Abstract
Sulfide is a toxic pollutant in the farming environment. Microbial removal of sulfide always faces various biochemical challenges, and the application of enzymes for agricultural environmental remediation has promising prospects. In this study, a strain of Cellulosimicrobium sp. was isolated: numbered strain L1. Strain L1 can transform S2−, extracellular enzymes play a major role in this process. Next, the extracellular enzyme was purified, and the molecular weight of the purified sulfur convertase was about 70 kDa. The sulfur convertase is an oxidase with thermal and storage stability, and the inhibitor and organic solvent have little effect on its activity. In livestock wastewater, the sulfur convertase can completely remove S2−. In summary, this study developed a sulfur convertase and provides a basis for the application in environmental remediation.
1. Introduction
Sulfide is one of the main environmental pollutants in wastewater from livestock and aquaculture. The main component of the unpleasant odor produced in poultry farms is H2S [1], which is produced by the decomposition sulfide of aerobic/anaerobic bacteria in livestock wastewater [2,3]. H2S can adversely affect organisms, such as poultry chronically exposed to >10 ppm H2S, and it can cause intestinal inflammation and reduce egg production [4,5]. Therefore, it is necessary to treat sulfide pollution in livestock wastewater.
Microbial removal has received extensive attention as an efficient and economical method. Microorganisms can convert harmful sulfur compounds into harmless products. The previous study has examined the potential for biofiltering with aerobic, chemotrophic [6], and phototrophic microbial consortia for the removal of sulfide from biogas [7]. However, 30–250 mg/L sulfide can inhibit the growth of microorganisms [8,9], and the sulfide transformation process by microorganisms is very time consuming in sewage [10,11]. Biological processes (such as activated sludge and trickling filters) are considered more beneficial than microbial processes because of their effectiveness; however, most of the emerging contaminants remain soluble in wastewater and cannot be eliminated [12]. The main advantage of the above methods is the production of various intracellular and extracellular enzymes that can degrade contaminants [13]. Similarly, the removal of sulfide by microorganisms relies heavily on the combined action of enzymes in the organism. Therefore, an exploration of the application of enzymes to treat pollutants in wastewater was conducted [14].
For example, lysozyme treatment of microorganisms causes the bacteria to release intracellular substances, thus improving the sewage treatment [15]. Garbage enzyme (GE), an organic solution rich in enzymes, has been applied in wastewater treatment [16]. Quinone oxidoreductase carbon matrix treats sulfide-rich tanning wastewater, and the removal rate of sulfide can reach 99% [17]. However, the application of enzymes to sulfide treatment in livestock wastewater is less studied. Based on this, the study was launched. In this study, the sulfur convertase was obtained and showed positive effects in livestock wastewater applications.
2. Materials and Methods
2.1. Enrichment and Isolation of Sulfur-Transforming Bacteria
To obtain sulfur conversion bacteria, wastewater (solid–liquid, separated and unfermented fresh wastewater, was collected from Guangze ecological pasture in Changchun, China) as a separation source. Next, 1 mL wastewater was added to 100 mL inorganic salt medium (NH4Cl 1 g/L, KH2PO4 0.5 g/L, K2HPO4 1.5 g/L, Na2HCO3 0.1 g/L, NaCl 1 g/L, MgCl2 0.2 g/L, sucrose 0.5 g/L), Na2S (purity: ≥98%; 500 g; CAS: 1313-84-4; Macklin; Shanghai, China) was used as a sulfur source. It was cultivated 5 times, and S2− concentration was set to 100, 300, 500, 700, and 1000 mg/L; each time was set to 7 days. The microbial suspension was cultured on the LB (10.0 g/L tryptone, 10.0 g/L NaCl, and 5.0 g/L yeast extract) plate. Colonies were picked and streaked on fresh LB plates 3 times, respectively, transferring the colonies to 5 mL LB, cultured 24 h. Next, the bacteria were transferred to an inorganic salt medium to assess the S2 conversion capacity. Based on conversion results, strains were selected for subsequent experiments.
2.2. Morphology and Identification of the Strain L1
To observe the colony morphology of bacteria, the cultured bacteria were streaked on the LB plate until single colonies appeared. The colony was picked for gram staining and observed in an optical microscope (PH100-3B41L-IPL, Phenix, Jiangxi, China). Next, selected colonies were added to LB culture for 24 h, and DNA was extracted from the cultured strain. The extracted DNA was used as a template for 16S rRNA amplification and submitted to Sangon Biotech (Shanghai, China) for sequencing. The obtained sequence was aligned with existing sequences in NCBI. After the alignment, the sequences with high scores were selected to construct the phylogenetic tree (MEGA X was used).
2.3. Location and Validation of S2− Transformation Active Components in the Strain L1
2.3.1. Location of S2− Transformation Active Components in Strain L1
To locate the main active components of the strain L1, sulfur conversion experiments were performed in the following groups: bacterial cultures (B cultures), bacterial cells (B cells), culture supernatant (CS, extracellular enzyme), and cell lysate (intracellular enzyme) of the strain L1 [18]. In brief, the strain L1 was incubated in the inorganic salt medium at 37 °C for 24 h; the obtained cultures were divided into 3 portions, with the first portion as B cultures. The second portion was centrifuged; the pellets were collected and washed 3 times with phosphate-buffered saline (PBS) solution as B cells. The third portion was centrifuged, and the supernatant was filtered through a sterile 0.22 um filter (JINTENG, Tianjin, China). The pellets were crushed on ice with an ultrasound cell crusher (VCX130PB, Sonics, Shanghai, China). After crushing, the supernatant was centrifuged and filtered to obtain cell lysate. S2− solution was added to 4 groups, respectively, (S2− final concentration set to 500 mg/L) for sulfur conversion experiments, with inorganic salt medium and PBS acting as controls [19].
