Abstract
Antimicrobial resistance (AMR) is one of the largest global concerns due to its influence in multiple areas, which is consistent with One Health’s concept of close interconnections between people, animals, plants, and their shared environments. Antibiotic-resistant bacteria (ARB) and antibiotic-resistance genes (ARGs) circulate constantly in various niches, sediments, water sources, soil, and wastes of the animal and plant sectors, and is linked to human activities. Sewage of different origins gets to the wastewater treatment plants (WWTPs), where ARB and ARG removal efficiency is still insufficient, leading to their transmission to discharge points and further dissemination. Thus, WWTPs are believed to be reservoirs of ARGs and the source of spreading AMR. According to a World Health Organization report, the most critical pathogens for public health include Gram-negative bacteria resistant to third-generation cephalosporins and carbapenems (last-choice drugs), which represent β-lactams, the most widely used antibiotics. Therefore, this paper aimed to present the available research data for ARGs in WWTPs that confer resistance to β-lactam antibiotics, with a particular emphasis on clinically important life-threatening mechanisms of resistance, including extended-spectrum β-lactamases (ESBLs) and carbapenemases (KPC, NDM).
1. Introduction
Antibiotics are widely used to prevent and treat infections in humans, animals, and plants, but their high and incorrect consumption have made them increasingly ineffective due to antimicrobial-resistant microorganisms emerging and spreading globally. Thus, antimicrobial resistance (AMR) was announced by the World Health Organization (WHO) as one of the top global public health threats facing humanity [1]. Some Gram-negative bacteria, such as carbapenem-resistant Pseudomonas aeruginosa, Acinetobacter baumannii, and Enterobacterales resistant to third-generation cephalosporins and carbapenems are considered to be of particular importance, and the WHO and Centers for Disease Control and Prevention (CDC) included them in the group of critical pathogens due to the fact that they are a major cause of nosocomial infections with high morbidity and mortality [2,3]. Systematic analysis estimated 4.95 million deaths associated with bacterial AMR in 2019 and indicated β-lactam-resistant (mainly to third generation cephalosporins and carbapenem) bacteria as the major cause of death [4]. Tremendously dangerous microorganisms accumulate various AMR mechanisms that leads to their multi-drug resistance (MDR), extensive-drug resistance (XDR), or even pan-drug resistance (PDR), leaving few, one, or no therapeutic options left, respectively. Consequently, infections caused by such bacteria carry an extremely high risk of death [5,6].
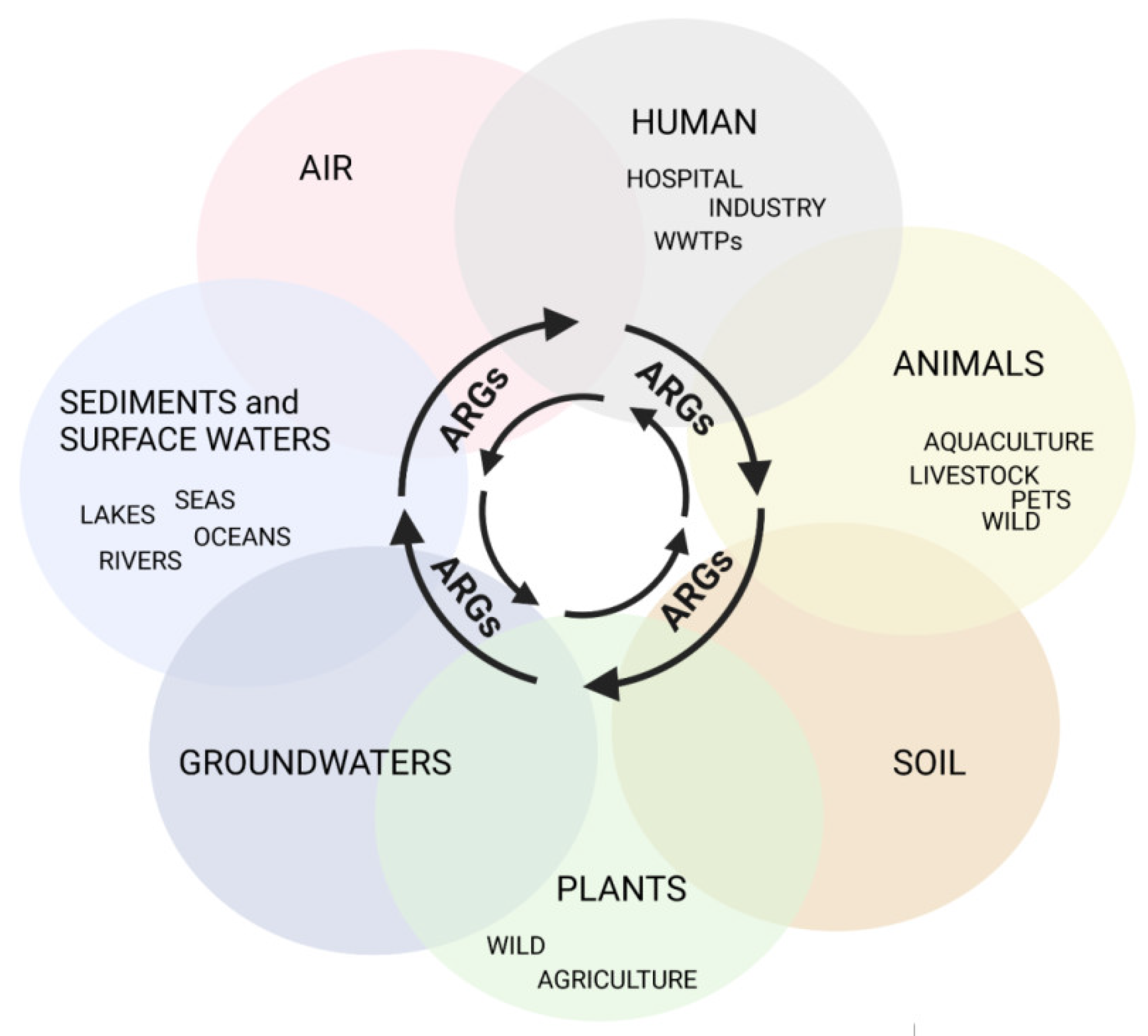
AMR is ubiquitous, associated with agriculture and livestock, medical, and veterinary settings, but it is also observed in many aquatic environments, which is in line with One Health’s concept (available online: https://www.cdc.gov/onehealth/index.html accessed on 23 May 2022) of close interconnections between people, animals, plants, and their shared environments (Figure 1) [7].
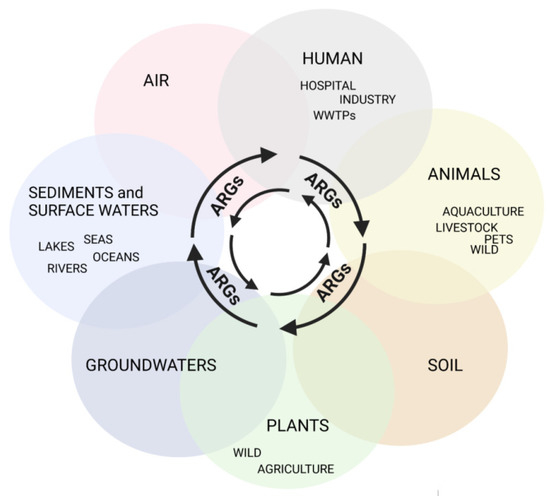
Figure 1.
Routes of ARG transmission in the total environment, created with BioRender (available online: https://biorender.com, accessed on 23 May 2022).
Many aspects related to geographic location, socio-economic level, climate, antibiotic consumption, and the technology of the treatment process affect the abundance of antimicrobial resistance genes (ARGs), the bacteria carrying them (antibiotic-resistant bacteria, ARB), and their dissemination in the environment [8,9]. One of the factors contributing to the scale and speed of AMR spreading is the fact that high amounts of antibiotics get into sewage and, consequently, into wastewater treatment plants (WWTPs). Although the applied technology and treatment methods are constantly being improved and developed, they are still insufficient to eliminate antibiotics, ARB, and ARGs completely. Moreover, the presence of antibiotics in sub-inhibitory concentrations creates conditions for selective pressure, and additionally, other factors present in sewage, such as pesticides, detergents, and heavy metals, stimulate the co-selection of resistant strains [10,11,12,13,14,15,16].
The genetic background of the AMR transmission process is of great importance. Resistance mechanisms are genetically based and linked with many genes localized on a bacterial chromosome or, what is more dangerous, on mobile genetic elements (MGEs). The genes encode enzymes, proteins that are involved in many processes, for example inactivating antibiotics or modifying their structure, alterating drug target sites, modifying the outer membrane structure that inhibits antibiotic penetration into the cell, or the active removal of the chemotherapeutics from the cell. Due to their location on MGEs, they pose a big risk to be transferred between bacteria of the same or different species through conjugation, transduction, or transformation [17,18]. Bacteria interacting with each other and exchanging genes by horizontal gene transfer (HGT) may lead to situations wherein previously sensitive and nonpathogenic strains may get resistance determinants and become virulent or reservoirs of ARGs for further transmission. These microorganisms, as well as resistance genes, may be discharged from WWTP systems into natural water bodies like lakes, rivers, and seas [19,20,21,22,23,24], which plays an important role in their further dissemination into human, animal and plant populations [25,26,27]. Therefore, it is believed that the WWTPs are reservoirs of ARGs, so-called “hotspots”, and one of the sources of spreading AMR, especially clinically relevant ARGs [28,29,30,31].
A great effort has been made to fight AMR and many global strategies have been taken, including developing new drugs and vaccines, improving the diagnostics of resistance mechanisms, the rational use of antibiotics, infection prevention and control, and developing new technological methods for the treatment and disinfection of wastewaters [32,33,34,35]. The monitoring of AMR and identifying the migration routes of bacteria with important mechanisms of resistance in the environment is also crucial and fundamental [36]. It may help to obtain knowledge about actual epidemiological situations, the origin of ARGs, mechanisms of spreading AMR, and transmission routes, which are essential for taking appropriate actions to prevent this phenomenon. Such surveillance studies concern the occurrence of not only resistant bacteria in ecosystems but also the occurrence of resistance genes that are easily and efficiently transmissible [16,30,31,34,37,38,39,40].
