Microbiol. Biotechnol. 2022, 38, 9. [Google Scholar] [CrossRef]
- McCauley, C. Assessment of Passive Treatment and Biochemical Reactors for Ameliorating Acid Mine Drainage at Stockton Coal Mine. Ph.D Thesis, Univesity of Canterbury, Christchurch, New Zealand, 2011; pp. 1–366. [Google Scholar]
- Angai, J.U.; Ptacek, C.J.; Pakostova, E.; Bain, J.G.; Verbuyst, B.R.; Blowes, D.W. Removal of Arsenic and Metals from Groundwater Impacted by Mine Waste Using Zero-Valent Iron and Organic Carbon: Laboratory Column Experiments. J. Hazard. Mater. 2022, 424, 127295. [Google Scholar] [CrossRef]
- Jørgensen, B.B.; Findlay, A.J.; Pellerin, A. The Biogeochemical Sulfur Cycle of Marine Sediments. Front. Microbiol. 2019, 10, 849. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Niu, Q.; Li, L.; Hu, Y.; Mribet, C.; Hojo, T.; Li, Y.-Y. A Gradual Change between Methanogenesis and Sulfidogenesis during a Long-Term UASB Treatment of Sulfate-Rich Chemical Wastewater. Sci. Total Environ. 2018, 636, 168–176. [Google Scholar] [CrossRef] [PubMed]
- St. James, A.R.; Richardson, R.E. Ecogenomics Reveals Community Interactions in a Long-Term Methanogenic Bioreactor and a Rapid Switch to Sulfate-Reducing Conditions. FEMS Microbiol. Ecol. 2020, 96, fiaa050. [Google Scholar] [CrossRef]
- Yu, N.; Guo, B.; Liu, Y. Shaping Biofilm Microbiomes by Changing GAC Location during Wastewater Anaerobic Digestion. Sci. Total Environ. 2021, 780, 146488. [Google Scholar] [CrossRef]
- Schmidtova, J. Microbial Processes and Carbon Utilization in High Sulfate Waters and Sediments. Ph.D. Thesis, University of British Columbia, Kelowna, BC, Canada, 2010; pp. 1–174. [Google Scholar]
- Satoh, H.; Oshima, K.; Suda, W.; Ranasinghe, P.; Li, N.; Gunawardana, E.G.W.; Hattori, M.; Mino, T. Bacterial Population Dynamics in a Laboratory Activated Sludge Reactor Monitored by Pyrosequencing of 16S RRNA. Microbes Environ. 2013, 28, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Dodamani, S.; Hattiholi, A.; Kurjogi, M. Microbial Communities: An Effective Tool for Cleaning Environment. In An Integration of Phycoremediation Processes in Wastewater Treatment; Elsevier: Amsterdam, The Netherlands, 2022; pp. 231–248. ISBN 978-0-12-823499-0. [Google Scholar]
- Sierra-Alvarez, R.; Hollingsworth, J.; Zhou, M.S. Removal of Copper in an Integrated Sulfate Reducing Bioreactor−Crystallization Reactor System. Environ. Sci. Technol. 2007, 41, 1426–1431. [Google Scholar] [CrossRef]
- Chen, J.L.; Ortiz, R.; Steele, T.W.J.; Stuckey, D.C. Toxicants Inhibiting Anaerobic Digestion: A Review. Biotechnol. Adv. 2014, 32, 1523–1534. [Google Scholar] [CrossRef]
- Paulo, L.M.; Stams, A.J.M.; Sousa, D.Z. Methanogens, Sulphate and Heavy Metals: A Complex System. Rev. Environ. Sci. Bio/Technol. 2015, 14, 537–553. [Google Scholar] [CrossRef] [Green Version]
- Loreto, C.D.; Monge, O.; Martin, A.R.; Ochoa-Herrera, V.; Sierra-Alvarez, R.; Almendariz, F.J. Effect of Carbon Source and Metal Toxicity for Potential Acid Mine Drainage (AMD) Treatment with an Anaerobic Sludge Using Sulfate-Reduction. Water Sci. Technol. 2021, 83, 2669–2677. [Google Scholar] [CrossRef]