2.3.2. Effects of Heat, SDS, and Proteinase K Treatments on S2− Conversion Activity by the Culture Supernatant of Strain L1
To verify the role of the L1 active site, the effect of proteinase K (PK), SDS, PK +SDS, and heat treatment on the S2− conversion of CS was investigated [18]. CS was exposed to 2 mg/mL PK, 50 mg/mL SDS, and both simultaneous treatments and incubated at 4 °C for 6 h each to investigate the effect on S2− conversion. The CS was boiled at 100 °C for 10 min and 1 h to explore the effect of heat treatment on S2− conversion. The PBS and untreated culture supernatants were used as controls, respectively.
2.4. Optimization of Enzyme Conversion Ability
To improve the conversion ability of the extracellular enzyme, the medium composition was optimized by single-factor experiments. The culture components were optimized using various carbon sources (Sucrose, glucose, CH3COONa, sodium citrate, mannitol) and nitrogen sources (NH4Cl, NH4H2PO4, ammonium tartrate, NaNO3, Urea, yeast). After the carbon and nitrogen source selection was completed, the appropriate concentration was explored. CS was prepared by medium with changed conditions, and after completion of the preparation, sulfur conversion experiments were performed, and the appropriate component was chosen based on conversion results.
2.5. Extraction and Purification of the Sulfur Convertase
2.5.1. Extraction and Purification of Sulfur Convertase
First, we prepared CS (crude enzyme), which was passed through a sterile 0.22 um filter (to completely remove bacterial interference). The beaker containing the crude enzyme solution was heated to 4 °C, and pre-ground (NH4)2SO4 powder was slowly added to make the solution saturation reach 20%, 40%, 60%, and 80% in turn. The protein content and enzyme activity of the precipitates were determined. The optimum precipitation interval S1 to S2 was calculated based on the enzymatic conversion activity. The enzyme solution obtained in the optimal precipitation interval was added to the activated 14 KD MW membrane and stood at 4 °C in PBS for 12 h, changing the PBS every 4 h to completely remove (NH4)2SO4 interference. The enzyme solution was concentrated by PEG20000, and the enzyme solution was further purified via SephadexG-75 gel filtration chromatography. The column was pre-equilibrated with PBS-NaN3 buffer, and the eluate was adjusted to a flow rate of 0.5 mL/min. After elution, the collection volume reached 5 mL, and the protein concentration and enzyme conversion activity were determined.
2.5.2. Determination of Protein Concentration and Molecular Weight
Protein concentration was determined by the BCA method. A Pierce BCA protein assay kit was purchased from Thermo Fisher Scientific (Rockford, IL, USA). The molecular weights at different stages of purification were estimated using SDS-PAGE [20]. Proteins were separated with 12% (w/v) acrylamide, and a protein marker mixture (Beyotime, Shanghai, China) was applied to calibrate the molecular weight. We performed staining with 0.25% Coomassie blue (R-250) and de-staining with 1% acetic acid, observing results with a gel documentation system (GenoSens 1880, Clinx Science Instruments Co., Ltd., Shanghai, China).
2.6. Effect of Physiochemical Factors on Sulfur Convertase Activity
2.6.1. Optimum Reaction Temperature and Thermal Stability of Enzymes
To explore the optimum reaction temperature for sulfur convertase, sulfur conversion experiments were conducted at 10–80 °C; the reaction time was 6 h.
To investigate the thermal stability of the enzyme, the enzymes were treated at different temperatures (10 °C, 20 °C, 30 °C, 40 °C, 50 °C, 60 °C, 70 °C, and 80 °C) for 2 h before the reaction, and then the sulfur conversion experiment was performed. The reaction was performed at 37 °C for 24 h.
2.6.2. Stability of Sulfur Convertase over Storage Time
The purified sulfur convertase was stored at 4 °C for 30 days, and the enzyme was taken out every 3 days for sulfur conversion experiments to detect the change in the activity [21].
2.6.3. Effect of Inhibitors and Organic Solvents on Enzyme Activity
To investigate the effect of inhibitors and organic solvents on enzyme activity, inhibitors including 0.1 and 1 mM PMSF, NaN3, 10, 50, and 100 mM EDTA; organic solvents including 10%, 20%, and 30% (v/v) methanol, ethanol, isopropanol, and DMSO; and the enzymes were treated with the above reagents separately for 2 h at 4 °C. After 2 h, S2− was added for conversion experiments.
2.6.4. Effect of Redox on Enzyme Activity
S2− conversion is an oxidative reaction, so the sulfur convertase may become an oxidative enzyme, followed by addition of reductant to verify this conjecture. The enzymes were treated with 1 mmol/L dithiothreitol (DTT), 1 mmol/L ascorbic acid, and 10 mmol/L β-mercaptoethanol (sulfhydryl protectors) for 2 h at 4 °C. Afterwards, S2− was added and the enzyme activity was measured after 24 h.
2.6.5. Zymolyte Competition Experiments
To verify the effect of different substrates on the S2− conversion, different concentrations of S2O32− (100, 500 mg/L) and SO42− (100, 500 mg/L) were added, and the enzyme activity was measured after 24 h. S2O32− concentration was determined by an improved iodometric method [22], and SO42− was determined by barium chromate spectrophotometry [23].
2.7. Sulfur Conversion Capacity of Enzymes under Livestock Wastewater
Sulfur conversion experiments were conducted in livestock wastewater (solid–liquid separated and unfermented fresh wastewater, collected from Guangze ecological pasture Changchun, China) to assess the availability of the sulfur convertase application. The experimental groups were as follows: group 1–strain L1, group 2¬¬¬–sulfur convertase. Each group added 1 mL to 100 mL livestock wastewater. The solution without addition of bacteria or enzymes was taken as the control. S2− was not added in the experimental group due to the S2− being present in the wastewater (75.7 ± 3.1 mg/L). The application ability of the sulfur convertase was evaluated by the changes in S2− concentration, ammonia nitrogen, total phosphorus, and COD. The water-quality indicators were measured by a multiparameter water-quality meter (TR6900, Shenzhen Tongao Co., Ltd., Shenzhen, China). COD was determined by the dichromate titration [24]; the ammonia nitrogen was determined by Nessler’s reagent spectrophotometry [25]; and the total phosphorus was determined by the spectrophotometric method of ammonium molybdate [26].