Zhuang et al. analyzed PubMed publications from the last 30 years (1990–2020) concerning reports of ARGs in the environment and showed that, on all continents, the highest frequency was related to genes encoding β-lactamases, enzymes that inactivate β-lactams, the most-used group of antibiotics [41]. Therefore, this paper aimed to present available research data on the identification of β-lactamase genes in WWTPs.
For this manual review of articles from the last decade, studies of β-lactamase genes in wastewater samples and from bacteria isolated from these type of samples were analyzed, including direct WWTP (i.e., influent, sewage sludge, effluent) and WWTP-related samples (i.e., air near bioreactors, discharge points). All of the research described below is summarized in detail in Table 1, where information about the type of tested samples, stages of the treatment process, methodology used, and detected variants are included. The reviewed studies were linked to municipal/urban WWTPs; however, if the authors involved additional information about the type of collected wastewater, it was noted.
2. β-Lactams and β-Lactamases—Background
Among the many antimicrobial drugs available, the group of β-lactams is one of the most important and most widely used in the treatment of bacterial infections, not only as the first choice, but above all as the last-choice drugs (available online https://www.ecdc.europa.eu/en/antimicrobial-consumption/surveillance-and-disease-data/database, accessed on 23 May 2022) [42]. β-lactams are classified based on chemical structure and the target of action. The common characteristic is the presence of the three carbon and one nitrogen ring (β-lactam ring). Depending on the modifications, different groups are distinguished. Generally, there are penicillins (natural penicillins, aminopenicillins, carboxypenicillins, and ureidopenicillins), cephalosporins (divided into five classes called generations), carbapenems, and monobactams.
All β-lactam antibiotics have a common mechanism of action, which is inhibition of the bacterial cell walls’ synthesis. They block the activity of bacterial enzymes, transpeptidases known as penicillin-binding proteins (PBPs), involved in the last stage of peptidoglycan synthesis, thus inducing a loss of viability and the lysis of bacterial cells. The modification of the PBPs’ structure may lead to a reduced affinity for β-lactams, which is the major pathway for β-lactam resistance among Gram-positive bacteria but is not very common for Gram-negative bacteria [5,43,44]. The other mechanisms of resistance, detected mainly in Gram-negative bacteria, are related to cell membrane modulations, including: (i) the reduction or loss of outer membrane porins that restrict the entry of antibiotic into the cell or (ii) the expression/overexpression of the efflux pump that allows the effective removal of the antibiotic from the cell. Examples are AcrAB-TolC-type pumps, described in clinical isolates of Klebsiella pneumoniae, and MexAB-OprM pumps, reported in P. aeruginosa [45]. Finally, the most common mechanism in Gram-negative bacteria, and relatively rarely found in Gram-positive bacteria, is the production of β-lactamases, enzymes that hydrolyze β-lactam antibiotics making them ineffective. These enzymes are critical, causing hard-treated human infections (urinary tract infections, bloodstream infections, wound infections, and pneumonia), especially caused by P. aeruginosa, A. baumannii, and Enterobacterales; thus, this paper focuses on them.
Two classification systems of β-lactamases are used. The structural one, based on the amino acid sequence of the enzyme, groups β-lactamases into 4 classes, A, B, C, and D, of which A, C, and D are β-lactamases with serine in the active center, while class B uses zinc cations as cofactors of the hydrolysis reaction (metallo-β-lactamases, MBL). Another classification scheme, functional, is based on substrate hydrolysis profiles and the inhibitor profile, distinguishing four main functional groups, 1–4. Group 1 includes cephalosporinases and cephamycinases, which are very weakly inhibited or uninhibited by clavulanic acid; group 2 is very extensive and diverse, with different substrate spectra, mostly inhibited by clavulanic acid. Group 3 additionally hydrolyzes carbapenems, but their activity is inhibited by EDTA, and group 4 are penicillinases weakly inhibited by clavulanic acid. Both classification systems of β-lactamases correlate well with each other. All of the enzymes that make up functional group 1 are structural class C; group 2 contains β-lactamases of classes A and D, and group 3 corresponds to class B [46,47].
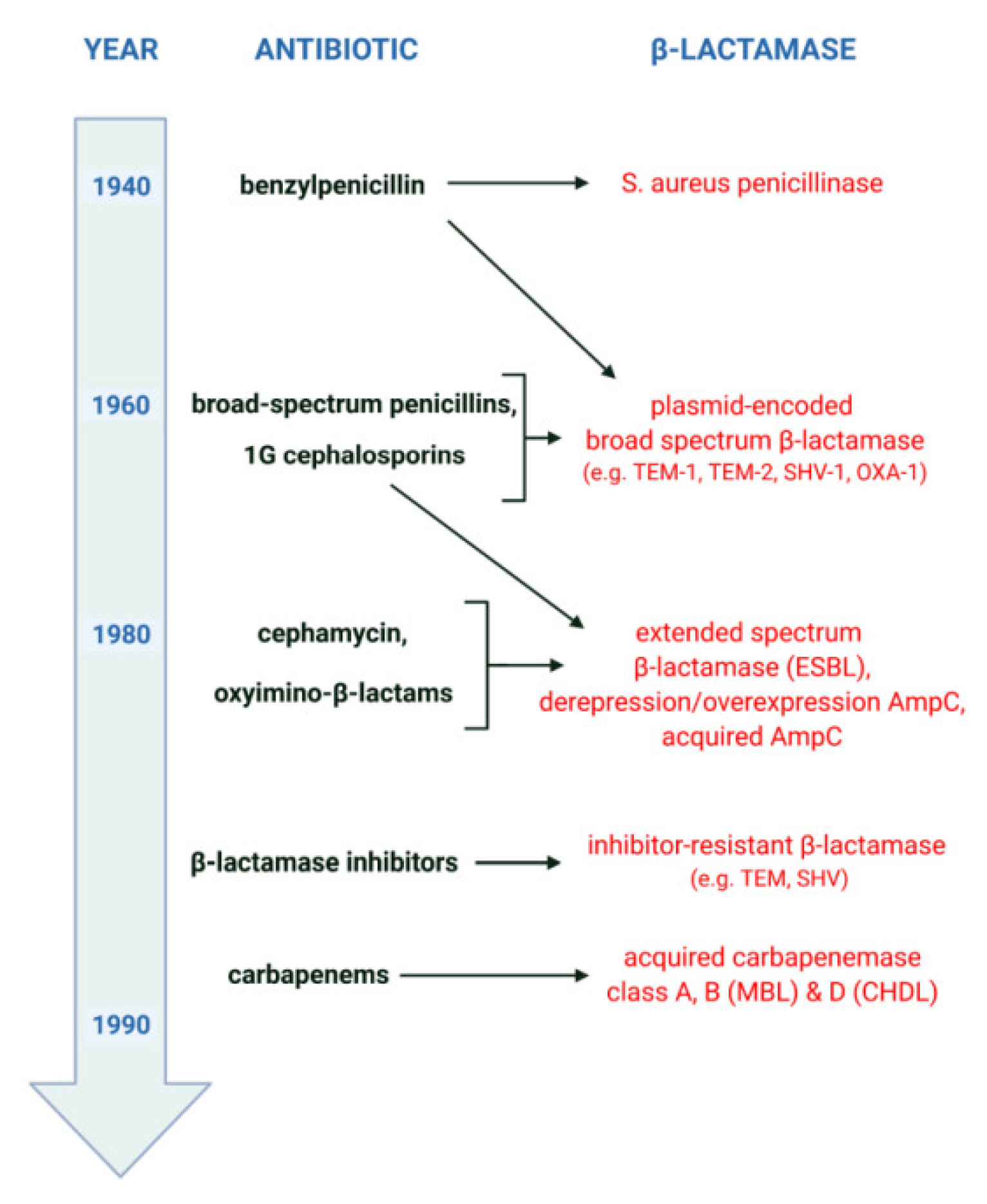
The general scenario of β-lactamase evolution was stimulated by the mass use of β-lactam antibiotics, as shown in Figure 2. It reveals a kind of “race” between pathogenic microorganisms and the pharmaceutical industry, which develops ever newer “generations” of β-lactams, as well as the adaptation of bacteria to environments in which the selection pressure of “older” and “newer” drugs accumulates. Shortly after the introduction of penicillins (benzylpenicillins) into therapy in the 1940s, the emergence and rapid growth of β-lactamase-producing strains of Staphylococcus aureus was observed. The first cephalosporins and broad-spectrum penicillins, used since the early 1960s, mainly against β-lactamase-producing S. aureus and/or Gram-negative bacilli, contributed to the emergence of new resistance mechanisms. Among other things, this resulted in the selection of Enterobacterales producing plasmid-encoded broad-spectrum β-lactamases. In turn, the intensive use of oxyimino-β-lactams since the early 1980s has led to the selection of new mechanisms of acquired resistance. This resistance is mainly related to the production of extended-spectrum β-lactamases (ESBLs) and acquired AmpC and includes phenomena such as the derepression or overexpression of AmpC. Finally, the bacterial response to the introduction of carbapenems has been the emergence of strains producing acquired carbapenemases such as MBLs and some class A and D enzymes.
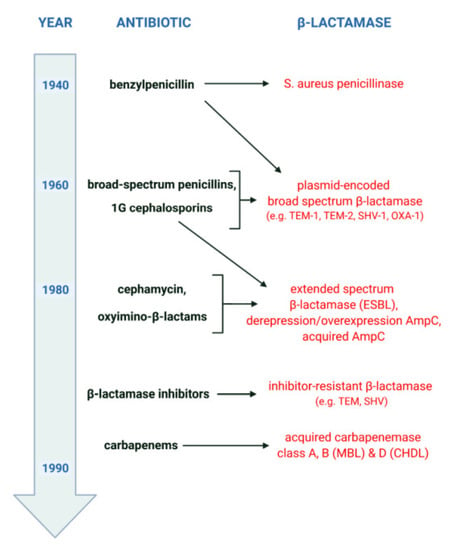
Figure 2.
Timeline of the evolution of β-lactamases, created with BioRender (available online: https://biorender.com, accessed on 23 May 2022).
All β-lactamases are encoded by bla genes and located on the bacterial chromosome or MGEs like plasmids, transposons, and integrons with gene cassettes. Bacteria can acquire ARGs by horizontal gene transfer, HGT, which enables the exchange of genetic material between commensals, environmental species, and pathogenic bacteria; therefore, HGT is considered the main method of antibiotic resistance dissemination [18].