- Lin, Y.; Grembi, J.A.; Goots, S.S.; Sebastian, A.; Albert, I.; Brennan, R.A. Advantageous Microbial Community Development and Improved Performance of Pilot-Scale Field Systems Treating High-Risk Acid Mine Drainage with Crab Shell. J. Hazard. Mater. 2021, 420, 126665. [Google Scholar] [CrossRef]
- Méndez-García, C.; Pelaez, A.I.; Mesa, V.; Sanchez, J.; Golyshina, O.V.; Ferrer, M. Microbial Diversity and Metabolic Networks in Acid Mine Drainage Habitats. Front. Microbiol. 2015, 6, 475. [Google Scholar] [CrossRef] [Green Version]
- Grettenberger, C.L.; Hamilton, T.L. Metagenome-Assembled Genomes of Novel Taxa from an Acid Mine Drainage Environment. Appl. Environ. Microbiol. 2021, 87. [Google Scholar] [CrossRef]
- Konopka, A.; Lindemann, S.; Fredrickson, J. Dynamics in Microbial Communities: Unraveling Mechanisms to Identify Principles. ISME J. 2015, 9, 1488–1495. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and Processes of Microbial Community Assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef] [Green Version]
- Aguinaga, O.E.; White, K.N.; Dean, A.P.; Pittman, J.K. Addition of Organic Acids to Acid Mine Drainage Polluted Wetland Sediment Leads to Microbial Community Structure and Functional Changes and Improved Water Quality. Environ. Pollut. 2021, 290, 118064. [Google Scholar] [CrossRef]
- Méndez, G.; Trueba, G.; Sierra-Alvarez, R.; Ochoa-Herrera, V. Treatment of Acid Rock Drainage Using a Sulphate-Reducing Bioreactor with a Limestone Precolumn. Environ. Technol. 2021, 1–12. [Google Scholar] [CrossRef]
- Hedin, R.S.; Watzlaf, G.R.; Nairn, R.W. Passive Treatment of Acid Mine Drainage with Limestone. J. Environ. Qual. 1994, 23, 1338–1345. [Google Scholar] [CrossRef] [Green Version]
- Rice, E.W.; Baird, R.B.; Eaton, A.D.; Clesceri, L.S. Standard Methods for the Examination of Water and Wastewater, 22nd ed.; American Public Health Association (APHA); American Water Works Association (AWWA); Water Environment Federation (WEF): Washington, WA, USA, 2012. [Google Scholar]
- Trüper, H.G.; Schlegel, H.G. Sulphur Metabolism in Thiorhodaceae I. Quantitative Measurements on Growing Cells OfChromatium Okenii. Antonie Leeuwenhoek 1964, 30, 225–238. [Google Scholar] [CrossRef]
- Barua, V.B.; Kalamdhad, A.S. Biochemical Methane Potential Test of Untreated and Hot Air Oven Pretreated Water Hyacinth: A Comparative Study. J. Clean. Prod. 2017, 166, 273–284. [Google Scholar] [CrossRef]
- Zambrano-Romero, A. Changes in Microbial Composition during the Removal of Copper and Zinc in a Bioreactor with a Limestone Pre-Column System. Master’s Thesis, Universidad San Francisco de Quito, Quito, Ecuador, 2018. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Copenhagen, Denmark, 2018. [Google Scholar]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package; R Package, 2015; Available online: https://www.worldagroforestry.org/publication/vegan-community-ecology-package-r-package-vegan-vers-22-1 (accessed on 14 December 2021).