2.8. Detection and Analysis Methods
Unless otherwise specified, the above sulfur conversion was conducted in a 2 mL EP tube, and S2− concentration was set to 500 mg/L, in triplicate. After the reaction, the enzyme activity was calculated based on the S2− concentration. S2− was determined by p-aminodimethylaniline spectrophotometry [27]. The graphs were prepared using GraphPad Prism (version 8.0, GraphPad: San Diego, CA, USA), data statistical analysis with SPSS software (applying one-way ANOVA, F-test, and Least Significant Difference test; version 25.0, SPSS: Armonk, NY, USA).
3. Results and Discussion
3.1. Isolation and Identification of Sulfur-Transforming Strain L1
Among the 12 bacterial strains isolated from wastewater, strain L1 showed the highest sulfide removal efficiency (Table 1). The control group also showed a 14.5% conversion rate, which may be due to the reaction of S2− with oxygen in the air. Therefore, strain L1 was chosen for further characterization. The colonies of strain L1 were yellow, round, and raised, with neat edges (Figure 1a). The strain was a short rod-shaped Gram-positive bacterium (Figure 1b).

Table 1.
S2− conversion rate of different strains isolated from wastewater.
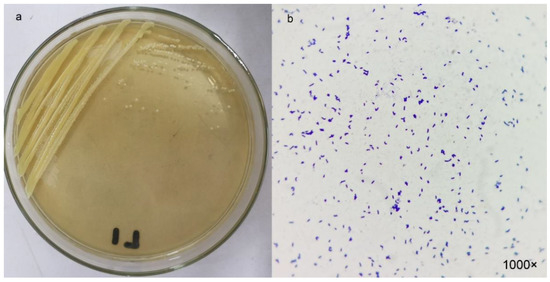
Figure 1.
(a) Colony and (b) bacterial morphology after Gram stain of strain L1.
Based on the gene sequence comparison, strain L1 showed the highest similarity to Cellulosimicrobium sp. (Figure 2), and the accession number (MZ687074) was submitted to GenBank. Previous studies showed that the Cellulosimicrobium sp. isolated from soil, marine sediment, and sewage can degrade 2,4,5-trichlorophenoxyacetic acid and biodiesel-oil [28,29,30]; however, studies on inorganic sulfur conversion are scarce. Therefore, further exploration was conducted.
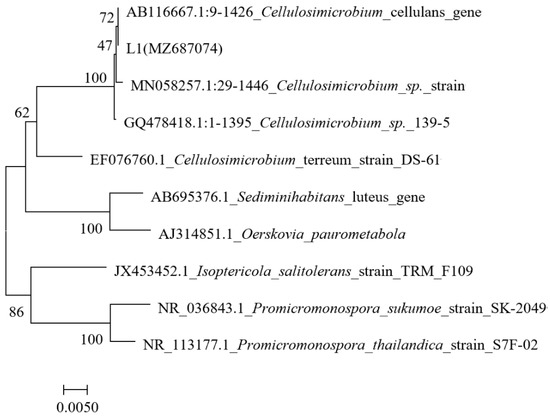
Figure 2.
Phylogenetic tree of strain L1 (the neighbor-joining method was used); the scale bars represent 0.005 substitutions per site.
3.2. Location of Active Component of S2− Conversion by Strain Cellulosimicrobium sp. L1
The results of transforming sulfur with different components of strain L1 are shown in Figure 3a. The S2− conversion rate of CS (60.37%) was higher than that of the other components after 24 h reaction. This shows that some extracellular substances of strain L1 play a significant role in S2− conversion. Hence, culture supernatants were used for further studies.
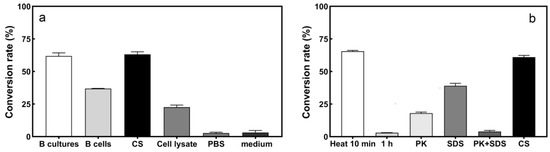
Figure 3.
Location of active components by strain L1 and the effect of different treatments on CS. (a) S2− conversion rate of the B cultures, B cells, CS, and cell lysate of strain L1; (b) the S2− conversion rate of CS under different treatments.
To confirm that the S2− transformation was enzymatic, the culture supernatant of strain L1 was exposed to heat, SDS, PK, and SDS + PK treatment, respectively. The results are shown in Figure 3b. The conversion rate of S2− did not decrease but increased by 6.04% after heating for 10 min. After heating for 1 h, the conversion rate of S2− basically disappeared. The conversion rate increased after heating for 10 min, which may be due to the heat-activating effect [31], whereas heating for 1 h could lead to denaturation and aggregation of proteins due to prolonged heat, resulting in loss of enzyme activity [19]. When the CS was exposed to SDS, the conversion rate decreased by 16.92%. When exposed to PK, the S2− conversion rate decreased 40.58%. When exposed to PK + SDS, the S2− conversion ability was lost. This could be due to SDS disrupting the spatial structure of proteins and PK disrupting the peptide bonds between amino acids in proteins, thereby inactivating or degrading proteins [32,33]. The above results show that CS plays a major role in the conversion, and the conversion of S2− is mainly catalyzed by the enzyme.
In previous studies, the conversion of S2− by microorganisms was mainly owed to enzymatic catalytic actions. Such as the use of intracellular enzymes, sulfide quinone oxidoreductase (SQR) and persulfide dioxygenase (PDO), produced by heterotrophic bacteria for sulfide oxidation [34]. Flavocytochrome c sulfide dehydrogenase (FSCD), sulfur oxidase Sox multi-enzyme complex, etc., can oxidize low-valent sulfur to high-valent sulfur containing inorganic compounds [35,36].
