Characterization of Planktochlorella nurekis Extracts and Virucidal Activity against a Coronavirus Model, the Murine Coronavirus 3
Abstract
1. Introduction
2. Materials and Methods
2.1. Obtention of the Microalgae Biomass
2.2. Chemical Characterization of Planktochlorella nurekis Extracts
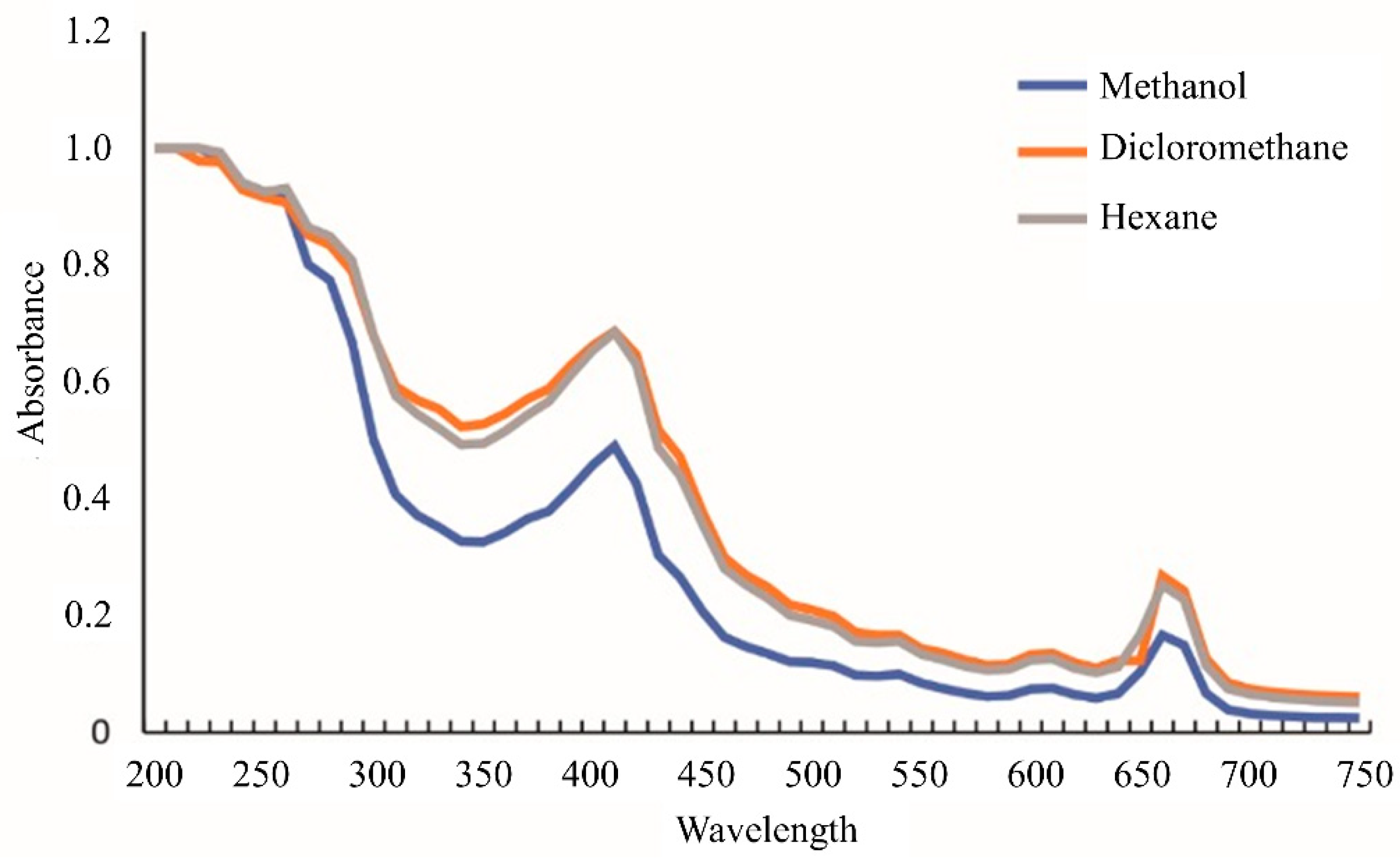
2.2.1. Ultraviolet-Visible Spectroscopy (UV-VIS) Profile
2.2.2. Nuclear Magnetic Resonance Spectroscopy (NMR)
2.2.3. Total Phenolic Content
2.2.4. β-Carotene Content
2.2.5. Amino Acids Characterization
2.2.6. Qualitative Analysis of the Extracts by Ultraperformance Liquid Chromatography-Mass Spectrometry (UPLC-MS)
2.3. Cytotoxicity Assay
2.4. Virucidal Assay
2.5. Statistical Analysis
3. Results and Discussion
3.1. Chemical Characterization of Planktochlorella nurekis Extracts
3.1.1. UV-VIS Profile
3.1.2. β-Carotene, Phenols, and Amino Acids Content
3.1.3. NMR Profile
3.2. Analysis of the Extracts by UPLC-MS
3.3. Cytotoxicity of Planktochlorella nurekis
3.4. Virucidal Activity of Planktochlorella nurekis
3.5. PCA Analysis of the P. nurekis Extracts
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bárcena, M.; Oostergetel, G.T.; Bartelink, W.; Faas, F.G.A.; Verkleij, A.; Rottier, P.J.M.; Koster, A.J.; Bosch, B.J. Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. USA 2009, 106, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Peiris, J.S.M. Coronaviruses. In Medical Microbiology: Eighteenth Edition; Elsevier: Amsterdam, The Netherlands, 2012; pp. 587–593. [Google Scholar] [CrossRef]
- Sharma, H.B.; Vanapalli, K.R.; Cheela, V.S.; Ranjan, V.P.; Jaglan, A.K.; Dubey, B.; Goel, S.; Bhattacharya, J. Challenges, opportunities, and innovations for effective solid waste management during and post COVID-19 pandemic. Resour. Conserv. Recycl. 2020, 162, 105052. [Google Scholar] [CrossRef] [PubMed]
- Denis, M.; Vandeweerd, V.; Verbeke, R.; Version, D.V. COVIPENDIUM: Information available to support the development of medical countermeasures and interventions against COVID-19 (Version 2020-05-19). In Transdisciplinary In-Sights; 2020; pp. 1–157. Available online: https://rega.kuleuven.be/if/corona_covid-19 (accessed on 25 February 2022).
- Jiang, F.; Deng, L.; Zhang, L.; Cai, Y.; Cheung, C.W.; Xia, Z. Review of the Clinical Characteristics of Coronavirus Disease 2019 (COVID-19). J. Gen. Intern. Med. 2020, 35, 1545–1549. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.-I.; Hsueh, P.-R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol. Immunol. Infect. 2020, 53, 365–367. [Google Scholar] [CrossRef] [PubMed]
- Körner, R.W.; Majjouti, M.; Alcazar, M.A.A.; Mahabir, E. Of Mice and Men: The Coronavirus MHV and Mouse Models as a Translational Approach to Understand SARS-CoV-2. Viruses 2020, 12, 880. [Google Scholar] [CrossRef] [PubMed]
