Simultaneous Detection of Four Main Foodborne Pathogens in Ready-to-Eat Food by Using a Simple and Rapid Multiplex PCR (mPCR) Assay
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Selection of a Co-Culture Medium
2.2.1. Effect of Various Culture Broths on Individual Growth
2.2.2. Evaluation of BPW Recovery Capacity
Preparation of the Inoculum
Effect of BPW on Individual and Co-Culture Growth
Effect of BPW on Co-Culture Growth, from Artificially Inoculated Food Matrix
2.3. PCR Detection
2.3.1. Preparation of DNA Template
Thermal Lysis Method
GenElute™ Bacterial Genomic DNA Kit
2.3.2. Primers
2.3.3. Simplex PCR
2.3.4. Multiplex PCR (mPCR)
2.3.5. Evaluation of Specificity
2.3.6. Evaluation of Sensitivity
Detection Limits of Simplex and mPCR, from Individual Cultures
Detection Limits from BPW Co-Culture Recovery
Detection Limits from BPW Co-Culture Recovery, with Artificially Inoculated Food Matrices
3. Results
3.1. Selection of a Co-Culture Medium
3.1.1. Effect of Various Culture Broths on Individual Growth
3.1.2. Effect of BPW on Individual and Co-Culture Growth
3.1.3. Effect of BPW on Co-Culture Growth from Artificially Inoculated Food Matrix
3.2. PCR Detection
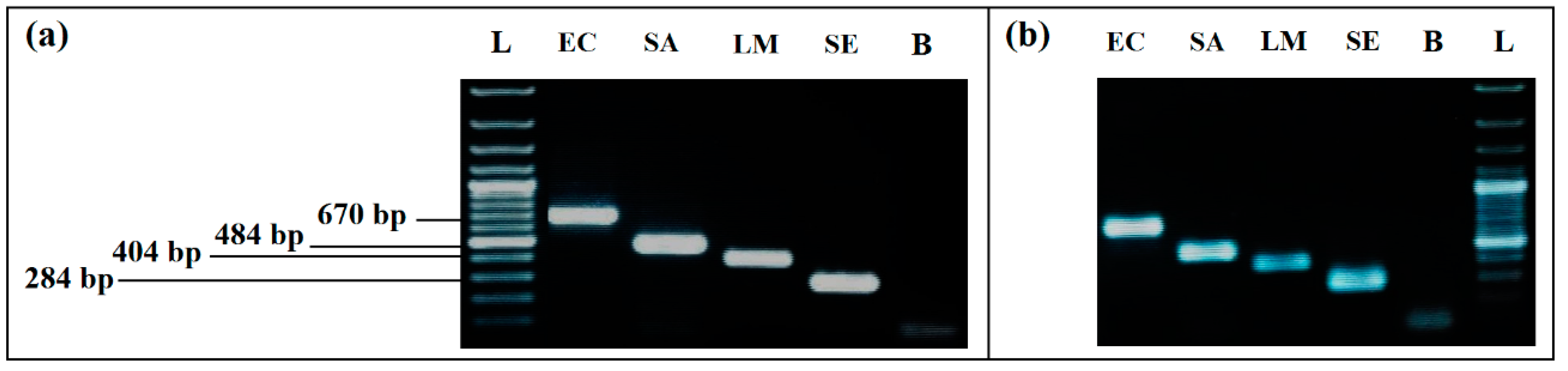
3.2.1. Simplex PCR
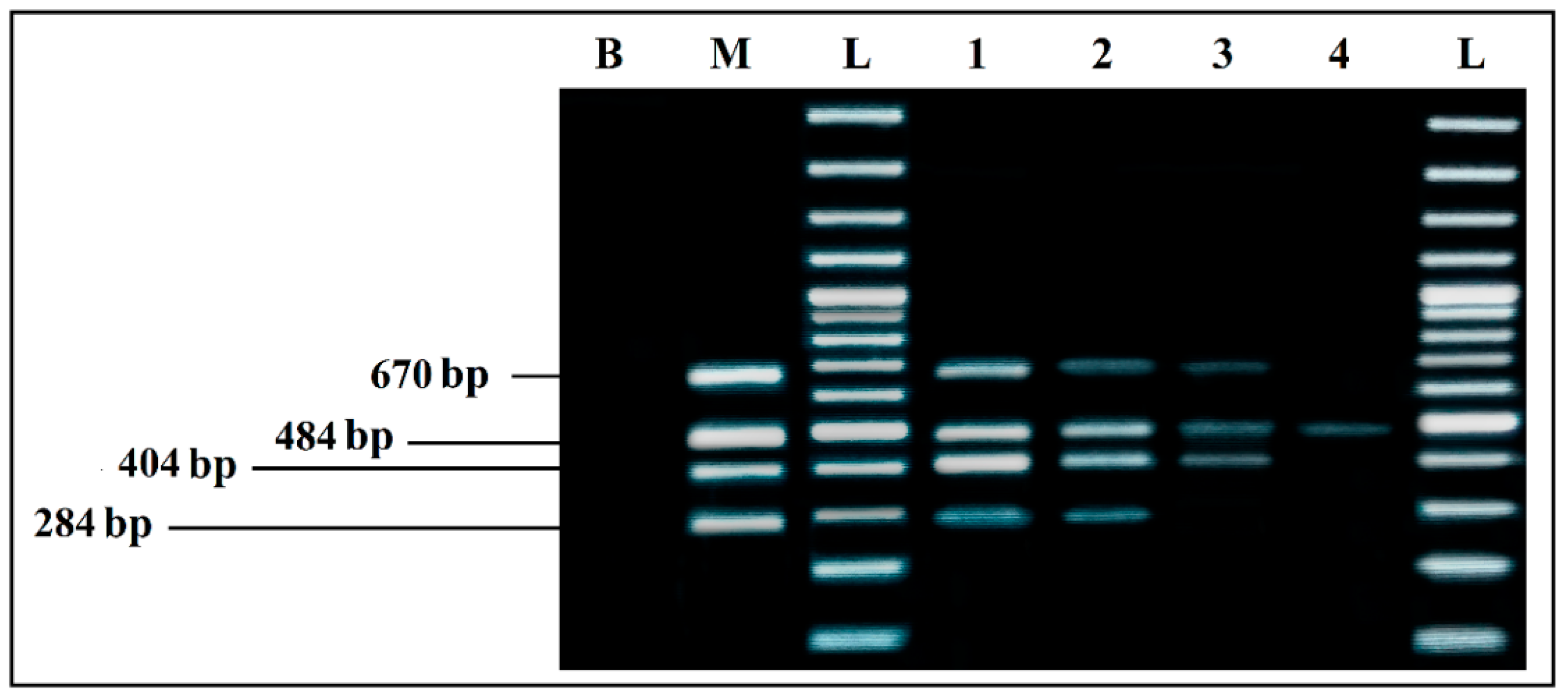
3.2.2. mPCR
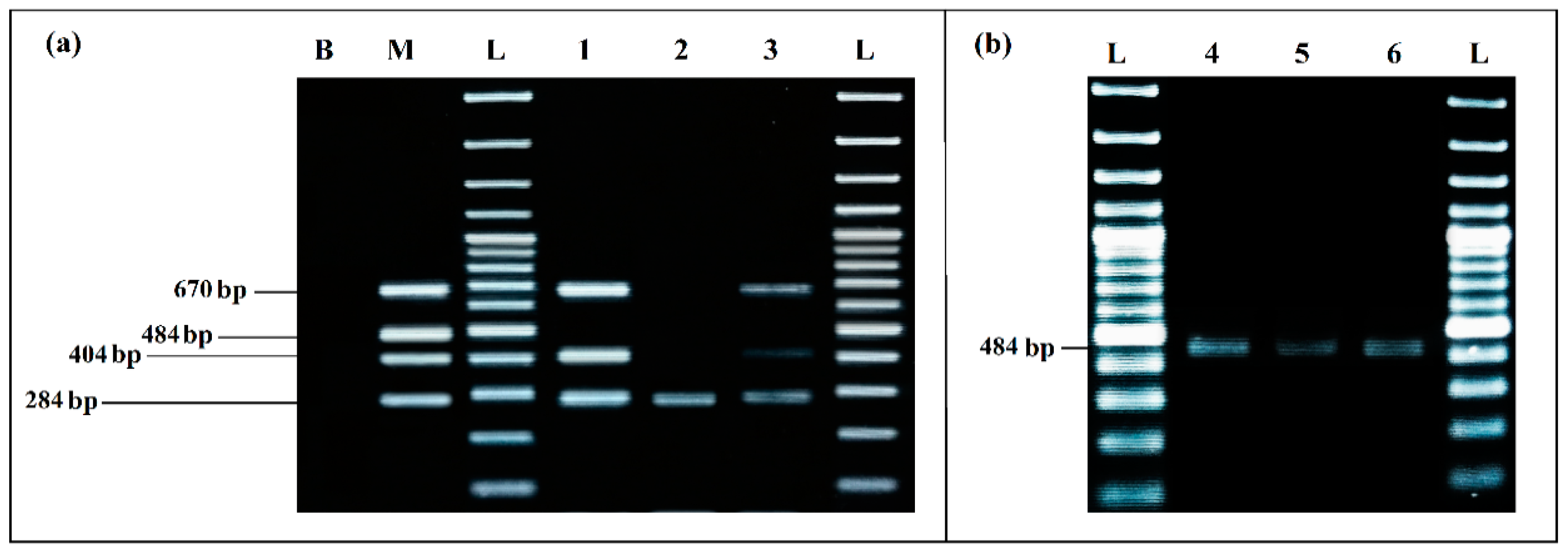