3.3. Optimization of Enzyme Conversion Activity
The effects of different carbon nitrogen sources on enzyme conversion activity are shown in Figure 4. Under different carbon sources, the maximum enzyme conversion activity (63.37%) was reached by adding sucrose, and when the sucrose concentration was 5 g/L, the maximum conversion activity (66.92%) was reached. However, the difference between the conversion activity of 5 g/L and 3 g/L (66.81%) was small, based on the principle of suitability for production and economy; 3 g/L was selected as the additional concentration.
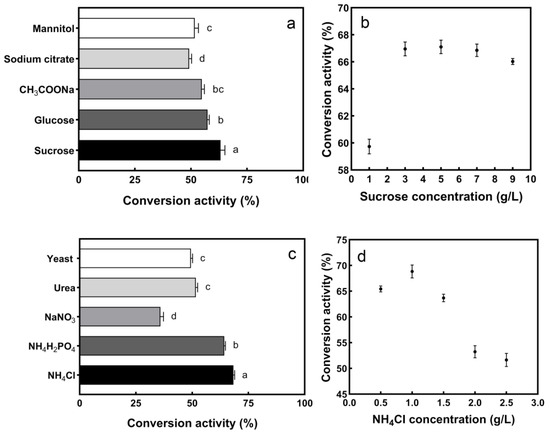
Figure 4.
Effect of different carbon nitrogen sources on enzyme conversion activity: (a) carbon source, (b) sucrose concentration, (c) nitrogen source, (d) NH4Cl concentration. The different letters indicate significant differences (p < 0.05) between each group.
Under different nitrogen sources, the highest conversion activity (68.93%) was achieved in the presence of NH4Cl, at 1 g/L. Therefore, the nitrogen source of the medium was adjusted to 1 g/L NH4Cl. Inorganic nitrogen sources such as NH4Cl and NH4H2PO4 are suitable for S2− conversion, which may be due to the easy uptake and low energy-dissipation of ammonium [37]. In summary, the most suitable carbon source is 3 g/L sucrose, and the most suitable nitrogen source is 1 g/L NH4Cl.
3.4. Extraction and Purification of Sulfur Convertase
3.4.1. Extraction and Purification of Sulfur Convertase
The purification results of sulfur convertase by ammonium-sulfate-graded precipitation are shown in Figure 5a. S2− conversion activity in the precipitate increased with the ammonium sulfate concentration. When the concentration of ammonium sulfate reached 60% to 80% (saturation), the activity was at its maximum. Therefore, the interval of 60% to 80% ammonium sulfate saturation was chosen to precipitate crude sulfur convertase.
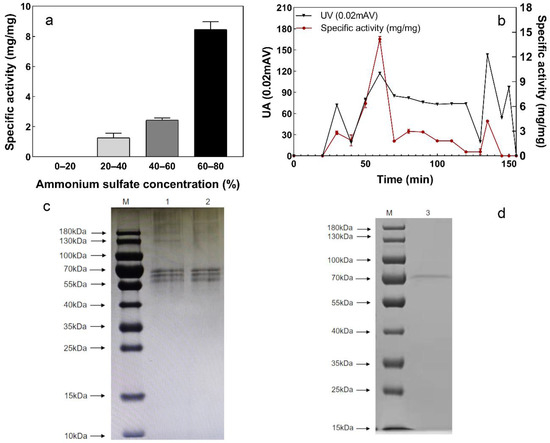
Figure 5.
Purification of sulfur convertase, (a) specific activity of precipitated enzymes at different ammonium sulfate concentrations, (b) precipitation time and specific activity of each protein; specific activity: the amount of sulfide (mg) converted per mg of protein for 24 h, (c,d) the SDS-PAGE images of strain L1, CS, and sulfur convertase M: protein marker; 1: strain L1; 2: CS; 3: sulfur convertase.
Next, the enzyme was further purified by SephadexG-75 gel filtration chromatography, and four protein peaks were observed (Figure 5b). The highest specific sulfur convertase activity was detected in the second peak (50–70 min). After two purifications, the specific activity of sulfur convertase increased from 2.59 to 14.29 mg/mg, a 5.52-fold purification (Table 2).

Table 2.
Steps of the purification of sulfur convertase.
3.4.2. Protein Molecular Weight of Sulfur Convertase
The molecular weights of purified sulfur convertase were estimated using SDS-PAGE (Figure 5c,d). Compared to strains L1 and CS, the purified sulfur convertase showed a single protein band of approximately 70 kDa, indicating that the purification achieved good effects.
3.5. Property Studies of the Purified Sulfur Convertase
3.5.1. Activity and Stability of Sulfur Convertase at Different Temperatures
From 10–80 °C, the sulfur convertase activity showed a trend of increasing, decreasing, increasing again, and then finally decreasing with increasing temperature. When the temperature reached 70 °C, the highest conversion activity was achieved (Figure 6a). The sulfur convertase can remain stable at 10–70 °C, with relative activity over 80%. The big difference in enzyme activity between the two groups at 50–60 °C may be due to the different contact times of the enzymes with the different temperatures. In previous studies, sulfite: acceptor oxidoreductase, the key enzyme for sulfide oxidation in strain LYH-3, reached maximum specific activity at 50 °C [38]; sulfur dioxygenase in Acidithiobacillus ferrooxidans showed the highest activity at 35 °C [39]; SQR in Urechis unicinctus had the highest enzyme activity at 37 °C [40]. The sulfur convertase in this study has a higher optimal reaction temperature, which may contribute to the practical application of the enzyme.