3. Methods of AMR and ARGs Analysis in Environmental Samples
The monitoring and evaluation of ARB in water environments use various methods, generally divided into two groups: culture-dependent and culture-independent. The first one is based on traditional microbiological methods used in clinical surveillance, requiring strains isolated from the environmental samples (determining: taxonomy, antibiotic susceptibility profiles, resistance mechanisms). To evaluate the level and mechanism of resistance carried by bacteria, the disk diffusion method and minimum inhibitory concentration (MIC) assays are used, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST; available online: www.eucast.org) and the Clinical & Laboratory Standards Institute (CLSI; available online: www.clsi.org). Analysis of the AMR patterns of strains may provide information about multidrug resistance. Bacterial conjugation assays are also conducted to confirm the transferability of selected genes. Time-consumption is the main limitation of such methods, because they require pure bacterial cultures, which may be troublesome or even unavailable for slow-growing bacteria. Additionally, breakpoints for antibiotic susceptibility tests may be applied to a narrow spectrum of pathogens detected in wastewater, only to clinical bacteria for which recommendations are available.
Therefore culture-independent, DNA-based methods were developed and, in recent years, have become extensively used. Molecular techniques, including nucleic acid amplification (polymerase chain reaction, PCR) and DNA sequencing, are successfully used for the analysis of direct environmental samples but are also widely used for the molecular analysis of isolated strains for the detection of genetic resistance determinants (ARGs, MGEs) and/or molecular typing methods to define genetic relatedness between isolates with clinical and environmental origin (multi-locus sequence typing, MLST; phylogrouping; pulsed-field gel electrophoresis, PFGE). Some studies focus on defining the efficiency of the treatment process; therefore, quantitative PCR (qPCR) is used to determine the number of selected gene copies/mL (absolute abundance) and/or the number of copies normalized to 16S rRNA copies (relative abundance). The developing metagenomics approaches that use various techniques of molecular biology deserve special attention. Metagenomics allows us to explore the biodiversity of a population of microorganisms and the identification of the present genes, as well as detecting new ones and determining their functions and analyzing their origin and the transfer and dissemination of ARGs between species [16,30,48,49,50,51,52]. Most results of the metagenomics approaches in sewage contain the data of the resistance genes present in different stages of the treatment process; correlations with various factors, like heavy metals, MGEs, and antibiotics, on the ARGs’ occurrence and abundance; and their transfer and removal efficiency in different types of treatment processes and disinfection. The intensification of metagenomics research concerning AMR in WWTPs has been significant in recent years; however, due to the different approaches, different goals of the research, variety of tested samples, and types of WWTPs, the obtained results may be difficult to compare; thus, the procedures should be standardized. However, the analysis of the data gives an overall picture and information on general trends concerning the spread of antibiotic resistance [36,53].
4. Clinically Significant β-Lactam Resistance Genes in Wastewater Treatment Plants—The Occurrence and Distribution
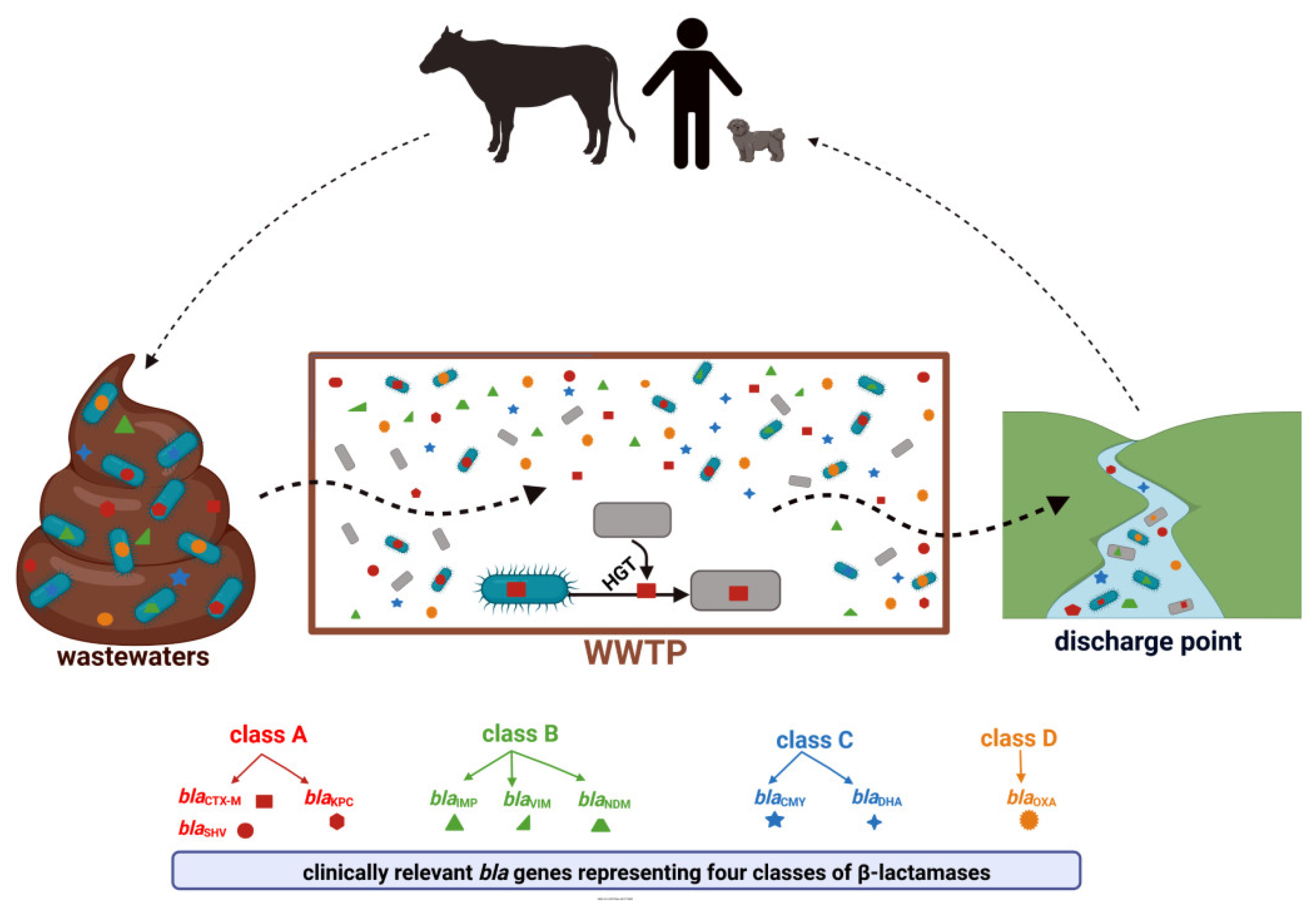
According to the β-lactamase database (available online: BLDB; http://bldb.eu/ accessed on 23 May 2022), these enzymes constitute a very heterogeneous group with more than 7000 genetic variants identified. Within each of the four classes (A, B, C, and D), β-lactamases of particular clinical importance can be distinguished. These are detected consistently in environmental niches, including WWTPs (Figure 3) [41].
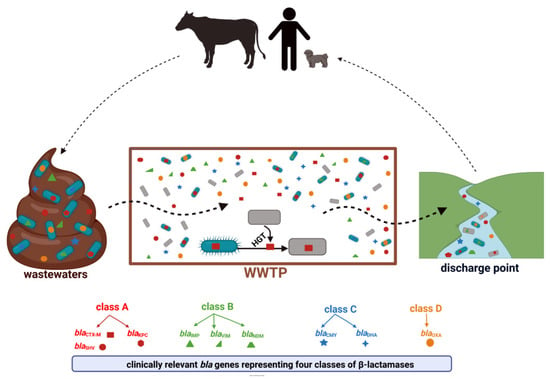
Figure 3.
WWTP as a hotspot for the transmission of clinically relevant β-lactam resistance genes, created with BioRender available online: https://biorender.com, accessed on 23 May 2022). Descriptions of the enzymes included in the Figure 3 can be found in Section 4.1, Section 4.2, Section 4.3 and Section 4.4.
4.1. Class A β-Lactamases
Class A β-lactamases are serine proteases that hydrolyze, on various levels, penicillins, monobactams, cephalosporins, and carbapenems and may be inhibited by β-lactamase inhibitors (e.g., clavulanic acid, sulbactam, tazobactam). It is the most diverse group, consisting of the enzymes with various spectra of hydrolysis, generally divided into: (i) a group with a narrow spectrum, e.g., carbenicillin-hydrolyzing β-lactamase (CARB) and Pseudomonas aeruginosa β-lactamase (PSE); (ii) a group with extended spectrum (ESBL) enzymes that originated from the first group but modified due to point mutations within the genes encoding them, which results in broadening their spectrum of hydrolyzing, e.g., cefotaximase-München-lactamase (CTX-M), Temoniera-lactamase (TEM), and sulfhydryl variable-lactamase (SHV); and (iii) a group with extremely extended spectrum including carbapenems—antibiotics of the last resort, e.g., Guiana extended-spectrum (GES), Klebsiella pneumoniae carbapenemase (KPC), Serratia marcescens enzyme (SME), and Serratia fonticola carbapenemase A (SFC-1) [54]. Among all, ESBL- and carbapenemase KPC-producing bacteria attracted the largest amount of clinical concern. Both TEM- and SHV-type ESBLs were described throughout the United States (US) and Europe in the late 1980s and 1990s, with specific variants noted to be regional in distribution [55,56]. The prevalence of these enzymes has now diminished at the same time as the worldwide dissemination of isolates producing CTX-M-type β-lactamases [57,58]. Once limited to hospital settings, ESBL-producing isolates quickly expanded into nursing homes and community settings as well [59,60]. The propagation of Enterobacterales-possessing ESBLs has had a significant impact on the choice of empirical antimicrobial therapy, driving the use of carbapenems in many institutions and resulting in increased resistance to carbapenems [61]. KPC carbapenemase has been extensively reported in K. pneumoniae, and it is endemic in the US but also in Latin America, China, Israel, and some European countries, such as Greece and Italy [62,63,64].