3.2.3. Evaluation of Specificity
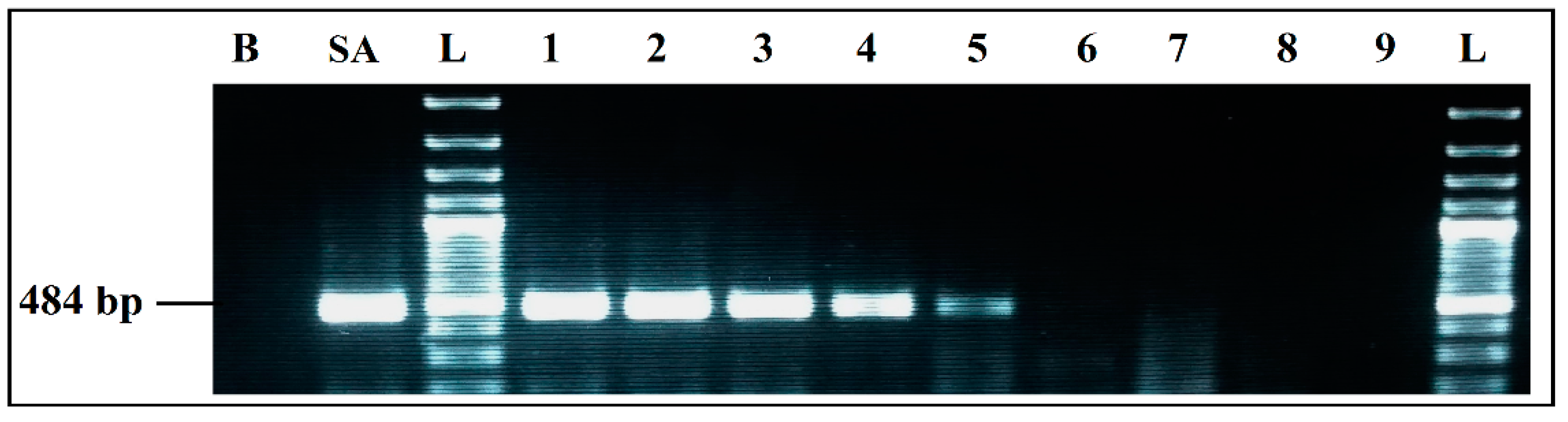
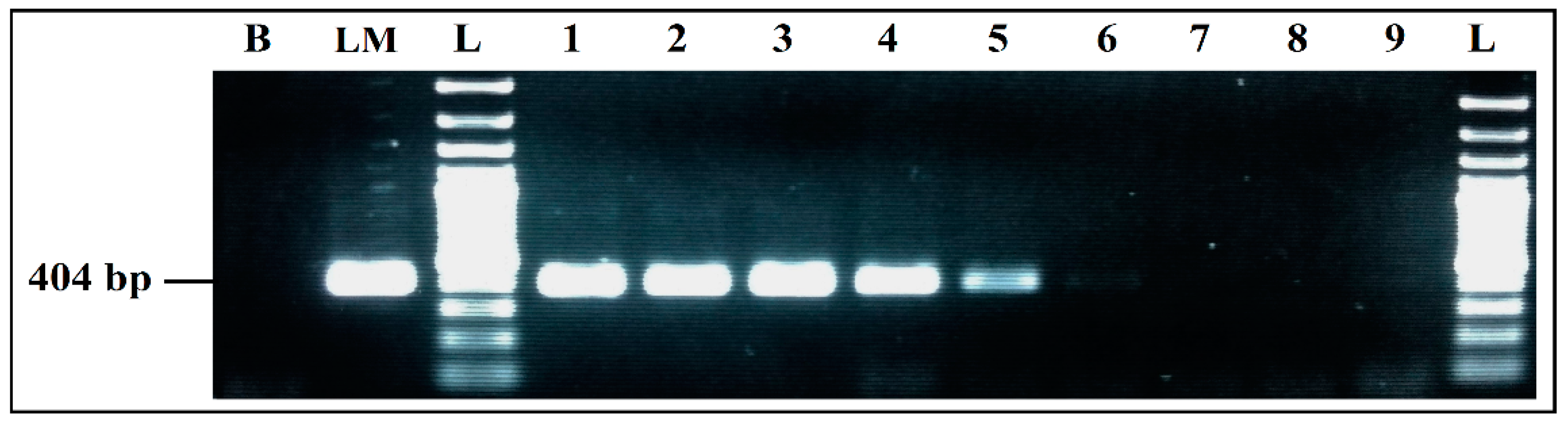
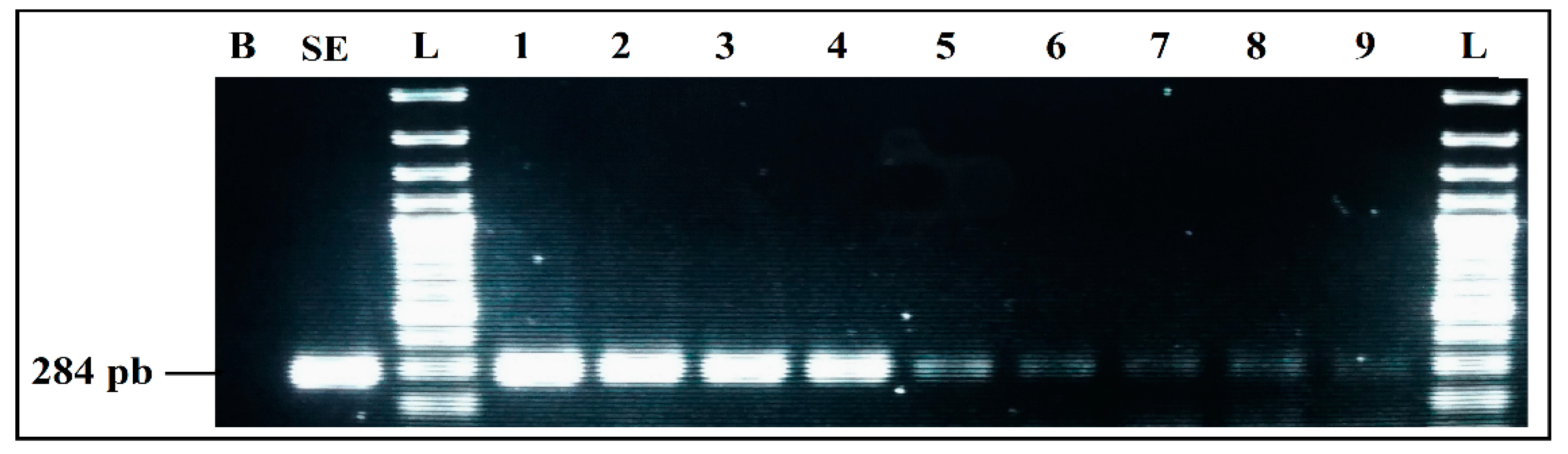
3.2.4. Evaluation of Sensitivity
Detection Limits of Simplex and mPCR, from Individual Cultures
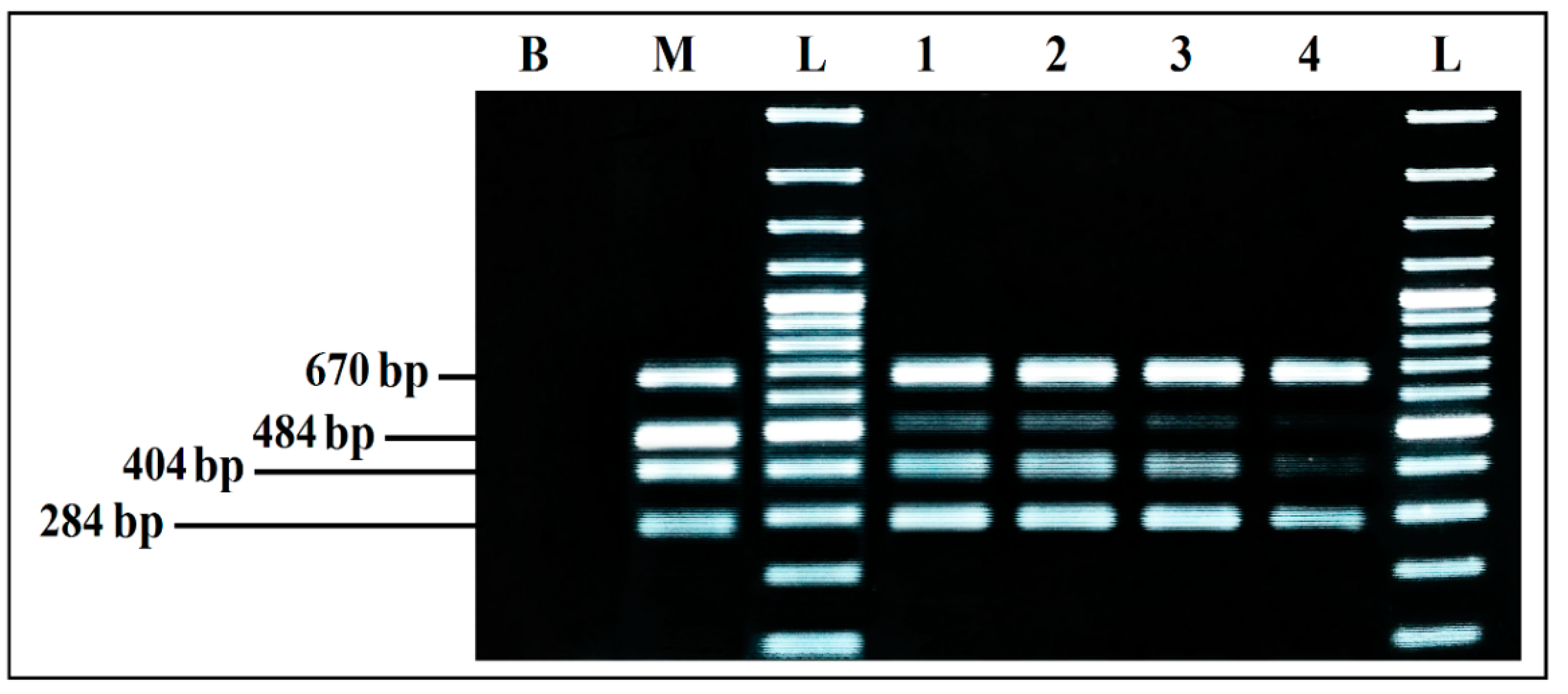
Detection Limits for Co-Culture in BPW
Detection Iimits from BPW Co-Culture from Artificially Inoculated Food Matrix
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BGBLB | Brilliant Green Bile Lactose Broth |
| BPA | Baird Parker Agar Base |
| BPW | Buffered Peptone Water |
| CECT | Spanish Type Culture Collection |
| CFU | Colony Forming Units |
| dNTPs | Deoxynucleotide triphosphates |
| F/R | Forward/Reverse |
| FB | Fraser Enrichment Broth |
| GC | Giolitti-Cantoni Broth |
| ISO | International Organization for Standardization |
| LB | Luria-Bertani Broth |
| mPCR | Multiplex PCR |
| NB | Nutrient Broth |
| PAL | Palcam Agar |
| PCR | Polymerase Chain Reaction |
| RV | Rappaport Vassiliadis Broth |
| TBX | Tryptone Bile Glucuronic Agar |
| UNE-EN | Spanish Association for Standardisation |
| XLD | Xylose-Lysine-Deoxycholate Agar |
References