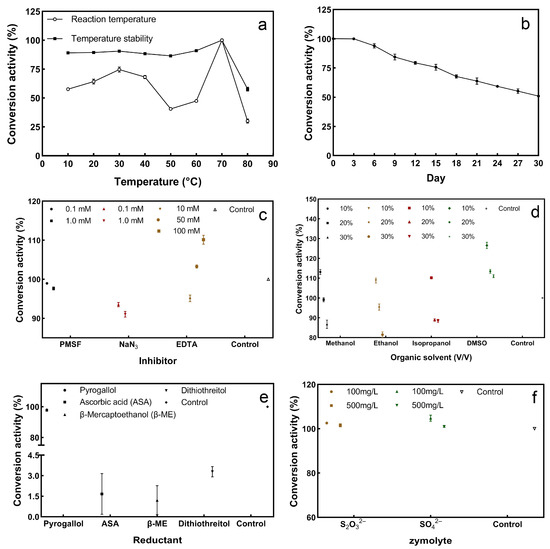
Figure 6.
Effect of physiochemical factors on sulfur convertase activity: (a) the change in sulfur-converting-enzyme activity and stability at 10–80 °C; the maximum enzyme activity was set at 100%, and this was used as a basis to calculate the relative enzyme activity at the other reaction temperatures, (b) sulfur convertase stability over storage time, (c) effect of inhibitors on sulfur convertase, (d) effect of organic solvents on sulfur convertase, (e) effect of reductants on sulfur convertase, (f) effect of zymolytes on sulfur convertase; the conversion activity of the control group was set to 100%, and this was used as a basis to calculate the relative activity of the enzyme under inhibitors, organic solvents, reductants, and zymolytes.
3.5.2. Sulfur Convertase Stability over Storage Time
Stability over storage time is also one of the factors affecting the application of enzymes. As the storage time increases, the relative activity of the enzyme gradually decreases. On the 30th day, the conversion activity of the sulfur convertase still exceeded 50% (Figure 6b). This indicates that the enzyme has good storage stability. Previous studies have shown that low storage stability limits the practical application of enzymes [41].
3.5.3. Effect of Inhibitors and Organic Solvents on Sulfur Convertase Activity
The stability of enzymes can affect their application [42]. Therefore, this section combines the application environment to study the effects of inhibitors and organic solvents on enzyme stability. The effect of inhibitors on the conversion activity is shown in Figure 6c. The three inhibitors had less effect, and the conversion activity was above 90%. Inhibitors also can be used to analyze which active residues are present in an enzyme and to classify the enzyme [43]. PMSF is a serine inhibitor and showed weak inhibition on the enzyme, indicating that the enzyme may not belong to the serine enzyme group [44]. Among the three inhibitors, NaN3 exhibited the highest inhibition. NaN3 can block the electron transfer ability of the cytochrome oxidase [45]. Therefore, this enzyme may be a member of the cytochrome oxidase family. The effect of EDTA on enzyme activity showed that low concentration inhibited and high concentration promoted, indicating that the sulfur convertase may not be a metalloprotease [46]. EDTA is a metal chelator; low concentrations inhibited the enzyme activity, which could be due to EDTA chelating the metal ions required for the conversion reaction. The promotion could be due to the high concentration of EDTA removing the impure metal ions completely, thus reducing the effect on the enzyme and promoting conversion.
Under different organic solvents, the effects of methanol, ethanol, and isopropanol on the enzyme showed the same trend, with low concentrations slightly promoted and high concentrations inhibited. Different concentrations of DMSO promoted enzymatic activity, but the promotion weakened with the increase in DMSO concentration (Figure 6d). DMSO can interact with the hydrophobic group of the protein to denature the protein [44], so it is speculated that the enzyme may not have a hydrophobic group in its active center. Overall, organic solvents have less effect on sulfur convertase activity.
3.5.4. Effect of Redox on Sulfur Convertase Activity
Pyrogallic acid did not affect the relative conversion activity of the enzyme, whereas ascorbic acid, β-mercaptoethanol, and dithiothreitol largely inhibited the relative conversion activity of the enzyme (Figure 6e). Three reducing agents are also sulfhydryl protectors that reduce the disulfide bonds in the protein [47]. It is speculated that disulfide bonds are present in the enzyme, and they may play a key role in the sulfur conversion process.
3.5.5. Effect of Zymolyte Competition on Sulfur Convertase Activity
S2O32− and SO42− are oxidation products of S2− [35]; therefore, two substances were added to explore the effect on sulfur conversion. Different concentrations of S2O32− and SO42− had little effect on the conversion (Figure 6f). This indicates that the two substances may have no feedback regulation or weak regulation on S2−conversion.
3.6. Sulfur Conversion of Enzymes under Livestock Wastewater Conditions
The conversion results and the change in wastewater indicators are shown in Table 3. Previous studies have shown that bacteria can effectively remove contaminants [48], and the same results were obtained in the present experiment—strain L1 can effectively improve the conversion of sulfur, and S2− was completely removed from livestock wastewater under the action of enzymes. Ammonia, total phosphorus, and COD were also reduced. Anaerobic digestion is the main technology used in wastewater treatment. In the previous studies, anaerobic digestion was used to treat slaughterhouse wastewater with 95.90% COD (600 mg/L) removal [49] and 98.4% COD (100–600 mg/L) removal for nitrogen fertilizer wastewater [50]. Compared with the current study, sulfur convertase removed more COD, but the removal rate was much lower than in the previous studies. This may be due to the different original COD of the wastewater. Compared to strain L1, sulfur convertase is more suitable for sulfur removal in wastewater conditions. Previous studies also showed that enzymatic bioremediation is superior to microbial bioremediation because it has a high bioconversion rate, a shorter remediation time, and low environmental risk [42,51]. High S2− concentrations can inhibit the growth of microorganisms [8]. Moreover, H2S is produced at all stages of oil production, and H2S can be hazardous to workers’ health [52]; the rapid conversion properties of sulfur convertase may be suitable for this. In this study, the enzyme maintained stable sulfur conversion activity and also remediated wastewater, which laid the foundation for future practical application of enzymes in the environment.