- Züst, R.; Cervantes-Barragán, L.; Kuri, T.; Blakqori, G.; Weber, F.; Ludewig, B.; Thiel, V. Coronavirus Non-Structural Protein 1 Is a Major Pathogenicity Factor: Implications for the Rational Design of Coronavirus Vaccines. PLoS Pathog. 2007, 3, e109. [Google Scholar] [CrossRef]
- Gonzalez, J.M.; Gomez-Puertas, P.; Cavanagh, D.; Gorbalenya, A.; Enjuanes, L. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 2003, 148, 2207–2235. [Google Scholar] [CrossRef]
- Besednova, N.; Andryukov, B.; Zaporozhets, T.; Kryzhanovsky, S.; Fedyanina, L.; Kuznetsova, T.; Zvyagintseva, T.; Shchelkanov, M. Antiviral Effects of Polyphenols from Marine Algae. Biomedicines 2021, 9, 200. [Google Scholar] [CrossRef]
- Abdo, S.M.; Hetta, M.H.; El-Senousy, W.M.; Salah El Din, R.A.; Ali, G.H. Antiviral activity of freshwater algae. J. Appl. Pharm. Sci. 2012, 2, 21–25. [Google Scholar]
- Kim, M.; Yim, J.H.; Kim, S.-Y.; Kim, H.S.; Lee, W.G.; Kim, S.J.; Kang, P.-S.; Lee, C.-K. In vitro inhibition of influenza A virus infection by marine microalga-derived sulfated polysaccharide p-KG03. Antivir. Res. 2012, 93, 253–259. [Google Scholar] [CrossRef]
- Yim, J.H.; Kim, S.J.; Ahn, S.H.; Lee, H.K. Characterization of a novel bioflocculant, p-KG03, from a marine dinoflagellate, Gyrodinium impudicum KG03. Bioresour. Technol. 2007, 98, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Shih, S.-R.; Tsai, K.-N.; Li, Y.-S.; Chueh, C.-C.; Chan, E.-C. Inhibition of enterovirus 71-induced apoptosis by allophycocyanin isolated from a blue-green algaspirulina platensis. J. Med. Virol. 2003, 70, 119–125. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, B.R.; Giomarelli, B.; Barnard, D.L.; Shenoy, S.R.; Chan, P.K.S.; McMahon, J.B.; Palmer, K.E.; Barnett, B.W.; Meyerholz, D.K.; Wohlford-Lenane, C.L.; et al. Broad-Spectrum In Vitro Activity and In Vivo Efficacy of the Antiviral Protein Griffithsin against Emerging Viruses of the Family Coronaviridae. J. Virol. 2010, 84, 2511–2521. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Mendoza, S.; García-Silva, I.; González-Ortega, O.; Sandoval-Vargas, J.M.; Malla, A.; Vimolmangkang, S. The Potential of Algal Biotechnology to Produce Antiviral Compounds and Biopharmaceuticals. Molecules 2020, 25, 4049. [Google Scholar] [CrossRef]
- El-Baz, F.K.; El-Senousy, W.M.; El-Sayed, A.B.; Kamel, M.M. In vitro antiviral and antimicrobial activities of Spirulina platensis extract. J. Appl. Pharm. Sci. 2013, 3, 52–56. [Google Scholar] [CrossRef]
- Hasui, M.; Matsuda, M.; Okutani, K.; Shigeta, S. In vitro antiviral activities of sulfated polysaccharides from a marine microalga (Cochlodinium polykrikoides) against human immunodeficiency virus and other enveloped viruses. Int. J. Biol.Macromol. 1995, 17, 293–297. [Google Scholar] [CrossRef]