- Ruppitsch, W.; Pietzka, A.; Cabal, A.; Chakeri, A.; Schmid, D.; Lakicevic, B.; Lepuschitz, S.; Allerberger, F. Advances in foodborne outbreak investigation and source tracking using whole genome sequencing. Conf. Ser. Earth Environ. Sci. 2019, 333, 012010. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef]
- Wei, C.; Zhong, J.; Hu, T.; Xihong, Z. Simultaneous detection of Escherichia coli O157:H7, Staphylococcus aureus and Salmonella by multiplex PCR in milk. 3 Biotech 2018, 8, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Harvey, R.R.; Zakhour, C.M.; Gould, L.H. Foodborne Disease Outbreaks Associated with Organic Foods in the United States. J. Food Prot. 2016, 79, 1953–1958. [Google Scholar] [CrossRef]
- Chen, J.; Tang, J.; Bhunia, A.K.; Tang, C.; Wang, C.; Shi, H. Development of a multi-pathogen enrichment broth for simultaneous growth of five common foodborne pathogens. J. Gen. Appl. Microbiol. 2015, 61, 224–231. [Google Scholar] [CrossRef]
- Burgess, C.M.; Gianotti, A.; Gruzdev, N.; Holah, J.; Knøchel, S.; Lehner, A.; Margas, E.; Esser, S.S.; Sela, S.; Tresse, O. The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int. J. Food Microbiol. 2016, 221, 37–53. [Google Scholar] [CrossRef]
- Ding, T.; Suo, Y.; Zhang, Z.; Liu, D.; Ye, X.; Chen, S.; Zhao, Y. A Multiplex RT-PCR Assay for S. aureus, L. monocytogenes, and Salmonella spp. Detection in Raw Milk with Pre-enrichment. Front. Microbiol. 2017, 8, 989. [Google Scholar] [CrossRef]