- Kim, B.-R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R.E. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jost, L.; Chao, A.; Chazdon, R. Compositional Similarity and β (Beta) Diversity. In Biological Diversity Frontiers in Measurement and Assesment. Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Wolda, H. Similarity Indices, Sample Size and Diversity. Oecologia 1981, 50, 296–302. [Google Scholar] [CrossRef]
- Vítěz, T.; Novák, D.; Lochman, J.; Vítězová, M. Methanogens Diversity during Anaerobic Sewage Sludge Stabilization and the Effect of Temperature. Processes 2020, 8, 822. [Google Scholar] [CrossRef]
- Rabus, R.; Venceslau, S.S.; Wöhlbrand, L.; Voordouw, G.; Wall, J.D.; Pereira, I.A.C. Chapter Two—A Post-Genomic View of the Ecophysiology, Catabolism and Biotechnological Relevance of Sulphate-Reducing Prokaryotes. Adv. Microb. Physiol. 2015, 66, 55–321. [Google Scholar]
- Hattori, S. Syntrophic Acetate-Oxidizing Microbes in Methanogenic Environments. Microbes Environ. 2008, 23, 118–127. [Google Scholar] [CrossRef] [Green Version]
- Kolde, R. Pheatmap: Pretty Heatmaps. 2015. Available online: https://mran.microsoft.com/snapshot/2015-09-04/web/packages/pheatmap/pheatmap.pdf (accessed on 14 December 2021).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Haroon, M.F.; Hu, S.; Shi, Y.; Imelfort, M.; Keller, J.; Hugenholtz, P.; Yuan, Z.; Tyson, G.W. Anaerobic Oxidation of Methane Coupled to Nitrate Reduction in a Novel Archaeal Lineage. Nature 2013, 500, 567–570. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v4: Recent Updates and New Developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of Anaerobic Digestion Process: A Review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Parra, P.; Sierra-Alvarez, R.; Field, J.A. Treatment of Acid Rock Drainage Using a Sulfate-Reducing Bioreactor with Zero-Valent Iron. J. Hazard. Mater. 2016, 308, 97–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dinh, H.T.; Kuever, J.; Mußmann, M.; Hassel, A.W.; Stratmann, M.; Widdel, F. Iron Corrosion by Novel Anaerobic Microorganisms. Nature 2004, 427, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Kieu, H.T.Q.; Müller, E.; Horn, H. Heavy Metal Removal in Anaerobic Semi-Continuous Stirred Tank Reactors by a Consortium of Sulfate-Reducing Bacteria. Water Res. 2011, 45, 3863–3870. [Google Scholar] [CrossRef]
- Bao, Y.; Jin, X.; Guo, C.; Lu, G.; Dang, Z. Sulfate-Reducing Bacterial Community Shifts in Response to Acid Mine Drainage in the Sediment of the Hengshi Watershed, South China. Environ. Sci. Pollut. Res. 2021, 28, 2822–2834. [Google Scholar] [CrossRef]
- Calderón, D.; Ochoa-Herrera, V. Evaluación de Consumo de Sustrato y Toxicidad Microbiana de Cobre (Cu (II)) y Zinc (Zn (II)) En Bacterias Sulfato-Reductoras Presentes En Sedimentos Anaerobios. Ph.D Thesis, Universidad San Francisco de Quito, Quito, Ecuador, 2016; pp. 1–54. [Google Scholar]
- Jarrell, K.F.; Saulnier, M.; Ley, A. Inhibition of Methanogenesis in Pure Cultures by Ammonia, Fatty Acids, and Heavy Metals, and Protection against Heavy Metal Toxicity by Sewage Sludge. Can. J. Microbiol. 1987, 33, 551–554. [Google Scholar] [CrossRef]