- Santoyo, S.; Plaza, M.; Jaime, L.; Ibañez, E.; Reglero, G.; Señorans, F.J. Pressurized Liquid Extraction as an Alternative Process To Obtain Antiviral Agents from the Edible Microalga Chlorella vulgaris. J. Agric. Food Chem. 2010, 58, 8522–8527. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, X. Separation, antitumor activities, and encapsulation of polypeptide from Chlorella pyrenoidosa. Biotechnol. Prog. 2013, 29, 681–687. [Google Scholar] [CrossRef]
- Katharios, P.; Papadakis, I.E.; Prapas, A.; Dermon, C.R.; Ampatzis, K.; Divanach, P. Mortality control of viral encephalopathy and retinopathy in 0+ grouper Epinephelus marginatus after prolonged bath in dense Chlorella minutissima culture. Bull. Eur. Assoc. Fish Pathol. 2005, 25, 28–31. [Google Scholar]
- Connan, S.; Stengel, D.B. Impacts of ambient salinity and copper on brown algae: 2. Interactive effects on phenolic pool and assessment of metal binding capacity of phlorotannin. Aquat. Toxicol. 2011, 104, 1–13. [Google Scholar] [CrossRef]
- Škaloud, P.; Němcová, Y.; Pytela, J.; Bogdanov, N.I.; Bock, C.; Pickinpaugh, S.H. Planktochlorella nurekis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta), a novel coccoid green alga carrying significant biotechnological potential. Fottea 2014, 14, 53–62. [Google Scholar] [CrossRef]
- Safafar, H.; Van Wagenen, J.M.; Møller, P.; Jacobsen, C. Carotenoids, Phenolic Compounds and Tocopherols Contribute to the Antioxidative Properties of Some Microalgae Species Grown on Industrial Wastewater. Mar. Drugs 2015, 13, 7339–7356. [Google Scholar] [CrossRef]
- Aono, Y.; Asikin, Y.; Wang, N.; Tieman, D.; Klee, H.; Kusano, M. High-Throughput Chlorophyll and Carotenoid Profiling Reveals Positive Associations with Sugar and Apocarotenoid Volatile Content in Fruits of Tomato Varieties in Modern and Wild Accessions. Metabolites 2021, 11, 398. [Google Scholar] [CrossRef]
- Rajeshwari, K.R.; Rajashekhar, M. Biochemical composition of seven species of cyanobacteria isolated from different aquatic habitats of Western Ghats, Southern India. Braz. Arch. Biol. Technol. 2011, 54, 849–857. [Google Scholar] [CrossRef][Green Version]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef]
- Pombal, S.; Hernández, Y.; Diez, D.; Mondolis, E.; Mero, A.; Morán-Pinzón, J.; Guerrero, E.I.; Rodilla, J.M. Antioxidant activity of carvone and derivatives against superoxide ion. Nat. Prod. Commun. 2017, 12, 653–655. [Google Scholar] [CrossRef]
- Tekiner, M.; Kurt, M.; Ak, I.; Kurt, A. Determination of absorption coefficient of Chlorella vulgaris and Arthrospira maxima in water. AIP Conf. Proc. 2018, 1935, 120002. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Buschmann, C. Chlorophylls and Carotenoids: Measurement and Characterization by UV-VIS Spectroscopy. Curr. Protoc. Food Anal. Chem. 2001, 1, F4.3.1–F4.3.8. [Google Scholar] [CrossRef]
- Schmitz, C.; Pizzatto Dos Passos, A.; Bauer, C.M.; Cunha, J.; Mattioni, B.; Maraschin, M. Comparison of five methods for lipid extraction from the Phaeodactylum tricornutum microalga and determination of fucoxanthin and fatty acids profiles. Adv. Biotechnol. Microbiol. 2019, 14, 1–8. [Google Scholar] [CrossRef]