- Okafor, A.C.; Ogbo, F.C. Occurrence and enumeration of multiple bacterial pathogens in edible snails from Southeast Nigeria. J. Food Sci. Technol. 2019, 7, 23–30. [Google Scholar] [CrossRef][Green Version]
- Leclerc, V.; Dufour, B.; Lombard, B.; Gauchard, F.; Garin-Bastuji, B.; Salvat, G.; Brisabois, A.; Poumeyrol, M.; Buyser, M.L.; Besse, N.G.; et al. Pathogens in meat and milk products: Surveillance and impact on human health in France. Livest. Prod. Sci. 2002, 76, 195–202. [Google Scholar] [CrossRef]
- Elizaquível, P.; Aznar, R. A multiplex RTi-PCR reaction for simultaneous detection of Escherichia coli O157:H7, Salmonella spp. and Staphylococcus aureus on fresh, minimally processed vegetables. Food Microbiol. 2008, 25, 705–713. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Zhou, W.W.; Zhou, Y.; Wang, X.F.; Xu, J.F. Response surface methodology to design a selective co-enrichment broth of Escherichia coli, Salmonella spp. and Staphylococcus aureus for simultaneous detection by multiplex PCR. Microbiol. Res. 2012, 167, 405–412. [Google Scholar] [CrossRef]
- Harris, L.J.; Farber, J.N.; Beuchat, L.R.; Parish, M.E.; Suslow, T.V.; Garrett, E.H.; Busta, F.F. Outbreaks associated with fresh produce: Incidence, growth, and survival of pathogens in fresh and fresh cut produce. Compr. Rev. Food Sci. Food Saf. 2003, 2, 78–141. [Google Scholar] [CrossRef]