4.1.1. Class A β-Lactamases—Occurrence and Variability in WWTPs-Linked Samples
Due to the global spread of class A β-lactamases, it is a commonly, or even predominantly, detected group in WWTPs (Table 1). In a multi-national study of WWTPs from Denmark, Spain, and the United Kingdom (UK) with high-throughput qPCR used, these β-lactamases was leading, accounting for 70% of all detected bla genes [65]. Among them, the most relevant were two groups linked with ESBL and KPC enzymes. It is noteworthy that, among the ESBL group, the most common in clinical settings and in various wastewater sources is CTX-M encoded by blaCTX-M, carried mainly by Enterobacterales [66]. In this review, blaCTX-M was detected in the majority of included studies and, in many, had the highest prevalence [67,68,69,70,71,72,73,74,75,76,77,78,79,80,81]. However, blaSHV and/or blaTEM were found frequently as well [15,82,83,84,85]. In some studies, blaTEM was predominant, e.g., in an Irish study [86], as well as in Colombia [87], Poland and Portugal [9,88], Belgium [89], the US [90], and Africa [91]. Another significant group representing the KPC family encoded by blaKPC genes was detected in numerous WWTPs from European [65,69,89,92,93,94,95,96,97,98,99,100,101], as well as from American [90,102,103,104], African [72,91,105], and Asian countries [106,107]. Moreover, analysis of reviewed articles, especially those using developed techniques as high-throughput qPCR, whole-genome sequencing, or metagenomics, shows a high variety of detected genes of the discussed β-lactamases, not only representing blaCTX-M, blaSHV, blaTEM, and blaKPC families, but also others less frequently associated with public health, i.e., BEL, cfxA, GES, PER, SME, VEB, and others [65,92,96,102,107,108,109,110,111].
Environmental studies based on the analysis of bacterial strains during the treatment process most often concern the most critical pathogens posing the greatest threat, mainly Enterobacterales. In the reviewed literature, the predominantly tested and detected species among this bacteria family were Escherichia coli and K. pneumoniae [67,68,71,72,74,77,78,79,81,83,85,99,112,113,114,115,116,117,118,119,120,121,122,123,124,125]; however, different species of Citrobacter spp., Enterobacter spp., Pseudomonas spp., Aeromonas spp., or others were noted as well [71,76,80,87,92,102,104,109,110,121,126,127,128].
It is noteworthy that antibiotic susceptibility testing of the studied ESBL-producing strains isolated from the WWTPs confirms a high percentage of multi-drug resistance. It was also noted that these bacteria may survive the treatment process and that the WWTPs were unable to eradicate them completely. Generally, the number of MDR isolates decreased during the treatment, but for some, their proportion was still significant in effluents, in some even higher than in influent samples [70,71,73,86,88,92,94,97,99,118,119,123,125,129,130]. Moreover, analyzing downstream river or marine samples where final effluents are released, MDR isolates carrying ESBL enzymes were commonly detected [20,79,88,92,130].
Molecular typing concerning bacteria isolated from WWTPs confirmed high genetic relatedness between bacteria from WWTPs and human- and animal-associated sources, as well as the presence of clinically important lineages such as pandemic ST131 E. coli in WWTPs-related samples. Liedhegner et al. compared E. coli isolated from samples of various environmental compartments from one geographic area (clinical samples, hospital wastewater, and WWTP). The data including antibiotic resistance, virulence, and ESBL gene profiles confirmed high phenotypic and genotypic similarity across strains of these different origins and demonstrated potential health risks related to ESBL transmission [125]. An interesting study conducted by Raven et al. showed genetic relatedness between E. coli isolated from 20 WWTPs in the UK, livestock farms, retail meat, and isolates responsible for human blood infections. The genomic analysis of i.e., ESBL-producing isolates revealed that the three most common sequence types (STs) associated with bloodstream infections (ST131, ST73, and ST95) and the specific and most common for livestock (ST10) were found in wastewater samples [120]. In many other studies, human-associated, multidrug-resistant, and highly virulent clone ST131 E. coli was detected in WWTP samples as well [75,87,113,131,132,133,134].
4.1.2. Class A β-Lactamases—Removal during the Treatment Process
Concerning the removal of class A β-lactam ARGs, there is no universal target panel in qPCR studies; however, it has been noted that, although the WWTPs could effectively eliminate examined genes, their abundance was still reported in effluents and receiving water bodies. For example, in the study of Schages et al., strains harboring blaCTX-M were isolated from the effluent [123], as well as in a Japanese study wherein strains possessing ARGs belonging to the blaCTX-M-1, blaCTX-M-9, blaTEM, and blaSHV families survived even after sterilization [124]. Other studies reported similar results of the ARGs’ presence in effluent samples [108,135,136,137,138]. In Polish research from Kozieglowy, it was noticed that the wastewater treatment process leads to a significant increase in the relative abundance of blaTEM and blaGES genes, while the abundance of blaKPC decreased. Finally, the removal efficiency of ARGs was the least for blaGES (94.8%) and blaCTX-M (95.3%), while for other genes, it was >98% [69]. In another study, the presence of blaKPC was completely eliminated even after the first mechanical procedure [93]. In a Chinese survey comparing bacteria carrying blaCTX-M, blaSHV, and blaTEM, isolated from influent and effluent, higher prevalence was noted in influent samples, except for blaCTX-M, which was more frequently detected in effluent samples [129]. Significant differences between influent and effluent were described in a Romanian investigation and concerned blaSHV-100, -145, which were decreased during treatment [85]. Interestingly, Neudorf et al. analyzed 3 WWTPs in Arctic Canada and noted a decrease of blaTEM abundance in two sites with a passive system and no significant changes for a third WWTP with a mechanical system. Moreover, no differences were found for blaCTX-M in all treatment plants [139]. A Spanish study by Rodriguez-Mozaz et al. demonstrated an increased frequency of blaTEM during the treatment process [140], while in a study of three WWTPs from Finland and Estonia, no significant changes were noted for blaCTX-M-32, unlike blaSHV-34, of which the relative concentration was increased in effluent samples but only in one tested WWTP [141]. Comparable data with similar blaCTX-M and blaTEM concentrations in influent and effluent samples were obtained in a study of five WWTPs in Tunisia; however, the abundance of the genes was higher in the effluent in a WWTP receiving additional hospital wastewater [142]. The occurrence of class A β-lactamases ARGs was also detected in downstream river samples whence final effluents were discharged, e.g., in a multi-national study including sixteen WWTPs from ten European countries [101], in a study conducted by Zieliński et al., wherein the predominant blaTEM was noted in receiving river water samples [15], and in a study performed by Osińska et al., wherein the presence of blaSHV and blaTEM in receiving river samples was confirmed [84].
WWTPs pose a health risk, not only because treated wastewater containing AMR genes or MDR bacteria are transferred into surface water bodies, but also because these pollutants are discharged into the air surrounding WWTPs through bioaerosol generated from bioreactors [15,68]. The study of the carriage of ESBL-producing Enterobacterales in WWTP workers and surrounding residents shows that these groups are much more like to acquire bacteria harboring the ESBL mechanism [25], thus confirming the direct influence of WWTPs on spreading ARGs into air. The contribution of WWTPs’ bioaerosols in ARGs and ARB propagation into air and different environments is commonly investigated [143,144,145,146]. For example, Gaviria-Figueroa et al. studied bioaerosol samples collected downwind from sludge aeration tanks and showed a significant presence of clinically relevant class A β-lactamases, along with other classes of these enzymes and different antibiotic groups [147].
4.2. Class B β-Lactamases
Class B β-lactamases consist of a wide variety of metallo-β-lactamases (MBLs), enzymes able to hydrolyze almost all β-lactams: penicillins, cephalosporins, clinically available β-lactamase inhibitors, and carbapenems, except monobactams. They use zinc ions for activity, hence the name “metallo-” and susceptibility to metallic ion chelators like EDTA. Numerous variants are distinguished and grouped into three subclasses, among which the most widespread MBLs are imipenem-resistant Pseudomonas (IMP), Verona integron-encoded metallo-β-lactamase (VIM), and New Delhi metallo-β-lactamase (NDM), all representing subclass B1 [54,148,149,150]. MBLs initially detected in P. aeruginosa are frequently found nowadays in K. pneumoniae and other Enterobacterales [62,63,64]. IMP carbapenemases mainly contribute to carbapenem resistance in Japan, as well as in other regions of Southeast Asia and Australia [151,152,153]. Although they have not spread extensively throughout the rest of the world, they are being reported more frequently in Middle-Eastern countries [154]. VIM MBLs are identified more frequently than IMP enzymes [155]. Initially, they spread rapidly throughout southern Europe with major outbreaks of VIM-producing P. aeruginosa reported in Italy and Greece in 2006, followed by outbreaks of VIM-producing K. pneumoniae [156,157]. Today, they are found globally, mainly in K. pneumoniae and E. cloacae complex strains [151]. Among the major types of MBLs, the NDM-type variants are especially associated with Enterobacterales. The first NDM was identified in 2008 in a K. pneumoniae isolate from a patient in Sweden who had arrived from India [158]. The Indian subcontinent, the Balkans, and the Middle-East/North Africa are considered to be the main NDM reservoirs [62,63]. An extremely wide spectrum of metallo-β-lactamases and the fact that isolates possessing MBL genes often simultaneously harbor other antibiotic resistance genes make these organisms an urgent public health threat. Although there is substantial geographic variability in the prevalence of MBL enzymes, they are noted worldwide and the speed of their dissemination is alarming, especially NDM enzymes [44,54,159,160,161].
4.2.1. IMP and VIM β-Lactamases in WWTPs-Linked Samples
As with the previously discussed ARGs, the environment plays a role in the transmission of blaIMP and blaVIM encoding MBLs enzymes with clinical importance, IMP and VIM, respectively (Table 1). Although the majority of reports focus on hospital wastewater, these genes were detected also in samples of wastewater treatment plants from the US [82,102,103,147], Canada [104], China [70,82], and Singapore [107] as well as from many European countries, such as Sweden [96,109], Switzerland [99], the UK [128], Germany [100,123,136,162], Poland [69,92,93,163], Slovakia [115], and Romania [94]. A multi-national study concerning urban WWTPs in Denmark, Spain, and the UK showed the permanent presence of blaVIM during the treatment process even in downstream river samples, in contrast to other tested genes, which were reduced under a detectable level [65]. Interesting results were presented by Khan et al., who compared Klebsiella oxytoca strains isolated from clinical sources (hospital wastewater) and the river receiving effluents from WWTP in Örebro, Sweden. Results obtained for two selected strains—the same antibiotic susceptibility patterns, antibiotic resistance gene profiles (i.e., blaVIM-1, blaOXA-10, blaACC-1), MLST type, furthermore phylogenetic relationship based on core genome single nucleotide polymorphism (SNP) analysis, and core genome MLST—suggest the transfer of K. oxytoca-producing carbapenemases from the hospital setting to the aquatic environment, which may pose a threat to the community [164].