- Szpyrka, E.; Broda, D.; Oklejewicz, B.; Podbielska, M.; Slowik-Borowiec, M.; Jagusztyn, B.; Chrzanowski, G.; Kus-Liskiewicz, M.; Duda, M.; Zuczek, J.; et al. A Non-Vector Approach to Increase Lipid Levels in the Microalga Planktochlorella nurekis. Molecules 2020, 25, 270. [Google Scholar] [CrossRef]
- Kumar, G.; Nguyen, D.D.; Sivagurunathan, P.; Kobayashi, T.; Xu, K.; Chang, S.W. Cultivation of microalgal biomass using swine manure for biohydrogen production: Impact of dilution ratio and pretreatment. Bioresour. Technol. 2018, 260, 16–22. [Google Scholar] [CrossRef]
- Hejazi, M.; Kleinegris, D.; Wijffels, R. Mechanism of extraction of ?-carotene from microalgaDunaliellea salina in two-phase bioreactors. Biotechnol. Bioeng. 2004, 88, 593–600. [Google Scholar] [CrossRef]
- Del Campo, J.A.; Rodríguez, H.; Moreno, J.; Vargas, M.Á.; Rivas, J.; Guerrero, M.G. Accumulation of astaxanthin and lutein in Chlorella zofingiensis (Chlorophyta). Appl. Microbiol. Biotechnol. 2004, 64, 848–854. [Google Scholar] [CrossRef]
- Craft, N.E.; Soares, J.H. Relative solubility, stability, and absorptivity of lutein and.beta.-carotene in organic solvents. J. Agric. Food Chem. 1992, 40, 431–434. [Google Scholar] [CrossRef]
- Barchan, A.; Bakkali, M.; Arakrak, A.; Pagán, R.; Laglaoui, A. The effects of solvents polarity on the phenolic contents and antioxidant activity of three Mentha species extracts. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 399–412. [Google Scholar]
- Boskou, D.; Tsimidou, M.; Blekas, G. Polar Phenolic Compounds. In Olive Oil: Chemistry and Technology: Second Edition; Elsevier: Amsterdam, The Netherlands, 2006; pp. 73–92. [Google Scholar] [CrossRef]
- Pantami, H.; Bustamam, M.A.; Lee, S.; Ismail, I.; Faudzi, S.M.; Nakakuni, M.; Shaari, K. Comprehensive GCMS and LC-MS/MS Metabolite Profiling of Chlorella vulgaris. Mar. Drugs 2020, 18, 367. [Google Scholar] [CrossRef] [PubMed]
- Nuzzo, G.; Gallo, C.; D’Ippolito, G.; Cutignano, A.; Sardo, A.; Fontana, A. Composition and Quantitation of Microalgal Lipids by ERETIC 1H NMR Method. Mar. Drugs 2013, 11, 3742–3753. [Google Scholar] [CrossRef]
- Hussein, H.A.; Kassim, M.N.I.; Maulidiani, M.; Abas, F.; Abdullah, M.A. Cytotoxicity and 1H NMR metabolomics analyses of microalgal extracts for synergistic application with Tamoxifen on breast cancer cells with reduced toxicity against Vero cells. Heliyon 2022, 8, 1–17. [Google Scholar] [CrossRef]
- Nzayisenga, J.; Sellstedt, A. Metabolomic Study of Heterotrophically Grown Chlorella sp. Isolated from Wastewater in Northern Sweden. Molecules 2021, 26, 2410. [Google Scholar] [CrossRef]
- Deyab, M.; Mofeed, J.; El-Bilawy, E.; Ward, F. Antiviral activity of five filamentous cyanobacteria against coxsackievirus B3 and rotavirus. Arch. Microbiol. 2019, 202, 213–223. [Google Scholar] [CrossRef]