- Moosa, S.; Harrison, S.T.L. Product Inhibition by Sulphide Species on Biological Sulphate Reduction for the Treatment of Acid Mine Drainage. Hydrometallurgy 2006, 83, 214–222. [Google Scholar] [CrossRef]
- Gonzalez-Estrella, J.; Puyol, D.; Sierra-Alvarez, R.; Field, J.A. Role of Biogenic Sulfide in Attenuating Zinc Oxide and Copper Nanoparticle Toxicity to Acetoclastic Methanogenesis. J. Hazard. Mater. 2015, 283, 755–763. [Google Scholar] [CrossRef]
- Yue, Z.; Yu, H.; Wang, Z. Anaerobic Digestion of Cattail with Rumen Culture in the Presence of Heavy Metals. Bioresour. Technol. 2007, 98, 781–786. [Google Scholar] [CrossRef]
- Mudhoo, A.; Kumar, S. Effects of Heavy Metals as Stress Factors on Anaerobic Digestion Processes and Biogas Production from Biomass. Int. J. Environ. Sci. Technol. 2013, 10, 1383–1398. [Google Scholar] [CrossRef] [Green Version]
- Alrawashdeh, K.A.b.; Gul, E.; Yang, Q.; Yang, H.; Bartocci, P.; Fantozzi, F. Effect of Heavy Metals in the Performance of Anaerobic Digestion of Olive Mill Waste. Processes 2020, 8, 1146. [Google Scholar] [CrossRef]
- Medírcio, S.N.; Leão, V.A.; Teixeira, M.C. Specific Growth Rate of Sulfate Reducing Bacteria in the Presence of Manganese and Cadmium. J. Hazard. Mater. 2007, 143, 593–596. [Google Scholar] [CrossRef]
- Neculita, C.-M.; Zagury, G.J.; Bussière, B. Passive Treatment of Acid Mine Drainage in Bioreactors Using Sulfate-Reducing Bacteria. J. Environ. Qual. 2007, 36, 1–16. [Google Scholar] [CrossRef]
- Iakovleva, E.; Mäkilä, E.; Salonen, J.; Sitarz, M.; Wang, S.; Sillanpää, M. Acid Mine Drainage (AMD) Treatment: Neutralization and Toxic Elements Removal with Unmodified and Modified Limestone. Ecol. Eng. 2015, 81, 30–40. [Google Scholar] [CrossRef]
- Tuomisto, H. A Diversity of Beta Diversities: Straightening up a Concept Gone Awry. Part 1. Defining Beta Diversity as a Function of Alpha and Gamma Diversity. Ecography 2010, 33, 2–22. [Google Scholar] [CrossRef]
- Wagner, B.D.; Grunwald, G.K.; Zerbe, G.O.; Mikulich-Gilbertson, S.K.; Robertson, C.E.; Zemanick, E.T.; Harris, J.K. On the Use of Diversity Measures in Longitudinal Sequencing Studies of Microbial Communities. Front. Microbiol. 2018, 9, 1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, K.E.; Martiny, J.B.H. Alpha-, Beta-, and Gamma-Diversity of Bacteria Varies across Habitats. PLoS ONE 2020, 15, e0233872. [Google Scholar] [CrossRef] [PubMed]
- ElNaker, N.A.; Elektorowicz, M.; Naddeo, V.; Hasan, S.W.; Yousef, A.F. Assessment of Microbial Community Structure and Function in Serially Passaged Wastewater Electro-Bioreactor Sludge: An Approach to Enhance Sludge Settleability. Sci. Rep. 2018, 8, 7013. [Google Scholar] [CrossRef]
- Pane, C.; Sorrentino, R.; Scotti, R.; Molisso, M.; Di Matteo, A.; Celano, G.; Zaccardelli, M. Alpha and Beta-Diversity of Microbial Communities Associated to Plant Disease Suppressive Functions of On-Farm Green Composts. Agriculture 2020, 10, 113. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Zhou, Y.; Yao, X.; Cai, J.; Liu, X.; Tang, X.; Zhang, Y.; Jang, K.-S.; Jeppesen, E. Decreasing Diversity of Rare Bacterial Subcommunities Relates to Dissolved Organic Matter along Permafrost Thawing Gradients. Environ. Int. 2020, 134, 105330. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Andrea, I.; Sanz, J.L.; Bijmans, M.F.M.; Stams, A.J.M. Sulfate Reduction at Low PH to Remediate Acid Mine Drainage. J. Hazard. Mater. 2014, 269, 98–109. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, J.; Hennebel, T.; Boon, N.; Verstraete, W. Methanosarcina: The Rediscovered Methanogen for Heavy Duty Biomethanation. Bioresour. Technol. 2012, 112, 1–9. [Google Scholar] [CrossRef]