- Al-Kharousi, Z.S.; Guizani, N.; Al-Sadi, A.M.; Al-Bulushi, I.M.; Shaharoona, B. Hiding in fresh fruits and vegetables: Opportunistic pathogens may cross geographical barriers. Int. J. Microbiol. 2016, 2016, 4292417. [Google Scholar] [CrossRef]
- Zeng, D.; Chen, Z.; Jiang, Y.; Xue, F.; Li, B. Advances and challenges in viability detection of foodborne pathogens. Front. Microbiol. 2016, 7, 1833. [Google Scholar] [CrossRef]
- Kothe, C.I.; Pessoa, J.P.; Malheiros, P.S.; Tondo, E.C. Assessing the growth of Staphylococcus aureus and Escherichia coli on fruits and vegetables. J. Infect. Dev. Ctries. 2019, 13, 480–486. [Google Scholar] [CrossRef]
- Rajabzadeh, S.; Bahreini, M.; Sharifmoghadam, M.R. A rapid method for separating and concentration of food-borne pathogens using elution from ready-to-eat vegetables. Iran. J. Microbiol. 2018, 10, 385–393. [Google Scholar] [PubMed]
- Centers for Disease Control and Prevention. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food-10 states, United States, 2005. Morb. Mortal. Wkly. Rep. 2006, 55, 392–395. [Google Scholar]
- World Health Organization. Food Safety and Foodborne Illness; World Health Organization: Geneva, Switzerland, 2007; Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 15 November 2021).