- Marinho, R.D.S.S.; Ramos, C.J.B.; Leite, J.P.G.; Teixeira, V.L.; Paixão, I.C.N.D.P.; Belo, C.A.D.; Pereira, A.B.; Pinto, A.M.V. Antiviral activity of 7-keto-stigmasterol obtained from green Antarctic algae Prasiola crispa against equine herpesvirus 1. J. Appl. Phycol. 2016, 29, 555–562. [Google Scholar] [CrossRef]
- Zhang, W.; Tan, N.G.J.; Fu, B.; Li, S.F.Y. Metallomics and NMR-based metabolomics of Chlorella sp. reveal the synergistic role of copper and cadmium in multi-metal toxicity and oxidative stress. Metallomics 2015, 7, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.; Collet, P.; Lardon, L.; Hélias, A.; Steyer, J.-P.; Bernard, O. Life-cycle assessment of microalgal-based biofuel. Biofuels Algae 2019, 2, 507–550. [Google Scholar] [CrossRef]
- Chen, H.; Zheng, Y.; Zhan, J.; He, C.; Wang, Q. Comparative metabolic profiling of the lipid-producing green microalga Chlorella reveals that nitrogen and carbon metabolic pathways contribute to lipid metabolism. Biotechnol. Biofuels 2017, 10, 153. [Google Scholar] [CrossRef] [PubMed]
- Carbone, D.; Pellone, P.; Lubritto, C.; Ciniglia, C. Evaluation of Microalgae Antiviral Activity and Their Bioactive Compounds. Antibiotics 2021, 10, 746. [Google Scholar] [CrossRef]
- Romero-Martínez, B.S.; Montaño, L.M.; Solís-Chagoyán, H.; Sommer, B.; Ramírez-Salinas, G.L.; Pérez-Figueroa, G.E.; Flores-Soto, E. Possible Beneficial Actions of Caffeine in SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 5460. [Google Scholar] [CrossRef]
- Yim, S.-K.; Kim, I.; Warren, B.; Kim, J.; Jung, K.; Ku, B. Antiviral Activity of Two Marine Carotenoids against SARS-CoV-2 Virus Entry In Silico and In Vitro. Int. J. Mol. Sci. 2021, 22, 6481. [Google Scholar] [CrossRef]
- Ghasemnejad-Berenji, M. Immunomodulatory and anti-inflammatory potential of crocin in COVID-19 treatment. J. Food Biochem. 2021, 45, e13718. [Google Scholar] [CrossRef]
- Al-Horani, R.A.; Kar, S. Potential Anti-SARS-CoV-2 Therapeutics That Target the Post-Entry Stages of the Viral Life Cycle: A Comprehensive Review. Viruses 2020, 12, 1092. [Google Scholar] [CrossRef]
- Afreen, R.; Tyagi, S.; Singh, G.P.; Singh, M. Challenges and Perspectives of Polyhydroxyalkanoate Production From Microalgae/Cyanobacteria and Bacteria as Microbial Factories: An Assessment of Hybrid Biological System. Front. Bioeng. Biotechnol. 2021, 9, 1–14. [Google Scholar] [CrossRef]
- Azizan, A.; Bustamam, M.S.A.; Maulidiani, M.; Shaari, K.; Ismail, I.S.; Nagao, N.; Abas, F. Metabolite Profiling of the Microalgal Diatom Chaetoceros Calcitrans and Correlation with Antioxidant and Nitric Oxide Inhibitory Activities via 1H NMR-Based Metabolomics. Mar. Drugs 2018, 16, 154. [Google Scholar] [CrossRef] [PubMed]
- Sarpal, A.S.; Teixeira, C.M.L.L.; Silva, P.R.M.; Monteiro, T.V.D.C.; da Silva, J.I.; da Cunha, V.S.; Daroda, R.J. NMR techniques for determination of lipid content in microalgal biomass and their use in monitoring the cultivation with biodiesel potential. Appl. Microbiol. Biotechnol. 2015, 100, 2471–2485. [Google Scholar] [CrossRef]