Table 3.
The change of wastewater indicators and S2−.
4. Conclusions
In this study, twelve strains were isolated from wastewater; strain L1 had the highest sulfur conversion capacity, and L1 was identified as Cellulosimicrobium sp. The extracellular enzyme of the L1 showed a stronger sulfur conversion capacity; 3 g/L sucrose and 1 g/L NH4Cl can promote enzyme activity. After purification, the activity of sulfur convertase increased 5.52-fold; temperature, storage time, inhibitors, organic solvents, and conversion products have little effect on the enzyme, but reductants can dramatically reduce conversion activity. In wastewater conditions, the enzyme also showed stable sulfur conversion capacity. In conclusion, the current work focuses on the development and availability evaluation of the sulfur convertase, which will be identified and predicted in the future, and its mechanism of action will be studied in depth.
Author Contributions
Conceptualization, Writing—original draft: X.L.; Methodology: W.Z. and M.L.; Investigation: X.D.; Editing and Supervision: C.G.; Project administration: Y.G., L.W., W.B., and Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Strategic Priority Research Program of the Chinese Academy of Science (XDA28080400), the Science and Technology Development Plan Project of Jilin Province (20190201296JC), the Science and Technology program of Tibet Autonomous Region (XZ202101ZD0002N-04), and the China Agriculture Research System of MOF and MARA (CARS-37).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Acknowledgments
Thanks to our country.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gruzdev, E.V.; Latygolets, E.A.; Beletsky, A.V.; Grigoriev, M.A.; Mardanov, A.V.; Kadyrbaev, M.K.; Ikkert, O.P.; Karnachuk, O.V.; Ravin, N.V. The Microbial Community of Poultry Farm Waste and Its Role in Hydrogen Sulfide Production. Microbiology 2021, 90, 507–511. [Google Scholar] [CrossRef]
- Pham, C.H.; Saggar, S.; Berben, P.; Palmada, T.; Ross, C. Removing Hydrogen Sulfide Contamination in Biogas Produced from Animal Wastes. J. Environ. Qual. 2019, 48, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Yun, H.-S.; Kim, Y.-S.; Kim, J.-G. Pollutant-Removing Biofilter Strains Associated with High Ammonia and Hydrogen Sulfide Removal Rate in a Livestock Wastewater Treatment Facility. Sustainability 2021, 13, 7358. [Google Scholar] [CrossRef]
- Hu, X.; Chi, Q.; Wang, D.; Chi, X.; Teng, X.; Li, S. Hydrogen sulfide inhalation-induced immune damage is involved in oxidative stress, inflammation, apoptosis and the Th1/Th2 imbalance in broiler bursa of Fabricius. Ecotoxicol. Environ. Saf. 2018, 164, 201–209. [Google Scholar] [CrossRef]
- Saksrithai, K.; King, A.J.J.A.R.N. Controlling hydrogen sulfide emissions during poultry productions. J. Anim. Res. Nutr. 2018, 3, 2. [Google Scholar] [CrossRef]
- Su, J.J.; Chang, Y.C.; Chen, Y.J.; Chang, K.C.; Lee, S.Y. Hydrogen sulfide removal from livestock biogas by a farm-scale bio-filter desulfurization system. Water Sci. Technol. 2013, 67, 1288–1293. [Google Scholar] [CrossRef]
- Da Silva, M.L.B.; Mezzari, M.P.; Ibelli, A.M.G.; Gregory, K.B. Sulfide removal from livestock biogas by Azospirillum-like anaerobic phototrophic bacteria consortium. Int. Biodeterior. Biodegrad. 2014, 86, 248–251. [Google Scholar] [CrossRef]
- Pokorna, D.; Zabranska, J. Sulfur-oxidizing bacteria in environmental technology. Biotechnol. Adv. 2015, 33, 1246–1259. [Google Scholar] [CrossRef] [PubMed]
- Watsuntorn, W.; Ruangchainikom, C.; Rene, E.; Lens, P.; Chulalaksananukul, W.J.B.T. Comparison of sulphide and nitrate removal from synthetic wastewater by pure and mixed cultures of nitrate-reducing, sulphide-oxidizing bacteria. Bioresour. Technol. 2019, 272, 40–47. [Google Scholar] [CrossRef]
- Watsuntorn, W.; Ruangchainikom, C.; Rene, E.R.; Lens, P.N.L.; Chulalaksananukul, W. Hydrogen sulfide oxidation under anoxic conditions by a nitrate-reducing, sulfide-oxidizing bacterium isolated from the Mae Um Long Luang hot spring, Thailand. Int. Biodeterior. Biodegrad. 2017, 124, 196–205. [Google Scholar] [CrossRef]
- Hansen, M.J.; Pedersen, C.L.; Søgaard Jensen, L.H.; Guldberg, L.B.; Feilberg, A.; Nielsen, L.P. Removal of hydrogen sulphide from pig house using biofilter with fungi. Biosyst. Eng. 2018, 167, 32–39. [Google Scholar] [CrossRef]
- Servos, M.R.; Bennie, D.T.; Burnison, B.K.; Jurkovic, A.; McInnis, R.; Neheli, T.; Schnell, A.; Seto, P.; Smyth, S.A.; Ternes, T.A. Distribution of estrogens, 17β-estradiol and estrone, in Canadian municipal wastewater treatment plants. Sci. Total Environ. 2005, 336, 155–170. [Google Scholar] [CrossRef]
- Pudi, A.; Rezaei, M.; Signorini, V.; Andersson, M.P.; Baschetti, M.G.; Mansouri, S.S. Hydrogen sulfide capture and removal technologies: A comprehensive review of recent developments and emerging trends. Sep. Purif. Technol. 2022, 298, 121448. [Google Scholar] [CrossRef]