4.2.2. New Delhi Metallo-β-Lactamase (NDM) in WWTPs-Linked Samples
According to epidemiological data, NDMs seem to pose the greatest threat among class B β-lactamases. Genes encoding them were noted in many aquatic environments, including animal production wastewaters, industrial, domestic sewage, tap water, surface water, and groundwater. However, hospital wastewater is considered to be a major source of blaNDM variants [165,166,167]. As the geographical origin of NDM-producing bacteria is India, multiple publications detecting blaNDM, especially in hospital sewage, come from India [168,169,170], together with other Asian [108,171,172,173] and African countries [105,174]. Interesting results were reported by Marathe et al., who studied hospital wastewater from Mumbai, India. Shotgun metagenomics revealed the presence of β-lactamase genes encoding clinically important MBLs, such as NDM, VIM, and IMP with blaNDM as the most common carbapenemase-encoding gene. Additionally, 27 unique MBL genes not known yet were detected, which showed the huge potential of the metagenomic approach [175]. However, NDM-lactamases in Asian countries were not only detected in hospital sewage samples (Table 1). Analysis of rivers and sewage treatment plants in five Indian states also showed an abundance of blaNDM [77]. Similarly obtained data from southwest China showed a wide distribution of blaNDM in hospital sewage, WWTP effluent, and river samples. Interestingly, the gene was found in many different bacterial species belonging to Enterobacterales, genus Acinetobacter, and Pseudomonas [176]. The data from northern China [177,178] and Saudi Arabia [179] also confirm the presence of blaNDM in WWTP samples. blaNDM has spread globally, and several variants were noted not only in India and China but in many other countries in various water samples, including those from WWTPs and the surface waters of WWTP discharge points in the UK [128], Belgium [89], Switzerland [99], Germany [100], Poland [69,163], the Czech Republic [180], Romania [85,94], Spain [98], Africa [91,105], and the US [90,102,103]. Interesting results concern the Irish study conducted by Mahon et al. They examined the genetic relationship between NDM-possessing E. coli and K. pneumoniae (separately) cultivated from three locally linked sources: sewage samples from the collection system, freshwater streams, and clinical isolates. E. coli were considered indistinguishable, and K. pneumoniae were very closely related. These results confirm that water sewage plays an important role in the resistance transfer process [181]. Another analysis by Walsh et al. concerning public tap water and seepage water from sites around New Delhi also indicates that the environment has an undeniable influence on the propagation of NDM resistance [182].
Data regarding the wastewater treatment process show a different level of the transmission of bacteria with the NDM mechanism during the treatment process and the effectiveness of blaNDM reduction. In a Polish urban WWTP from Kozieglowy, Makowska et al. studied β-lactamase genes in the genomes of ESBL-producing and carbapenem-resistant coliforms isolated from each stage of the treatment process. They found that blaNDM and blaVIM were present in all stages and that the highest frequency was recorded in isolates from effluent compared to raw sewage, which indicates that the treatment process in the mechanical–biological treatment plant is insufficient in eliminating blaNDM and the organisms carrying them [69]. Similarly, data from two WWTPs in north China show the persistent and prevailing presence of blaNDM even after disinfection [177] and the propagation of blaNDM from a WWTP into its receiving river [178]. Other studies measuring absolute (copies/mL) and relative (copies/16S) abundance of blaNDM in influent and effluent also confirm deficient reduction [98,183]. However, Divyashree et al., who studied treated and untreated effluents from hospital samples in Mangalore, South India, showed the absence of blaNDM in treated effluents [184]. A Polish study also showed a complete reduction of blaNDM in the treatment process, even after the initial treatment stage [93], similar to a multi-center study from Denmark, Spain, and the UK [65].
4.3. Class C β-Lactamases
β-lactamases belonging to class C (AmpC) confer resistance to broad-spectrum β-lactams including penicillins, monobactams, and, most of all, cephalosporins (except fourth and fifth generations). Three mechanisms of resistance are noted: (i) chromosomal resistance induced by β–lactams; (ii) derepression due to mutations in AmpC regulatory genes, which results in overexpression and the production of the enzyme at a very high level; and (iii) the presence of plasmid-mediated AmpC genes (pAmpC) that are easily transmissible, even between different species, thus posing the highest health risk among class C β-lactamases. The first pAmpC variant was identified in 1989 from K. pneumoniae isolated in South Korea [185]. Several families of plasmid-encoded AmpC variants were reported within the next decade, i.e., ACC, CIT (variants CMY, LAT, BIL), DHA, EBC (variants ACT, MIR), FOX, and MOX, differing in bacterial species of origin. The most commonly found among the strains responsible for human infections are ACC, CMY, and DHA enzymes encoded by blaACC, blaCMY, and blaDHA genes, respectively. Clinically relevant bacteria producing pAmpC enzymes are mainly Citrobacter spp., Salmonella spp., and Shigella spp., but they were also found in other Enterobacterales, including K. pneumoniae, Enterobacter aerogenes, Proteus mirabilis, Morganella morganii, and K. oxytoca [44,47,186,187].
Class C β-Lactamases in WWTPs-Linked Samples
Similar to the clinical surveillance of pAmpC, environmental studies concerning wastewaters and WWTPs report the predominance of genes encoding CMY and DHA enzymes (Table 1). Kwak et al. conducted an antimicrobial resistance analysis of E. coli in urban and hospital wastewaters. They noticed that, among β-lactam-resistant ARB, almost all (97%) were confirmed to possess ESBL or pAmpC, and among pAmpC, all were detected as carrying the blaCMY-2 variant [116]. This variant, as well as others representing the CMY and DHA families, were detected in many other European studies of WWTPs from Germany [123,135,136], Romania [85], Sweden [96,109], Portugal [88,110], Poland [92,93], Slovakia [115], and Spain [188], as well as in studies conducted in Africa [127], North America [80,90,102,147,189], South America [87], and Asia [77,107]. Interestingly, Yim et al. investigated samples for plasmid-mediated quinolone resistance genes from a WWTP in Canada and detected the presence of qnrB4-AmpC (blaDHA-1) genes in plasmids among Citrobacter freundii isolates. These were almost identical to those found in pathogenic Klebsiella isolates. Results of SNP analysis may suggest their dissemination from WWTP strains into clinical strains, which supports that WWTPs are a source of AMR spread [189].
In the reviewed studies, AmpC genes were detected in different stages of the treatment process, as well as in surface waters related to WWTPs. Alexander et al. conducted research on 20 critical points in aquatic systems, including WWTPs, and showed that, although the abundance at individual points and sampling periods over 2 years was variable, the presence of the AmpC genes was found in all sampling sites [162]. In another study, Su et al. analyzed the AmpC genes in Escherichia coli from two municipal WWTPs in China and noted that AmpC was detected in all treatment stages [190]. In s multi-national study, Yang et al. used shotgun metagenomics on activated sludge samples of 15 WWTPs from China, Singapore, the US, and Canada and detected the highest abundance of AmpC genes among all tested β-lactam resistance genes. They also found very high genetic diversity of AmpC genes [82]. Generally, metagenomic studies or studies using high throughput PCR are very useful in detecting multiple variants of genes encoding AmpC and representing different families, including, i.e., FOX, MOX, MIR, ACT, and ACC [65,93,96,102,104,107,109,123,147].
Although blaCMY and blaDHA are the most often detected and prevalent pAmpC genes, in some studies, other variants are predominant. For example, Amador et al. showed that, among the AmpC-producing Enterobacterales isolated from Portuguese WWTP samples, the dominant was blaEBC, followed by blaFOX and blaCIT [88]; Piotrowska et al. analyzed Aeromonas spp. strains isolated from urban WWTPs in Warsaw, Poland, and found blaFOX to be the most abundant, followed by blaMOX and blaACC [97]. For comparison, Fadare and Okoh studied Enterobacterales isolated from the effluents of two WWTPs in South Africa and reported that the most predominant were blaCIT and blaACC, whereas blaFOX was detected in only one isolate [72].
Due to the lower frequency and speed of spread compared to other β-lactam resistance mechanisms, AmpC enzymes do not represent such a high risk. However, they are present in WWTP samples including effluents, and as a result of plasmid-localized and HGT present during the treatment process, this group may still pose a health risk and needs to be monitored.
4.4. Class D β-Lactamases
According to the BLDB, class D β-lactamases, known as oxacillinases, include more than 1,000 enzymes divided into 19 groups, among which the OXA group is the most numerous and clinically relevant. Among these, carbapenem-hydrolyzing class D enzymes (CHDLs) pose the greatest risk [47]. The substrate spectrum of the variants and level of hydrolyzing may significantly differ; however, all class D β-lactamases are not inhibited by β-lactam inhibitors, and they confer resistance to the amino-, carboxy-, and ureidopenicillins [191]. Although not classical ESBLs, as defined by inhibition by clavulanate, several of the OXA-type β-lactamase variants, such as OXA-11 and OXA-14 to OXA-20, are associated with an ESBL phenotype in that they confer resistance to some of the late-generation cephalosporins [192]. Within the OXA family, only a small fraction has a functional role as a carbapenemase. Among these are OXA-23, OXA-40, and the increasingly prevalent OXA-48, with its related variants, OXA-162, OXA-181, and OXA-232 [193]. The major enterobacterial class D carbapenemase, OXA-48, was first reported in a Turkish K. pneumoniae isolate in 2001 [194]. Thereafter, OXA-48 and related variants have been found in almost all Enterobacterales, mainly in K. pneumoniae and E. coli, that spread globally, causing endemic states in the Middle East, North Africa, India, and some European countries [62,63,64].