- Davey, P.T.; Hiscox, W.C.; Lucker, B.F.; O’Fallon, J.V.; Chen, S.; Helms, G.L. Rapid triacylglyceride detection and quantification in live micro-algal cultures via liquid state 1H NMR. Algal Res. 2012, 1, 166–175. [Google Scholar] [CrossRef]
- Guella, G.; Frassanito, R.; Mancini, I. A new solution for an old problem: The regiochemical distribution of the acyl chains in galactolipids can be established by electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2003, 17, 1982–1994. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.; Valentão, P.; Andrade, P.B. Biologically Active Oxylipins from Enzymatic and Nonenzymatic Routes in Macroalgae. Mar. Drugs 2016, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Maltsev, Y.; Maltseva, K. Fatty acids of microalgae: Diversity and applications. Rev. Environ. Sci. Bio/Technology 2021, 20, 515–547. [Google Scholar] [CrossRef]
- Kristjánsson, J.M.; Rolfsson, Ó. Virucidal activity of a proprietary blend of plant-based oils (Viruxal) against SARS-CoV-2 and influenza viruses—An in vitro study. bioRxiv 2021, 1–11. [Google Scholar] [CrossRef]
- Mofeed, J.; Deyab, M.; Mohamed, A.; Moustafa, M.; Negm, S.; El-Bilawy, E. Antimicrobial activities of three seaweeds extract against some human viral and bacterial pathogens. BIOCELL 2022, 46, 247–261. [Google Scholar] [CrossRef]
- Ferdous, U.T.; Yusof, Z.N.B. Insight into Potential Anticancer Activity of Algal Flavonoids: Current Status and Challenges. Molecules 2021, 26, 6844. [Google Scholar] [CrossRef]
- Ferreira, G.F.; Pessoa, J.G.B.; Pinto, L.F.R.; Filho, R.M.; Fregolente, L.V. Mono- and diglyceride production from microalgae: Challenges and prospects of high-value emulsifiers. Trends Food Sci. Technol. 2021, 118, 589–600. [Google Scholar] [CrossRef]
- Nascimento, T.C.D.; Pinheiro, P.N.; Fernandes, A.S.; Murador, D.C.; Neves, B.V.; de Menezes, C.R.; de Rosso, V.V.; Jacob-Lopes, E.; Zepka, L.Q. Bioaccessibility and intestinal uptake of carotenoids from microalgae Scenedesmus obliquus. LWT 2020, 140, 110780. [Google Scholar] [CrossRef]
- Banskota, A.H.; Stefanova, R.; Gallant, P.; Osborne, J.A.; Melanson, R.; O’Leary, S.J. Nitric oxide inhibitory activity of monogalactosylmonoacylglycerols from a freshwater microalgae Chlorella sorokiniana. Nat. Prod. Res. 2013, 27, 1028–1031. [Google Scholar] [CrossRef]
- Solinski, A.E.; Koval, A.B.; Brzozowski, R.S.; Morrison, K.R.; Fraboni, A.J.; Carson, C.E.; Eshraghi, A.R.; Zhou, G.; Quivey, R.G.; Voelz, V.A.; et al. Diverted Total Synthesis of Carolacton-Inspired Analogs Yields Three Distinct Phenotypes in Streptococcus mutans Biofilms. J. Am. Chem. Soc. 2017, 139, 7188–7191. [Google Scholar] [CrossRef] [PubMed]