- Rodríguez Couto, S.; Toca Herrera, J.L. Industrial and biotechnological applications of laccases: A review. Biotechnol. Adv. 2006, 24, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Lu, M.; Li, Q.; Zhang, J.; Zong, Y.; She, Z. Three-dimensional fluorescence excitation–emission matrix (EEM) spectroscopy with regional integration analysis for assessing waste sludge hydrolysis treated with multi-enzyme and thermophilic bacteria. Bioresour. Technol. 2014, 171, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Arun, C.; Sivashanmugam, P. Identification and optimization of parameters for the semi-continuous production of garbage enzyme from pre-consumer organic waste by green RP-HPLC method. Waste Manag. 2015, 44, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Mahesh, M.; Swarnalatha, S.; Gnanamani, A.; Sekaran, G. Preparation and characterization of sulfide: Quinone oxidoreductase immobilized carbon matrix for the treatment of sulphide rich post-tanning wastewater. Biocatal. Agric. Biotechnol. 2020, 23, 101457. [Google Scholar] [CrossRef]
- Raksha Rao, K.; Vipin, A.V.; Hariprasad, P.; Anu Appaiah, K.A.; Venkateswaran, G. Biological detoxification of Aflatoxin B1 by Bacillus licheniformis CFR1. Food Control 2017, 71, 234–241. [Google Scholar] [CrossRef]
- Zhang, J.; Qin, X.; Guo, Y.; Zhang, Q.; Ma, Q.; Ji, C.; Zhao, L. Enzymatic degradation of deoxynivalenol by a novel bacterium, Pelagibacterium halotolerans ANSP101. Food Chem. Toxicol. 2020, 140, 111276. [Google Scholar] [CrossRef]
- Verma, J.; Pandey, S. Characterization of partially purified alkaline protease secreted by halophilic bacterium Citricoccus sp. isolated from agricultural soil of northern India. Biocatal. Agric. Biotechnol. 2019, 17, 605–612. [Google Scholar] [CrossRef]
- Lu, M.; Gao, Z.; Xing, S.; Long, J.; Li, C.; He, L.; Wang, X. Purification, characterization, and chemical modification of Bacillus velezensis SN-14 fibrinolytic enzyme. Int. J. Biol. Macromol. 2021, 177, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Kilroy, W.P. The iodometric determination of dithionite, thiosulphate and sulphite in the presence of alkali and/or cyanide. Talanta 1980, 27, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; An, D.; Xiao, Q.; Chen, F.; Zhang, Y.; Weng, H.; Xiao, A.J.M.D. Convenient Agarose Preparation with Hydrogen Peroxide and Desulfation Process Analysis. Mar. Drugs 2021, 19, 297. [Google Scholar] [CrossRef] [PubMed]
- Dedkov, Y.M.; Elizarova, O.V.; Kel’ina, S.Y. Dichromate method for the determination of chemical oxygen demand. J. Anal. Chem. 2000, 55, 777–781. [Google Scholar] [CrossRef]
- Lin, K.; Zhu, Y.; Zhang, Y.; Lin, H. Determination of ammonia nitrogen in natural waters: Recent advances and applications. Trends Environ. Anal. Chem. 2019, 24, e00073. [Google Scholar] [CrossRef]
- Yang, C.-X.; Sun, X.-Y.; Liu, B.; Lian, H.-T. Determination of Total Phosphorus in Water Sample by Digital Imaging Colorimetry. Chin. J. Anal. Chem. 2007, 35, 850–853. [Google Scholar] [CrossRef]
- Cline, J.D. Spectrophotometric determination of hydrogen sulfide in natural waters 1. Limnol. Oceanogr. 1969, 14, 454–458. [Google Scholar] [CrossRef]
- Korobov, V.V.; Zhurenko, E.Y.; Galkin, E.G.; Zharikova, N.V.; Iasakov, T.R.; Starikov, S.N.; Sagitova, A.I.; Markusheva, T.V. Cellulosimicrobium sp. strain NPZ-121, a degrader of 2,4,5-trichlorophenoxyacetic acid. Microbiology 2018, 87, 147–150. [Google Scholar] [CrossRef]
- Bertel-Sevilla, A.; Cervantes-Ceballos, L.; Tirado-Ballestas, I.; Maldonado-Rojas, W.; Alzate-Restrepo, J.; Olivero-Verbel, J.J.E.t. Biodegradation of biodiesel-oil by Cellulosimicrobium sp. Isolated from Colombian Caribbean soils. Environ. Technol. 2020, 41, 2337–2349. [Google Scholar] [CrossRef] [PubMed]
- Bharagava, R.N.; Mishra, S. Hexavalent chromium reduction potential of Cellulosimicrobium sp. isolated from common effluent treatment plant of tannery industries. Ecotoxicol. Environ. Saf. 2018, 147, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Mohamed, E.A.; Lancine, S.; Yueju, Z.; Jonathan, S.; Fuguo, X.; Yan, W.; Hongping, Y.; Yang, L.J.T. Novel Aflatoxin-Degrading Enzyme from Bacillus shackletonii L7. Toxins 2017, 9, 36. [Google Scholar] [CrossRef]
- Sigman, D.S.; Mooser, G. Chemical Studies of Enzyme Active Sites. Annu. Rev. Biochem. 1975, 44, 889–931. [Google Scholar] [CrossRef] [PubMed]
- Larsen, A.; Moe, E.; Helland, R.; Gjellesvik, D.; Willassen, N.J.T.F.j. Characterization of a recombinantly expressed proteinase K-like enzyme from a psychrotrophic Serratia sp. FEBS J. 2006, 273, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Liu, H.; Xun, L. Cytoplasmic Localization of Sulfide:Quinone Oxidoreductase and Persulfide Dioxygenase of Cupriavidus pinatubonensis JMP134. Appl. Environ. Microbiol. 2017, 83, e01820-17. [Google Scholar] [CrossRef]