- Suo, B.; Wang, Y. Evaluation of a multiplex selective enrichment broth SEL for simultaneous detection of injured Salmonella, Escherichia coli O157:H7 and Listeria monocytogenes. Braz. J. Microbiol. 2013, 44, 737–742. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Omiccioli, E.; Amagliani, G.; Bandi, G.; Magnani, M. A new platform for real time PCR detection of Salmonella spp., L. monocytogenes and E. coli O157 in milk. Food Microbiol. 2009, 26, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Latha, C.; Anu, C.J.; Ajaykumar, V.J.; Sunil, B. Prevalence of Listeria monocytogenes, Yersinia enterocolitica, Staphylococcus aureus, and Salmonella enterica Typhimurium in meat and meat products using multiplex polymerase chain reaction. Vet. World 2017, 10, 927–931. [Google Scholar] [CrossRef][Green Version]
- Feng, K.; Hu, W.; Jiang, A.; Sarengaowa, X.Y.; Zou, Y.; Yang, L.; Wang, X. A dual filtration-based Multiplex PCR method for simultaneous detection of viable Escherichia coli O157:H7, Listeria monocytogenes, and Staphylococcus aureus on fresh-cut cantaloupe. PLoS ONE 2016, 11, e0166874. [Google Scholar] [CrossRef] [PubMed]
- Burnett, S.; Beuchat, L. Human pathogens associated with raw produce and unpasteurized juices, and difficulties in decontamination. J. Ind. Microbiol. Biotechnol. 2001, 27, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Rantsiou, K.; Iacumin, L.; Cantoni, C.; Comi, G. Direct identification in food samples of Listeria spp. and Listeria monocytogenes by molecular methods. Appl. Environ. Microbiol. 2002, 68, 6273–6282. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Hogan, J.; Smith, K.L. Development of a PCR-based assay to detect Shiga toxin producing Escherichia coli, Listeria monocytogenes, and Salmonella in milk. Food Microbiol. 2003, 20, 345–350. [Google Scholar] [CrossRef]
- Dwivedi, H.P.; Jaykus, L.A. Detection of pathogens in foods: The current state-of-the-art and future directions. Crit. Rev. Microbiol. 2011, 37, 40–63. [Google Scholar] [CrossRef]
- Nam, H.M.; Srinivasan, V.; Gillespie, B.E.; Murinda, S.E.; Oliver, S.P. Application of SYBR green real-time PCR assay for specific detection of Salmonella spp. in dairy farm environmental samples. Int. J. Food Microbiol. 2005, 102, 161–171. [Google Scholar] [CrossRef]
- Bai, J.; Shi, X.; Nagaraja, T.G. A multiplex PCR procedure for the detection of six major virulence genes in Escherichia coli O157:H7. J. Microbiol. Methods 2010, 82, 85–89. [Google Scholar] [CrossRef]
- Qin, H.; Shi, X.; Yu, L.; Li, K.; Wang, J.; Chen, J.; Yang, F.; Xu, H.; Xu, H. Multiplex real-time PCR coupled with sodium dodecyl sulphate and propidium monoazide for the simultaneous detection of viable Listeria monocytogenes, Cronobacter sakazakii, Staphylococcus aureus and Salmonella spp. in milk. Int. Dairy J. 2020, 108, 104739. [Google Scholar] [CrossRef]
- Qu, Y.; Bai, Y.; Liu, Y.; Zhou, C.; Zhou, X.; Zhang, D.; Shi, C.; Suo, Y. SSEL, a selective enrichment broth for simultaneous growth of Salmonella enterica, Staphylococcus aureus, Escherichia coli O157: H7, and Listeria monocytogenes. J. Food Saf. 2020, 40, 12837. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, X.; Wang, Q. Sensitive and specific detection of E. coli, Listeria monocytogenes, and Salmonella enterica serovar Typhimurium in milk by microchip electrophoresis combined with multiplex PCR amplification. Microchem. J. 2020, 157, 104876. [Google Scholar] [CrossRef]
- Shi, X.; Yu, L.; Lin, C.; Li, K.; Chen, J.; Qin, H. Biotin exposure–based immunomagnetic separation coupled with sodium dodecyl sulfate, propidium monoazide, and multiplex real-time PCR for rapid detection of viable Salmonella Typhimurium, Staphylococcus aureus, and Listeria monocytogenes in milk. Int. J. Dairy Sci. 2021, 104, 6588–6597. [Google Scholar] [CrossRef]
- Jacobse, C.N. The influence of commonly used selective agents on the growth of Listeria monocytogenes. Int. J. Food Microbiol. 1999, 50, 221–226. [Google Scholar] [CrossRef]
- Kim, H.; Bhunia, A.K. SEL, a selective enrichment broth for simultaneous growth of Salmonella enterica, Escherichia coli O157:H7, and Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 4853–4866. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.G.; Wu, H.; Liu, Y.Y.; Li, S.L.; Yang, X.Q.; Xiao, X.L. A multipathogen selective enrichment broth for simultaneous growth of Salmonella enterica serovar Enteritidis, Staphylococcus aureus, and Listeria monocytogenes. Can. J. Microbiol. 2010, 56, 585–597. [Google Scholar] [CrossRef] [PubMed]
- International Organization for Standardization. ISO 4832 (2006); Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Coliforms—Colony Count Techniqu; 6, 28004. AENOR: Génova, Italy; Madrid, Spain, 2006.
- Spanish Association for Standardization. UNE-EN ISO 6579-1 (2017); Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp.; 6, 28004. AENOR: Génova, Italy; Madrid, Spain, 2017.
- Spanish Association for Standardization. UNE-EN ISO 6888-3 (2003); Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Coagulase-positive Staphylococci (Staphylococcus aureus and Other Species)—Part 3: Detection and MPN Technique for Low Numbers; 6, 28004. AENOR: Génova, Italy; Madrid, Spain, 2003.
- Spanish Association for Standardization. UNE-EN ISO 11290-1 (2017); Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp.—Part 1: Detection Method; 6, 28004. AENOR: Génova, Italy; Madrid, Spain, 2018.
- Spanish Association for Standardization. UNE-EN ISO 7218 (2007); Microbiology of Food and Animal Feeding Stuffs—General Requirements and Guidance for Microbiological Examinations; 6, 28004. AENOR: Génova, Italy; Madrid, Spain, 2008.