4.4.1. OXA Family β-Lactamases Carried in ARB
The reviewed approaches concerning class D β-lactamases are focused on bacterial strains carrying blaOXA isolated from WWTP samples (Table 1). The majority of these studies confirm a blaOXA presence in isolates from both untreated and treated samples, and the prevalent variants are blaOXA-1 and blaOXA-48. Multiple examples come from European countries: a Czech study reported ESBL-producing Enterobacterales carrying blaOXA-1 and isolated from effluent; globally spread MDR clones of E. coli ST131 and K. pneumoniae ST321 and ST323 harboring large FIIK plasmids with multiple antibiotic-resistance genes were found among tested strains [113]; a Spanish study detected blaOXA-1 in strains isolated from effluents of two out of 21 tested WWTPs [76]; two German studies reported the presence of blaOXA-51 and blaOXA-48 in carbapenemase-producing bacteria [100] and blaOXA-58, blaOXA-48 and blaOXA-23 in bacterial strains isolated from influent, activated sludge and effluent [123]; four Polish studies identified blaOXA genes among ceftazidime- or meropenem-resistant bacterial strains [92], Aeromonas spp. strains isolated from raw sewage, activated sludge, and effluent [97], ESBL-producing Enterobacterales [68] and Acinetobacter spp. isolates [163]; an Austrian study of carbapenemase-producing Enterobacterales from activated sludge confirmed harboring blaOXA-48 [95]; and a study concerning the WWTP in Basel, Switzerland, where carbapenemase-resistant Enterobacterales and other Gram-negative bacteria isolated from municipal and hospital wastewater and WWTP receiving this sewage were compared, and identical isolates from the WWTP and wastewater samples were detected, including OXA-48-producing E. coli ST38 and Citrobacter spp. [99]. Similarly, a molecular epidemiology approach was conducted in a Romanian study. Surleac et al. detected variants of blaOXA in K. pneumoniae isolated from samples of three WWTPs [85], while Teban-Man et al. compared carbapenemase-producing K. pneumoniae isolated from the influent and effluent of two WWTPs with and without hospital input and found that blaOXA-48 was carried by strains isolated from raw and treated samples of WWTPs collecting hospital wastewater. In the second WWTP, the gene was observed only in strains from influent. Moreover, isolates harboring blaOXA-48 were genetically typed, which showed they belonged to sequence types of high-risk clones (ST258, ST101, ST147, ST2502). These clones were associated with clinical settings and reported to be multi-drug resistant [94]. In a study of a Swedish WWTP, Gram-negative bacteria harboring blaOXA were noted in influent, effluent, and recipient waters of the river and lake [109]. However, in a Portuguese study conducted by Araujo et al., blaOXA was detected only in strains isolated from raw sewage samples [110]. Another Portuguese investigation of ampicillin-resistant Enterobacterales isolated from influent and effluent showed different results; blaOXA was the most prevalent gene among tested ESBL-producing strains [88]. There are significantly fewer studies detecting blaOXA in the African region and they cover Algeria, where blaOXA-1 was detected [74]; Durban, South Africa, where cefotaxime-resistant E. coli were studied and blaOXA-1 was found as well [91]; Eastern Cape Province, South Africa, where blaOXA-1-like and blaOXA-48-like variants harbored by Enterobacterales isolated from effluents of WWTPs were noted [72]; and Tunisia, where C. freundii isolate carrying blaOXA-204 [121] and Enterobacterales strains possessing blaOXA-1 [127] were detected. American studies concerning WWTPs also confirm blaOXA presence in bacteria isolated from WWTP samples [80,90,102,104,119,125,133].
4.4.2. OXA Family β-Lactamases in Direct WWTP Samples—Occurrence and Removal
Multiple studies report the presence of blaOXA in direct WWTP samples and determine the concentration and relative abundance of selected gene variants to define the efficiency of the treatment process (Table 1). Comparable to previously discussed β-lactamases, bacteria producing OXA enzymes, as well as blaOXA, can be detected after the treatment process. For example, the study of two WWTPs in the Brussels region determined the relative abundance of blaOXA-48 in different stages of the treatment, as well as in samples of the river as the discharge point for the WWTP effluents. In that study, Proia et al. showed a significant increase of blaOXA-48 from influent to effluent and from upstream to downstream river samples [89]. Similarly, in Kozieglowy, a Polish WWTP, it was reported that the wastewater treatment process leads to a significant increase in the relative abundance of blaOXA-48 genes in the effluent [69], whereas in research from the Baltic Sea area, the relative abundance of blaOXA-58 was decreased in the effluent; however it was weakly significant and found only in one of the three studied WWTPs [141]. In the German study, the absolute abundance of selected blaOXA genes was determined, and when comparing raw and treated samples from WWTP, a significant decrease was reported regarding blaOXA-58 and blaOXA-48 but not blaOXA-23 [123]. Similar results were obtained in a multi-national study of WWTPs from ten European countries, where qPCR and absolute abundance were performed for selected blaOXA genes. It was noticeable that, among all tested β-lactamase genes, blaOXA-58 was found in all tested samples, had the highest absolute abundance, and was significantly reduced during treatment [101]. In three Swedish municipal sludge treatment plants, a metagenomics approach was conducted, and many variants of blaOXA were detected at all stages of the treatment process. Some of them, like blaOXA-48, were consistently enriched in treated sludge compared to primary sludge [96]. Other metagenomic approaches or using qPCR provide similar results—the presence of multiple blaOXA gene variants, including effluent samples [93,109,136,147], while others detected only single or a few variants [65,69,89,101,107,108,111,141,163]. Interestingly, in a Polish study, where blaOXA was detected as one of the prevalent tested genes in influent and effluent samples, comparative metagenomic analysis of DNA from WWTP samples and employees’ swabs revealed the presence of similar ARGs in both types of samples with significantly higher concentrations than in control samples [15]. Other studies that report the presence of blaOXA genes at different stages of the treatment process include the research of Yang et al., wherein activated sewage sludge from 15 WWTPs was tested, and three variants (blaOXA-1, blaOXA-2 and blaOXA-10) were detected [82], while in WWTP active sludge in South Carolina, in the US, a higher variability among blaOXA (seven variants) was noted [147]. Interesting results concerning the seasonal increase of blaOXA concentration between the summer and winter seasons were reported in the study of four small-scale domestic WWTPs. Furthermore, blaOXA in winter was prevalent among tested ARGs in raw sewage, as well as in effluent samples; additionally, the gene was detected in receiving river samples, in both the winter and summer seasons [84]. Results of a multi-national study, analyzing samples from Denmark, Spain, and the UK, indicated a country-specific presence for blaOXA-10 detected only in WWTPs from the UK [65].
The above data, showing the presence of blaOXA genes and bacteria harboring them in WWTPs and related samples, confirms that WWTPs are a hotspot for antibiotic-resistant gene transmission into not only the aquatic compartments of the environment but also to the atmospheric air, creating an additional health risk for the workers of WWTPs.

Table 1.
ARGs encoding class A, B, C, and D β-lactamases detected in WWTPs-linked samples.
Table 1.
ARGs encoding class A, B, C, and D β-lactamases detected in WWTPs-linked samples.
| Location | Gene Variant(s) Detected in WWTP Samples 1 | Sample Source(s) 2 | Type of Tested Samples 3 | Type of Methods 4 | Ref. | |||
|---|---|---|---|---|---|---|---|---|
| Class A | Class B | Class C | Class D | |||||
| Austria, Graz/Styria | CTX-M-15, -24 KPC-2 SHV-1, -26 TEM-1 | nd | FOX | OXA-48 | WWTP collecting DW and HW | CPE from SS | MM PCR seq-DNA | [95] |
| CTX-M-1, -3, -14, -15, -38 PER-1 SHV-1, -2, -11, -12 TEM-1 | nd | nd | nd | ESBL-producing Enterobacterales from SS | [81] | |||