- Sousa, F.; Pereira, J.; Marreiros, B.; Pereira, M.J.B.e.b.a.B. Taxonomic distribution, structure/function relationship and metabolic context of the two families of sulfide dehydrogenases: SQR and FCSD. BBA Bioenerg. 2018, 1859, 742–753. [Google Scholar] [CrossRef] [PubMed]
- Bagchi, A.; Roy, D.; Roy, P.J.J.o.B.S.; Dynamics. Homology modeling of a transcriptional regulator SoxR of the Lithotrophic sulfur oxidation (Sox) operon in alpha-proteobacteria. J. Biomol. Struct. Dyn. 2005, 22, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Aliabadi, N.; Aminzadeh, S.; Karkhane, A.A.; Haghbeen, K. Thermostable chitinase from Cohnella sp. A01: Isolation and product optimization. Braz. J. Microbiol. 2016, 47, 931–940. [Google Scholar] [CrossRef]
- Chen, L.; Li, W.; Zhao, Y.; Zhang, S.; Meng, L.; Zhou, Y. Characterization of sulfide oxidation and optimization of sulfate production by a thermophilic Paenibacillus naphthalenovorans LYH-3 isolated from sewage sludge composting. J. Environ. Sci. 2023, 125, 712–722. [Google Scholar] [CrossRef]
- Wang, H.; Liu, S.; Liu, X.; Li, X.; Wen, Q.; Lin, J.J. Identification and characterization of an ETHE1-like sulfur dioxygenase in extremely acidophilic Acidithiobacillus spp. Appl. Microbiol. Biotechnol. 2014, 98, 7511–7522. [Google Scholar] [CrossRef]
- Ma, Y.-B.; Zhang, Z.-F.; Shao, M.-Y.; Kang, K.-H.; Tan, Z.; Li, J.-L.J.M.B. Sulfide: Quinone oxidoreductase from echiuran worm Urechis unicinctus. Mar. Biotechnol. 2011, 13, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, X.; Dong, Y.; Hui, M.; Xu, L.; Li, H.; Lv, J.; Yang, L.; Cui, Y. Uric acid and creatinine biosensors with enhanced room-temperature storage stability by a multilayer enzyme matrix. Anal. Chim. Acta 2022, 1227, 340264. [Google Scholar] [CrossRef] [PubMed]
- Iyer, P.V.; Ananthanarayan, L. Enzyme stability and stabilization—Aqueous and non-aqueous environment. Process Biochem. 2008, 43, 1019–1032. [Google Scholar] [CrossRef]
- Shaikh, I.K.; Dixit, P.P.; Shaikh, T.M. Purification and characterization of alkaline soda-bleach stable protease from Bacillus sp. APP-07 isolated from Laundromat soil. J. Genet. Eng. Biotechnol. 2018, 16, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Nazari, L.; Mehrabi, M. Purification and characterization of an extracellular thermotolerant alkaliphilic serine protease secreted from newly isolated Bacillus sp. DEM07 from a hot spring in Dehloran, Iran. Biocatal. Agric. Biotechnol. 2019, 18, 101053. [Google Scholar] [CrossRef]
- Suzuki, Y.; Taguchi, K.; Hanyu, S.; Kure, T.; Enoki, Y.; Otagiri, M.; Sakai, H.; Matsumoto, K. Oxidized liposomal artificial red blood cells rescue azide-poisoned mice from lethal toxidrome by recovering cytochrome c oxidase activity. J. Drug Deliv. Sci. Technol. 2022, 71, 103282. [Google Scholar] [CrossRef]
- Zheng, L.; Yu, X.; Wei, C.; Qiu, L.; Yu, C.; Xing, Q.; Fan, Y.; Deng, Z. Production and characterization of a novel alkaline protease from a newly isolated Neurospora crassa through solid-state fermentation. LWT 2020, 122, 108990. [Google Scholar] [CrossRef]
- Guven, R.G.; Kaplan, A.; Guven, K.; Matpan, F.; Dogru, M. Effects of various inhibitors on β-galactosidase purified from the thermoacidophilic Alicyclobacillus acidocaldarius subsp. Rittmannii isolated from Antarctica. Biotechnol. Bioprocess Eng. 2011, 16, 114–119. [Google Scholar] [CrossRef]
- Yu, Y.; Yin, H.; Peng, H.; Lu, G.; Dang, Z.J.J.O.H.M. Proteomic mechanism of decabromodiphenyl ether (BDE-209) biodegradation by Microbacterium Y2 and its potential in remediation of BDE-209 contaminated water-sediment system. J. Hazard. Mater. 2020, 387, 121708. [Google Scholar] [CrossRef] [PubMed]
- Adou, K.E.; Kouakou, A.R.; Ehouman, A.D.; Tyagi, R.D.; Drogui, P.; Adouby, K. Coupling anaerobic digestion process and electrocoagulation using iron and aluminium electrodes for slaughterhouse wastewater treatment. Sci. Afr. 2022, 16, e01238. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, H.; Chen, Z.; Wei, D.; Song, Y.; Ma, Y.; Zhang, H. Achieving biogas production and efficient pollutants removal from nitrogenous fertilizer wastewater using combined anaerobic digestion and autotrophic nitrogen removal process. Bioresour. Technol. 2021, 339, 125659. [Google Scholar] [CrossRef]
- Miri, S.; Davoodi, S.; Brar, S.; Rouissi, T.; Sheng, Y.; Martel, R.J.B.t. Psychrozymes as novel tools to biodegrade p-xylene and potential use for contaminated groundwater in the cold climate. Bioresour. Technol. 2021, 321, 124464. [Google Scholar] [CrossRef] [PubMed]
- Elmawgoud, H.A.; Elshiekh, T.M.; Abdelkreem, M.; Khalil, S.A.; Alsabagh, A.M. Optimization of petroleum crude oil treatment using hydrogen sulfide scavenger. Egypt. J. Pet. 2019, 28, 161–164. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).