- McDaniels, A.E.; Rice, E.W.; Reyes, A.L.; Johnson, C.H.; Haugland, R.A.; Stelma, G.N., Jr. Confirmational identification of Escherichia coli, a comparison of genotypic and phenotypic assays for glutamate decarboxylase and β Dglucuronidase. Appl. Environ. Microbiol. 1996, 62, 3350–3354. [Google Scholar] [CrossRef]
- Xu, Y.P.; Cheng, W.; Shao, Y.C.; Chen, F.S. Detection of Salmonella spp., Escherichia coli and Staphylococcus aureus by multiplex PCR. Chin. J. Microbiol. Immunol. 2006, 33, 89–94. [Google Scholar]
- Wu, S.J.; Chan, A.; Kado, C.I. Detection of PCR amplicons from bacterial pathogens using microsphere agglutination. J. Microbiol. Methods 2004, 56, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Rahn, K.; De Grandis, S.A.; Clarke, R.C.; McEwen, S.A.; Galan, J.E.; Ginocchio, C.; Curtiss, R., III; Gyles, C.L. Amplification of an invA gene sequence of Salmonella enterica by polymerase chain reaction as a specific method of detection of Salmonella. Mol. Cell. Probes 1992, 6, 271–279. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Goli, M.; Ezzatpanah, H.; Ghavami, M.; Chamani, M.; Aminafshar, M.; Toghiani, M.; Eghbalsaied, S. The effect of multiplex-PCR-assessed major pathogens causing subclinical mastitis on somatic cell profiles. Trop. Anim. Health Prod. 2012, 44, 1673–1680. [Google Scholar] [CrossRef]
- Abd El-Salam, A.F.; Abd El-Ghany, Z.M.; El-Tahan, M.H. The comparison between different enrichment broth media and selective solid media for growing of Salmonella typhimurium and Listeria monocytogenes. JASB 2010, 7, 351–363. [Google Scholar] [CrossRef]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Suo, B. A new 7-plex PCR assay for simultaneous detection of shiga toxin-producing Escherichia coli O157 and Salmonella Enteritidis in meat products. JCF 2011, 6, 441–447. [Google Scholar] [CrossRef]
- Alarcoän, B.; Garciäa-Canas, V.; Cifuentes, A.; Gonzaälez, R.; Aznar, R. Simultaneous and Sensitive Detection of Three Foodborne Pathogens by Multiplex PCR, Capillary Gel Electrophoresis, and Laser-Induced Fluorescence. J. Agric. Food Chem. 2004, 52, 7180–7186. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.S.; Cox, N.A. Universal preenrichment broth for the simultaneous detection of Salmonella and Listeria in foods. J. Food Prot. 1992, 55, 256–259. [Google Scholar] [CrossRef]
- Dailey, R.C.; Martin, K.G.; Smiley, R.D. The effects of competition from non-pathogenic foodborne bacteria during the selective enrichment of Listeria monocytogenes using buffered Listeria enrichment broth. Food Microbiol. 2014, 44, 173–179. [Google Scholar] [CrossRef][Green Version]
- Oberhofer, T.R.; Frazier, W.C. Competition of Staphylococcus aureus with other organisms. J. Milk Food Technol. 1961, 24, 172–175. [Google Scholar] [CrossRef]
- Herman, K.M.; Hall, A.J.; Gould, L.H. Outbreaks attributed to fresh leafy vegetables, United States, 1973–2012. Epidemiol. Infect. 2015, 143, 3011–3021. [Google Scholar] [CrossRef]
- Zilelidou, E.; Manthou, E.; Skandamis, P. Growth differences and competition between Listeria monocytogenes strains determine their predominance on ham slices and lead to bias during selective enrichment with the ISO protocol. Int. J. Food Microbiol. 2016, 235, 60–70. [Google Scholar] [CrossRef]
- Kim, J.; Demeke, T.; Clear, R.M.; Patrick, S.K. Simultaneous detection by PCR of Escherichia coli, Listeria monocytogenes and Salmonella typhimurium in artificially inoculated wheat grain. Int. J. Food Microbiol. 2006, 111, 21–25. [Google Scholar] [CrossRef]
- Germini, A.; Masola, A.; Carnevali, P.; Marchelli, R. Simultaneous detection of Escherichia coli O175: H7, Salmonella spp., and Listeria monocytogenes by multiplex PCR. Food Control 2009, 20, 733–738. [Google Scholar] [CrossRef]
- Yuan, Y.; Xu, W.; Zhai, Z.; Shi, H.; Luo, Y.; Chen, Z.; Huang, K. Universal Primer-Multiplex PCR Approach for Simultaneous Detection of Escherichia coli, Listeria monocytogenes, and Salmonella spp. in Food Samples. J. Food Sci. 2009, 74, 446–452. [Google Scholar] [CrossRef]
- Ma, K.; Deng, Y.; Bai, Y.; Xu, D.; Chen, E.; Wu, H.; Li, B.; Gao, L. Rapid and simultaneous detection of Salmonella, Shigella, and Staphylococcus aureus in fresh pork using a multiplex real-time PCR assay based on immunomagnetic separation. Food Control 2014, 42, 87–93. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, L.; Xu, H.; Aguilar, Z.P.; Liu, C.; Gan, B.; Xiong, Y.; Lai, W.; Xu, F.; Wei, H. Detection of non-emetic and emetic Bacillus cereus by propidium monoazide multiplex PCR (PMA-mPCR) with internal amplification control. Food Control 2014, 35, 401–406. [Google Scholar] [CrossRef]