| Austria | CTX-M TEM | nd | nd | nd | 5 WWTPs | ESBL-producing E. coli from SS | MM PCR | [112] |
| Belgium, Brussels Capital Region | CTX-M KPC TEM | NDM-1 | nd | OXA-48 | 2 WWTPs collecting HW | samples of IN, EF, RR and HW | qPCR | [89] |
| Czech Republic, Brno | CTX-M -1, 14b, -15 TEM-1 | nd | nd | OXA-1 | WWTP collecting HW | ESBL-producing Enterobacterales from EF | MM PCR seq-DNA molecular typing (MLST, PFGE) | [113] |
| Czech Republic, Moravian-Silesian Region | nd | NDM-1 | nd | nd | 2 WWTPs | samples from the nitrification and sedimentation tanks and bacteria isolated from them | MM qPCR WGS | [180] |
| France | CTX-M-1, -14, -15, -27 SHV-12 | nd | nd | nd | WWTP collecting HW, rainwater | E. coli from IN, AS and EF | MM PCR seq-DNA molecular typing (MLST, PFGE) | [75] |
| Germany, Bielefeld-Heepen | CTX-M-4, -27, -32 GES-3 PER-2 SHV-34 TEM-1 TLA-2 VEB-1 | IMP-2, -5, -9, -11, -13 VIM-4 | AmpC CMY-5, -9, -10, -13 | NPS-1, -2 OXA -1, -2, -5, -9, -10, -12, -18, -20, -22, -27, -29, -40, -45,-46, -48, -50, -54, -55, -58, -60, -61, -75 | WWTP | strains from SS and EF, resistant to selected antibiotics | MM PCR seq-DNA | [136] |
| Germany, District of Kleve | CTX-M-1, -9 GES | VIM | ACT CMY-2 DHA FOX MIR | OXA-23, -48, -58 | WWTP collecting DW, HW and IW | samples of IN, SS and EF, imipenem-, cefotaxime- or colistin-resistant strains | MM PCR, qPCR seq-DNA | [123] |
| Germany, North-Rhine Westphalia | IMI KPC | GIM VIM NDM | nd | OXA-48, -51 | WWTPs collecting HW | ESBL-producing bacteria and CPE from HW, IN, EF, RR and rural wastewater | MM PCR molecular typing | [100] |
| Germany, South Region | nd | VIM-1 | AmpC | nd | 4 WWTPs collecting HW | samples of IN, EF and HW, receiving surface waters, groundwater and rain overflow | qPCR | [162] |
| Germany | CTX-M TEM | nd | CMY-2 | nd | 7 WWTPs with various inflow | IN and EF samples |
qPCR seq-DNA | [135] |
| Ireland | CTX-M-1, -15 SHV-12 TEM-1l-like, -12, -116 | nd | nd | nd | 2 WWTPs | coliform strains from EF | MM PCR seq-DNA | [86] |
| Italy, The Oltrepò Pavese Plain | CTX-M-1, -14, -15, -28, -138 KPC-2 SHV-5 TEM-1 | nd | nd | nd | 4 WWTPs | cefotaxime-resistant Enterobacterales from WWTP, RR and groundwaters | MM PCR seq-DNA molecular typing (MLST, PFGE) | [78] |
| Poland, Kozieglowy | CTX-M KPC SHV TEM | NDM VIM | nd | OXA-1, -48 | WWTP | samples of IN and EF, ESBL-producing and carbapenems-resistant coliforms | MM PCR qPCR seq-DNA | [69] |
| Poland, Olsztyn | CTX-M-1, -3, -9, -15 SHV-5 TEM-1, -47, -49 | nd | nd | OXA-1 | WWTP collecting HW | samples of IN, SS, EF, RR and the air near WWTP Enterobacterales from the samples | MM PCR seq-DNA | [68] |
| SHV TEM | nd | nd | OXA | samples of IN, SS, EF, RR, nasal and throat employees’ swabs | metagenomics qPCR | [15] | ||
| nd | IMP-1 VIM-2 NDM | nd | OXA-23, -24, -51, -58 | Acinetobacter spp. from IN, SS, EF and RR | metagenomics qPCR | [163] | ||
| Poland, Warsaw | CTX-M-15, 27/98 GES-7 KPC PER-1/5, -3, -4 SHV-11, -12 TEM VEB | nd | ACC FOX-1, -2-like, -3, -4-like, -9, -10, -10-like, -13-like MOX-10/11, -4/8 | OXA | WWTP collecting DW, MW and HW | Aeromonas spp. from IN, SS and EF | MM PCR seq-DNA | [97] |
| CTX-M-1-like, -3-like, -15-like, -27-like GES KPC-2-like ORN PER-1/5 SHV-11-like, -12-like TEM-1-like, -12, -30, -47/68, -116 | VIM-1-like, -2-like | ACT CMY-2-like, -4, -39, -40, -42/146/145, -65/75/89/113, -139, -157 FOX-15 MOX-13 | OXA | ceftazidime- or meropenem- resistant Gram-negative bacteria from IN, SS and EF | [92] | |||
| Poland, Warmia and Mazury District | SHV TEM | nd | nd | OXA | 4 domestic WWTPs | samples of IN, EF, and RR | qPCR | [84] |
| Poland, Warmia and Mazury District/Silesia District | Multiple variants i.e.,: AER BEL CARB Cfx GES TEM VEB | Multiple variants i.e., IMP VIM NDM | Multiple variants i.e.,: CMY FOX | Multiple variants i.e.,: LCR NPS OXA-23, -24, -48, -58 | 2 WWTPs collecting HW | samples of IN, SS and EF | metagenomics | [93] |
| Portugal, Coimbra | CTX-M -1, -9 TEM | nd | ACT MIR-1 FOX-1, -5 DHA-1, -2 CMY-2, -17, -12, -18, -21, -23 LAT-1, -3 BIL-1 | OXA | WWTP collecting MW, HW and IW | Enterobacterales resistant to ampicillin from IN, EF, HW and RR | MM PCR | [88] |
| Northern Portugal | BEL-1 GES-5 TEM-1b | nd | CMY-101 DHA-1 | OXA-1 | WWTP collecting DW and HW | Gram-negative bacteria resistant to meropenem from IN, SS and EF | MM PCR qPCR seq-DNA molecular typing (rep-PCR, PFGE phylogrouping) WGS | [110] |
| Northern Portugal | CTX-M-1, -14, -15, -27, -32 SHV-1, -27 TEM-1 | nd | nd | nd | WWTP | ESBL-producing and cefotaxime-resistant Enterobacterales from different stages of treatment | MM PCR seq-DNA | [118] |
| Romania, Cluj County | KPC-2 | NDM-1, -6 VIM-2 | nd | OXA-48 | 2 WWTPs, with and without hospital contribution | carbapenemase-producing K. pneumoniae from IN and EF | MM PCR molecular typing (MLST, phylogrouping) | [94] |
| Romania, Bucharest/Galati/Taˆrgovişte | CTX-M-15 KPC-2 SHV-1, -11, -12, -33, -100, -101, -106, -107, -145, -158, -187 TEM-1, -150 | NDM-1 | CMY-4 DHA-1 | OXA-1, -9, -10, -48, -162 | 3 WWTPs HW | ESBL- and carbapenemase- producing K. pneumoniae from IN and EF | MM WGS | [85] |
| Slovakia, Kosice | CTX-M-1, -2 | IMP | CMY-2 | OXA-1 | WWTP | ESBL-producing E. coli from IN and EF | MM PCR molecular typing | [115] |
| Spain, Catalonia | KPC | NDM | nd | nd | WWTP | samples of IN, EF, hospital EF, RR, sediment and biofilm | qPCR | [98] |
| Spain, Girona | TEM | nd | nd | nd | WWTP | samples of IN, EF, HW and RR | qPCR | [140] |
| Spain, Navarra | CTX-M-1, -14, -15, -55 SHV-12 TEM-1, 42, -145 | nd | nd | nd | 21 WWTPs | cefpodoxime-resistant Enterobacterales from EF | MM PCR seq-DNA | [76] |
| nd | nd | ACC DHA EBC | nd | WWTPs | β-lactam-resistant bacteria from IN, EF and RR | MM PCR seq-DNA | [188] | |
| Sweden, Stokholm/Uppsala/Lidingö | CTX-M GES KPC PER SHV SME TEM VEB | CAR IMP IND VIM | ACC ACT CFE CMY-1, -2 DHA FOX MIR MOX | OXA-1, -2, -10, 20, -23, -24, -48, -50, -51, -58, -60, -63 | 3 WWTPs collecting MW, HW, IW and storm water | samples of IN, SS and EF | metagenomics | [96] |
| Sweden, Örebro | CTX-M-1, -9 GES PER-1 SFO-1 SHV VEB | IMP-5, -12 | ACC-1, -3 ACT-1, -5/7 CFE-1 CMY-10 DHA FOX LAT MIR MOX | OXA -2, -10, -50, -51, -58 | WWTP collecting DW, HW and IW | Gram-negative bacteria from IN, EF, HW, RR and lake water | MM qPCR | [109] |
| Sweden, Stockholm | CTX-M-1, -9 | nd | CMY-2 | nd | WWTP | E. coli from IN, EF and HW | MM PCR | [116] |
| Switzerland, Basel | KPC-2 | NDM-1, -5 VIM-1 | nd | OXA-48, -181 | WWTP collecting MW and HW | CPE and Gram- negative bacteria from IN, EF, HW and RR | MM PCR seq-DNA molecular typing (MLST, phylogrouping) | [99] |
| The UK | CTX-M-15 LEN-25-like OXY-6 SHV-12 TEM-1 | IMP-1 NDM-1-like, -5 | nd | OXA-1, -17, -48, -181 | 20 WWTPs | carbapenem-resistant Gram-negative strains isolated from treated and untreated samples | MM WGS | [128] |
| CTX-M-1, -14, -15, -27 | nd | nd | nd | ESBL-producing E. coli from treated and untreated samples | MM metagenomic | [120] | ||
| Canada, Arnpior/Ottawa/Toronto | CARB CTX-M GES KPC OXY PER SHV TEM | cphA IMP VIM PAM | ACT CepH FOX, MOX | OXA-2, -10 | 3 WWTPs | carbapenem-resistant strains from IN | MM PCR, WGS | [104] |
| Canada, Alberta/Calgary | CTX-M-15 | nd | AmpC | OXA-1 | 13 WWTPs | multidrug-resistant E. coli from IN and EF | MM PCR WGS molecular typing | [133] |
| Canada, Baffin Island (Pond Inlet/Clyde river/Iqaluit) | CTX-M TEM | nd | nd | nd | 3 WWTPs | IN and EF samples | qPCR | [139] |
| Guadeloupe/North America | CTX-M-1, -8, -14, -15, -27 TEM-1-like, -3 VEB-1 | nd | CMY-2, -8 | OXA-1-like | 2 WWTPs | Enterobacterales from IN, EF, RR and sea waters, with a focus on ESBL- and AmpC-producers | MM PCR seq-DNA phylogrouping | [80] |
| The US, Colorado | CTX-M TEM | nd | nd | OXA-1 | WWTP | ESBL- and KPC-producing E. coli from IN and EF | MM PCR seq-DNA WGS | [119] |
| The US, South Carolina | BES-1 CTX-M-1 GES KPC SHV TLA-1 VEB | ccrA IMP-5, -12 | ACT-1 CMY-10 FOX LAT MIR MOX | OXA-2, -10, -23, -24, -51, -58, -60 | WWTP | samples of SS and bioaerosol collected downwind from sludge aeration tanks and upwind from WWTP | MM qPCR seq-DNA | [147] |
| The US, Washington | CTX-M KPC TEM | NDM-1 | CMY-2 | OXA-48 | 2 WWTPs | samples of IN, SS, EF, RR and irrigation water | PCR | [90] |