- Santoyo, S.; Jaime, L.; Plaza, M.; Herrero, M.; Rodriguez-Meizoso, I.; Ibañez, E.; Reglero, G. Antiviral compounds obtained from microalgae commonly used as carotenoid sources. J. Appl. Phycol. 2012, 24, 731–741. [Google Scholar] [CrossRef]
- Ishikawa, C.; Tafuku, S.; Kadekaru, T.; Sawada, S.; Tomita, M.; Okudaira, T.; Nakazato, T.; Toda, T.; Uchihara, J.-N.; Taira, N.; et al. Antiadult T-cell leukemia effects of brown algae fucoxanthin and its deacetylated product, fucoxanthinol. Int. J. Cancer 2008, 123, 2702–2712. [Google Scholar] [CrossRef]
- Reynolds, D.; Huesemann, M.; Edmundson, S.; Sims, A.; Hurst, B.; Cady, S.; Beirne, N.; Freeman, J.; Berger, A.; Gao, S. Viral inhibitors derived from macroalgae, microalgae, and cyanobacteria: A review of antiviral potential throughout pathogenesis. Algal Res. 2021, 57, 102331. [Google Scholar] [CrossRef]
- Gómez-García, M.; Puente, H.; Argüello, H.; Mencía-Ares, Ó.; Rubio, P.; Carvajal, A. In vitro Assessment of Antiviral Effect of Natural Compounds on Porcine Epidemic Diarrhea Coronavirus. Front. Veter Sci. 2021, 8, 652000. [Google Scholar] [CrossRef]
- Brown, A.; Strobel, G.; Hanrahan, K.; Sears, J. Antiviral Activity of the PropylamylatinTM Formula against the Novel Coronavirus SARS-CoV-2 In Vitro using direct injection and gas assays in Virus suspensions. Viruses 2021, 13, 415. [Google Scholar] [CrossRef]
| Biomarkers | Dichloromethane | Hexane | Methanol |
|---|---|---|---|
| Carotenoids (µg/g) | 16.65 ± 3.04 a | 10.10 ± 0.91 b | 7.14 ± 0.49 c |
| Polyphenols (mg/g) | 40.96 ± 3.58 a | 29.17 ± 3.20 b | 84.91 ± 4.13 c |
| Cysteine (mg/g) | 15.94 ± 0.89 a | 16.21 ± 0.68 a | 15.67 ± 0.88 a |
| Phenylalanine (mg/g) | 30.06 ± 1.54 a | 30.52 ± 1.50 a | 29.59 ± 1.51 a |
| Histidine (mg/g) | 14.63 ± 1.23 a | 15.00 ± 0.94 a | 14.25 ± 1.21 a |
| Isoleucine (mg/g) | 164.54 ± 9.80 a | 167.48 ± 7.46 a | 161.58 ± 9.64 a |
| Leucine (mg/g) | 9.18 ± 2.05 a | 9.79 ± 1.56 a | 8.56 ± 2.02 a |
| Proline (mg/g) | 31.57 ± 3.08 a | 32.49 ± 2.34 a | 30.63 ± 3.03 a |
| Serine (mg/g) | 73.13 ± 6.15 a | 74.98 ± 6.15 a | 71.27 ± 6.05 a |
| Threonine (mg/g) | 99.13 ± 6.15 a | 100.98 ± 6.15 a | 97.27 ± 6.05 a |
| Tryptophan (mg/g) | 44.24 ± 2.05 a | 44.86 ± 1.56 a | 43.62 ± 2.02 a |
| Valine (mg/g) | 94.56 ± 6.94 a | 119.40 ± 5.29 b | 54.96 ± 6.83 c |
| Compound Name | Concentration (µM) | |
|---|---|---|
| Hexane Extract | Dichloromethane Extract | |
| 2-Hydroxybutyrate | 0.3 | 11.1 |
| 3-Hydroxyisobutyrate | 4.2 | 15.2 |
| Acetic acid | 0.2 | 0.6 |
| Citric acid | 0.8 | 10.2 |
| Ethanol | 0.7 | 0.7 |
| Glycerol | 0.1 | N/A |
| Formic acid | 0.9 | 11.4 |
| L-Glutamic acid | 1.0 | 1.1 |
| Hypoxanthine | 0.6 | 4.0 |
| L-Tyrosine | 0.1 | 1.1 |
| L-Alanine | 4.3 | N/A |
| L-Threonine | 1.6 | 12.1 |
| L-Lactic acid | 1.1 | 12.8 |
| L-Aspartic acid | 1.5 | 8.5 |
| Pyruvic acid | N/A | 1.2 |
| Succinic acid | N/A | 12.2 |
| Pyroglutamic acid | N/A | 6.7 |
| 3-Hydroxybutyric acid | 0.6 | 2.0 |