- Lackner, N.; Hintersonnleitner, A.; Wagner, A.O.; Illmer, P. Hydrogenotrophic Methanogenesis and Autotrophic Growth of Methanosarcina Thermophila. Archaea 2018, 2018, 4712608. [Google Scholar] [CrossRef] [Green Version]
- Serrano-Silva, N.; Sarria-Guzmán, Y.; Dendooven, L.; Luna-Guido, M. Methanogenesis and Methanotrophy in Soil: A Review. Pedosphere 2014, 24, 291–307. [Google Scholar] [CrossRef]
- Tate, K.R. Soil Methane Oxidation and Land-Use Change—From Process to Mitigation. Soil Biol. Biochem. 2015, 80, 260–272. [Google Scholar] [CrossRef]
- Ferry, J.G. Methanosarcina Acetivorans: A Model for Mechanistic Understanding of Aceticlastic and Reverse Methanogenesis. Front. Microbiol. 2020, 11, 1806. [Google Scholar] [CrossRef]
- Ochoa-Herrera, V.; Field, J.A.; Luna-Velasco, A.; Sierra-Alvarez, R. Microbial Toxicity and Biodegradability of Perfluorooctane Sulfonate (PFOS) and Shorter Chain Perfluoroalkyl and Polyfluoroalkyl Substances (PFASs). Environ. Sci. Processes Impacts 2016, 18, 1236–1246. [Google Scholar] [CrossRef]
- Waite, D.W.; Chuvochina, M.; Pelikan, C.; Parks, D.H.; Yilmaz, P.; Wagner, M.; Loy, A.; Naganuma, T.; Nakai, R.; Whitman, W.B.; et al. Proposal to Reclassify the Proteobacterial Classes Deltaproteobacteria and Oligoflexia, and the Phylum Thermodesulfobacteria into Four Phyla Reflecting Major Functional Capabilities. Int. J. Syst. Evol. Microbiol. 2020, 70, 5972–6016. [Google Scholar] [CrossRef]
- Cruz-Morales, P.; Orellana, C.A.; Moutafis, G.; Moonen, G.; Rincon, G.; Nielsen, L.K.; Marcellin, E. Revisiting the Evolution and Taxonomy of Clostridia, a Phylogenomic Update. Genome Biol. Evol. 2019, 11, 2035–2044. [Google Scholar] [CrossRef] [Green Version]
- Besaury, L.; Bodilis, J.; Delgas, F.; Andrade, S.; De la Iglesia, R.; Ouddane, B.; Quillet, L. Abundance and Diversity of Copper Resistance Genes CusA and CopA in Microbial Communities in Relation to the Impact of Copper on Chilean Marine Sediments. Mar. Pollut. Bull. 2013, 67, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Gillan, D.C. Metal Resistance Systems in Cultivated Bacteria: Are They Found in Complex Communities? Curr. Opin. Biotechnol. 2016, 38, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Giordani, A.; Rodriguez, R.P.; Sancinetti, G.P.; Hayashi, E.A.; Beli, E.; Brucha, G. Effect of Low PH and Metal Content on Microbial Community Structure in an Anaerobic Sequencing Batch Reactor Treating Acid Mine Drainage. Miner. Eng. 2019, 141, 105860. [Google Scholar] [CrossRef]
- Müller, A.L.; Kjeldsen, K.U.; Rattei, T.; Pester, M.; Loy, A. Phylogenetic and Environmental Diversity of DsrAB-Type Dissimilatory (Bi)Sulfite Reductases. ISME J. 2015, 9, 1152–1165. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).