- Markoulatos, P.; Siafakas, N.; Moncany, M. Multiplex polymerase chain reaction: A practical approach. J. Clin. Lab. Anal. 2002, 16, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Garrido, A.; Chapela, M.J.; Román, B.; Fajardo, P.; Vieites, J.M.; Cabado, G. In-house validation of a multiplex real-time PCR method for simultaneous detection of Salmonella spp., Escherichia coli O157:H7 and Listeria monocytogenes. Int. J. Food Microbiol. 2013, 164, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, A.; Mukhopadhyay, U.K. Novel multiplex PCR approaches for the simultaneous detection of human pathogens: Escherichia coli 0157:H7 and Listeria monocytogenes. J. Microbiol. 2007, 68, 193–200. [Google Scholar] [CrossRef] [PubMed]

| Strains | Primers | Sequences | Size | References |
|---|---|---|---|---|
| E. coli | GADA/F | ACCTGCGTTGCGTAAATA | 670 bp | McDaniels et al., 1996 [41] |
| GADA/R | GGGCGGGAGAAGTTGATG | |||
| S. aureus | Nuc/F | CTTTAGCCAAGCCTTGACGAAC | 484 pb | Xu et al., 2006 [42] |
| Nuc/R | AAAGGGCAATACGCAAAGAGGT | |||
| L. monocytogenes | LM404/F | ATCATCGACGGCAACCTCGGAGAC | 404 bp | Wu et al., 2004 [43] |
| LM404/R | CACCATTCCCAAGCTAAACCAGTGC | |||
| S. enterica | SalinvA139 | GTGAAATTATCGCCACGTTCGGGCAA | 284 bp | Rahn et al., 1992 [43] |
| SalinvA141 | TCATCGCACCGTCAAAGGAACC |
| Bacteria | Selective Enrichment (BGBLB, RV, GC, FB) | NB | BPW | LB | SSSLE |
|---|---|---|---|---|---|
| E. coli | 9.30 × 107 | 1.48 × 109 | 1.19 × 109 | 2.73 × 108 | 5.20 × 105 |
| S. enterica | 6.83 × 107 | 3.29 × 108 | 4.10 × 108 | 4.26 × 107 | 0 |
| S. aureus | 1.74 × 108 | 7.70 × 1010 | 1.07 × 109 | 7.88 × 108 | 0 |
| L. monocytogenes | 8.65 × 109 | 8.15 × 108 | 9.97 × 107 | 3.49 × 108 | 0 |
| Bacteria | Initial Inoculum | Individual Culture | Co-Culture |
|---|---|---|---|
| E. coli | 103 CFU/mL | 8.55 × 108 | 7.70 × 108 |
| S. aureus | 3.27 × 108 | 1.70 × 106 | |
| L. monocytogenes | 2.10 × 107 | 4.60 × 105 | |
| S. enterica | 2.20 × 108 | 1.60 × 107 | |
| E. coli | 102 CFU/mL | 7.50 × 108 | 5.65 × 108 |
| S. aureus | 4.60 × 108 | 2.70 × 105 | |
| L. monocytogenes | 1.75 × 107 | 1.38 × 105 | |
| S. enterica | 1.28 × 108 | 9.95 × 106 | |
| E. coli | 101 CFU/mL | 5.15 × 108 | 4.20 × 108 |
| S. aureus | 1.32 × 108 | 4.20 × 105 | |
| L. monocytogenes | 1.50 × 107 | 6.85 × 104 | |
| S. enterica | 1.08 × 108 | 9.75 × 106 |
| Bacteria | Without Matrix | With Lettuce Matrix | With Minced Meat Matrix |
|---|---|---|---|
| E. coli | 7.70 × 108 | 2.00 × 108 | 5.20 × 107 |
| S. aureus | 1.70 × 106 | 8.75 × 106 | 4.45 × 107 |
| L. monocytogenes | 4.60 × 105 | 1.30 × 106 | 4.85 × 105 |
| S. enterica | 1.60 × 107 | 1.32 × 108 | 5.50 × 107 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boukharouba, A.; González, A.; García-Ferrús, M.; Ferrús, M.A.; Botella, S. Simultaneous Detection of Four Main Foodborne Pathogens in Ready-to-Eat Food by Using a Simple and Rapid Multiplex PCR (mPCR) Assay. Int. J. Environ. Res. Public Health 2022, 19, 1031. https://doi.org/10.3390/ijerph19031031
Boukharouba A, González A, García-Ferrús M, Ferrús MA, Botella S. Simultaneous Detection of Four Main Foodborne Pathogens in Ready-to-Eat Food by Using a Simple and Rapid Multiplex PCR (mPCR) Assay. International Journal of Environmental Research and Public Health. 2022; 19(3):1031. https://doi.org/10.3390/ijerph19031031
Chicago/Turabian StyleBoukharouba, Aya, Ana González, Miguel García-Ferrús, María Antonia Ferrús, and Salut Botella. 2022. "Simultaneous Detection of Four Main Foodborne Pathogens in Ready-to-Eat Food by Using a Simple and Rapid Multiplex PCR (mPCR) Assay" International Journal of Environmental Research and Public Health 19, no. 3: 1031. https://doi.org/10.3390/ijerph19031031
APA StyleBoukharouba, A., González, A., García-Ferrús, M., Ferrús, M. A., & Botella, S. (2022). Simultaneous Detection of Four Main Foodborne Pathogens in Ready-to-Eat Food by Using a Simple and Rapid Multiplex PCR (mPCR) Assay. International Journal of Environmental Research and Public Health, 19(3), 1031. https://doi.org/10.3390/ijerph19031031