| Creatinine | 3.7 | N/A |
| L-Glutamine | N/A | 13.3 |
| L-Leucine | 0.7 | 7.5 |
| L-Methionine | N/A | 1.9 |
| L-Valine | 9.6 | 14.3 |
| Acetone | 30.3 | 100.2 |
| Isobutyric acid | 19.0 | 17.8 |
| 1,5-Anhydrosorbitol | 1.0 | 0.1 |
| Dimethylsulfone | 0.2 | 1.0 |
| EDTA | 0.1 | N/A |
| Extracts | CC50 a µg/mL | CI95 b µg/mL |
|---|---|---|
| Hexane | 330.4 | 270.1 to 404.3 |
| Dichloromethane | 53.19 | 30.94 to 91.45 |
| Methanol | 73.30 | 58.74 to 91.46 |
| Conc (µg/mL) | ||||||
|---|---|---|---|---|---|---|
| Extract | 3.12 | 6.25 | 12.5 | 25.00 | 50.00 | |
| Hexane | 24 ± 2 °C | NR | NR | −7.0 ± 1.2 | −7.0 ± 1.4 | −7.0 ± 1.2 |
| Dichloromethane | −6.0 ± 0.5 | −6.0 ± 0.3 | −6.0 ± 0.2 | −8.0 ± 0.3 | −8.0 ± 0.4 | |
| Methanol | −6.0 ± 1.5 | −6.0 ± 0.5 | −6.0 ± 0.8 | −6.0 ± 0.3 | −6.0 ± 0.4 | |
| Hexane | 35 ± 2 °C | NR | NR | NR | NR | NR |
| Dichloromethane | NR | NR | −8.0 ± 0.3 | −8.0 ± 0.3 | −8.0 ± 0.4 | |
| Methanol | −6.0 ± 1.5 | −6.0 ± 0.5 | −6.0 ± 0.8 | −6.0 ± 0.3 | −6.0 ± 0.4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graff Reis, J.; Dai Prá, I.; Michelon, W.; Viancelli, A.; Piedrahita Marquez, D.G.; Schmitz, C.; Maraschin, M.; Moura, S.; Thaís Silva, I.; de Oliveira Costa, G.; et al. Characterization of Planktochlorella nurekis Extracts and Virucidal Activity against a Coronavirus Model, the Murine Coronavirus 3. Int. J. Environ. Res. Public Health 2022, 19, 15823. https://doi.org/10.3390/ijerph192315823
Graff Reis J, Dai Prá I, Michelon W, Viancelli A, Piedrahita Marquez DG, Schmitz C, Maraschin M, Moura S, Thaís Silva I, de Oliveira Costa G, et al. Characterization of Planktochlorella nurekis Extracts and Virucidal Activity against a Coronavirus Model, the Murine Coronavirus 3. International Journal of Environmental Research and Public Health. 2022; 19(23):15823. https://doi.org/10.3390/ijerph192315823
Chicago/Turabian StyleGraff Reis, Jacqueline, Isabella Dai Prá, William Michelon, Aline Viancelli, David Guillermo Piedrahita Marquez, Caroline Schmitz, Marcelo Maraschin, Sidnei Moura, Izabella Thaís Silva, Geovanna de Oliveira Costa, and et al. 2022. "Characterization of Planktochlorella nurekis Extracts and Virucidal Activity against a Coronavirus Model, the Murine Coronavirus 3" International Journal of Environmental Research and Public Health 19, no. 23: 15823. https://doi.org/10.3390/ijerph192315823
APA StyleGraff Reis, J., Dai Prá, I., Michelon, W., Viancelli, A., Piedrahita Marquez, D. G., Schmitz, C., Maraschin, M., Moura, S., Thaís Silva, I., de Oliveira Costa, G., Tizziani, T., Sandjo, L. P., Rodríguez-Lázaro, D., & Fongaro, G. (2022). Characterization of Planktochlorella nurekis Extracts and Virucidal Activity against a Coronavirus Model, the Murine Coronavirus 3. International Journal of Environmental Research and Public Health, 19(23), 15823. https://doi.org/10.3390/ijerph192315823