| The US, Wisconsin | CTX-M-1 and -9 group TEM | nd | nd | OXA | WWTP | cefotaxime-resistant E. coli from IN, EF and HW | MM PCR molecular typing WGS | [125] |
| The US (New Jersey, Maryland, Ohio, Texas, Colorado, California) | CTX-M GES KPC TEM | VIM NDM | nd | OXA | 7 WWTPs with various inflow | E. coli from EF | MM PCR seq-DNA molecular typing (phylogrouping, sequence typing) | [103] |
| The US | CARB-2 CTX-M-15 GES-5 KPC-2, -3 OKP-B-2, -7 ORN-1b OXY-1, -5 PLA-2 SHV-11, -12 TEM-1, -1a, -1b | VIM-1 NDM-1, -5, -7 | AmpC ACT-1 CMY-66, -79 FOX-5 MIR-3, -6, -9, -15 | OXA-1, -2, -9, -105 | 50 WWTPs | carbapenemase-producing bacteria from EF and surface water of WWTP discharge | MM WGS | [102] |
| Colombia, Antioquia | CTX-M-1, -2, -8/25, -9 SHV TEM | nd | LAT/BIL/CMY group ACT/MIR group DHA | nd | WWTP collecting DW, HW and IW | β-lactam-resistant Gram negative bacilli from IN, SS and EF, with focus on E. coli | MM PCR seq-DNA molecular typing (PFGE, MLST) | [87] |
| Brazil, Curitiba | CTX-M-1, -2, -8, -9, -15 SHV-12 GES-5 | nd | nd | nd | WWTP | cefotaxime-resistant Gram-negative bacteria from IN, SS, EF, hospital, sanitary effluent and RR | MM PCR seq-DNA | [79] |
| Brazil, São Paulo | CTX-M-8, -15 SHV-28 | nd | nd | nd | 5 WWTPs | ampicillin-resistant Enterobacterales from IN | MM PCR seq-DNA molecular typing (phylogrouping E. coli, MLST) | [117] |
| India, Jasola Vihar, New Delhi | CTX-M-15, -152, -205 SHV TEM-1 | nd | nd | nd | WWTP | ESBL-producing bacteria from EF, lentic water bodies and slaughterhouse | MM PCR seq-DNA | [71] |
| India, New Dehli | nd | NDM-1 | nd | nd | 12 WWTPs | coliforms bacteria from EF | MM PCR seq-DNA | [183] |
| India, Jaipur | Multiple variants i.e.,: Cfx-A2, -A3 GES-15 VEB-1 | nd | nd | NPS-1 OXA-209 | 4 WWTPs collecting HW | samples of IN, SS and EF | metagenomics | [111] |
| India, State of Bihar, Goa, Karnataka, Tamil Nadu and Telangana | CTX-M-15, -55 SHV-12 TEM-1, -1b | NDM-1, -5, -7 | CMY-2, -6, -42 | OXA-1, -9, -10 | 5 WWTPs | ESBL- and carbapenem- producing E. coli from WWTPs and rivers | MM PCR seq-DNA, molecular typing WGS | [77] |
| Singapore | cfxA6 TEM VEB-1a | nd | AmpC | OXA-198, -333, -347 | WWTP | samples of IN, EF, HW and surface waters | metagenomics | [108] |
| Singapore | AER-1 CARB-3, -(5-9), -12 Cfx-A2, -A3 CTX-M-1, -15, -19, -34, -147 KPC-1, -10, -11, -13, -16 LEN-19, -21 OKP-A, -B PER-1, -3, -4, -7 PSE-1, -4 ROB-1 SHV-4, -12, -39, -51, -53, -167 VEB-(2–8) multiple variants of GES and TEM groups | GOB-1 IMP-31 LRA | ACT-2, -3, -16, -19, -20 DHA-6, -5, -6, -7 FOX-2, -4, -5, -7, -8, -9 MIR-1, -2, -6, -8 MOX-(1–7) PDC-2, -5 multiple variants of CMY group | LCR OXA-278 | WWTP | samples of IN, SS and EF | metagenomics | [107] |
| China, Guangdong Province | nd | nd | AmpC | nd | 2 WWTPs | E. coli from WWTPs | MM PCR | [190] |
| China, Harbin | CTX-M | nd | nd | nd | 4 WWTPs | samples of IN, SS and EF | PCR qPCR | [137] |
| China, Tianjin | KPC-2 GES-1 | nd | nd | nd | WWTP collecting DW and IW | EF samples | qPCR | [106] |
| nd | NDM-1 | nd | nd | EF and RR samples | qPCR | [178] | ||
| China, Wuxi | CTX-M SHV TEM | nd | nd | nd | 3 WWTPs collecting DW and IW | IN and EF samples, cultivable heterotrophic bacteria and total coliforms | MM qPCR seq-DNA | [129] |
| China | CTX-M TEM | VIM | nd | nd | 3 WWTPs | multiple antibiotic-resistant Escherichia spp. from WWTPs, HW and livestock manure | MM PCR seq-DNA | [70] |
| China | nd | NDM-1 | nd | nd | 2 WWTPs collecting DW and IW | samples of IN, SS and EF | MM PCR seq-DNA | [177] |
| Japan, Tokyo | CTX-M-1 group, -2 group and -9 group SHV group TEM group | nd | nd | nd | WWTP | fecal coliforms from different stages of treatment process | MM PCR seq-DNA | [124] |
| Japan | CTX-M-1, -2, -3, -8, -14, -15, -27, -55, -64, -65, -123, -174 | nd | nd | nd | 4 WWTPs | cefotaxime-resistant E. coli from IN | MM PCR seq-DNA molecular typing (MLST, phylogrouping) WGS | [122] |
| United Arab Emirates, Dubai | SHV TEM | nd | nd | nd | WWTP | ESBL-producing Enterobacterales from SS | MM PCR | [83] |
| Saudi Arabia, Jeddach | nd | NDM-1 | nd | nd | WWTP | ESBL- and carbapenemase- producing bacteria from IN | MM qPCR WGS | [179] |
| South Africa, Durban | CTX-M TEM | nd | nd | nd | WWTP collecting DW, HW and IW | ESBL-producing E. coli from IN, SS, EF and RR | MM PCR | [130] |
| CTX-M KPC-2 TEM | NDM-1 | nd | OXA-1 | coliforms bacteria from IN and EF focused on E. coli | MM PCR | [91] | ||
| South Africa, Mgungundlovu District | CTX-M-3, -15, -28 SHV-28 TEM-1, -116, -181, -213, -215 | nd | nd | nd | 4 WWTPs collecting DW, HW and IW | ESBL-producing E. coli from IN and EF | MM PCR seq-DNA | [73] |
| South Africa, Eastern Cape Province, Amathole District | PSE-1 TEM | nd | nd | nd | 2 WWTPs | Aeromonas spp. from WWTPs | MM PCR | [126] |
| South Africa, Eastern Cape Province, Amathole and Chris Hani District | CTX-M-1, -2, -9 GES KPC PER SHV TEM | nd | ACC CIT DHA EBC MOX | OXA-1-like, -48-like | 2 WWTPs collecting DW, IW, run-off waters and residential sewage | Enterobacterales from EF | MM PCR | [72] |
| South Africa, Eastern Cape Province, Amathole, Chris Hani and Sarah Baartman District | KPC | NDM-1 | nd | nd | 4 WWTPs | Enterobacterales from EF, HW and surface waters, with focus on Klebsiella spp. | MM PCR | [105] |
| Algeria, Boumerdes | CTX-M-3, -15 TEM-1 | nd | nd | OXA-1 | WWTP collecting DW, HW and IW | cefotaxime-resistant strains from IN and EF | MM PCR seq-DNA, molecular typing (MLST, phylogrouping) | [74] |
| Tunisia | CTX-M-1, 14a, -15 TEM-1a, -1b | nd | CMY-2 | OXA-1 | 8 WWTPs | cefotaxime-resistant Enterobacterales from IN, EF, MW, effluents of MW and IW, RR and surface waters not connected to WWTP | MM PCR seq-DNA molecular typing of E.coli (MLST, phylogrouping, PFGE, virulence genotyping) | [127] |
| Tunisia | CTX-M-1, -3, -14, -15, -27 | nd | nd | OXA-204 | 2 WWTPs | ESBL-producing Enterobacterales from WWTP and various animal samples | MM PCR seq-DNA, molecular typing (MLST, phylogrouping, PFGE) | [121] |
| Tunisia, Monastir Governorate | CTX-M TEM | nd | nd | nd | 5 WWTPs collecting DW, HW and IW | IN and EF samples | qPCR | [142] |
| Australia, Queensland | CTX-M TEM | nd | nd | nd | 2 WWTPs | ESBL-producing E. coli from IN and HW | MM PCR molecular typing | [67] |
| Multinational study: Denmark, Spain, the UK | cfxA BEL CARB CTX-M-1, -3, -15 GES KPC LEN OXY-1, -2 SFO SHV-11 SPM TEM TLA VEB | IMP VIM NDM | AmpC ACC CMY DHA FOX MIR | OXA-10 | 3 WWTPs collecting DW and HW | samples of IN, SS, EF and RR | qPCR seq-DNA | [65] |
| Multinational study: Finland, Estonia | CTX-M-32 SHV-34 | nd | nd | OXA-58 | 3 WWTPs | IN and EF samples | qPCR | [141] |
| Multinational study: France, Italy, Norway, Portugal, Germany, Netherlands, Cyprus, Turkey, Austria and the UK | CTX-M-15, -32 KPC-3 TEM | nd | nd | OXA-48, -58 | 16 WWTPs | samples of EF and corresponding receiving water bodies | qPCR | [101] |
| Multinational study: China, Singapore, the US, Canada | TEM-1 | IMP | AmpC | OXA-1, -2, -10 | 15 WWTPs | SS samples | PCR qPCR | [82] |
1 nd—no data. 2 WWTP—wastewater treatment plant, DW—domestic wastewater, MW—municipal wastewater, HW—hospital wastewater, IW— industrial wastewater. 3 CPE—carbapenem-resistant Enterobacterales, IN—influent, EF—effluent, SS—sewage sludge, RR—receiving river waters. 4 MM—microbiological methods, PCR—specific PCR, qPCR—quantitative PCR, seq-DNA—sequencing DNA, WGS—whole genome sequencing, MLST—multilocus sequence typing, PFGE—pulsed-field gel electrophoresis.
5. Conclusions
AMR is a serious and urgent problem, and it is clear that the environment plays a key role in the process of transmission and propagation of ARGs and ARB with life-threatening clinical consequences. The multitude of publications confirms that β-lactamases genes encoding especially ESBLs (TEM, SHV, CTX-M) and KPC, NDM, and OXA carbapenemases, which pose one of the greatest health risks, are widely found in WWTPs and disseminated to further portions of the environment. Molecular analysis shows repeatedly high genetic relatedness between environmental and clinical isolates, e.g., ST131 E. coli. Generally, different kinds of sewage treatment processes do not eliminate these ARGs completely. Furthermore, some data indicate an increased level of β-lactam ARGs in effluent or even the presence of the genes and bacteria harboring them in samples after additional disinfection treatments.
Due to β-lactam ARGs’ potential to transfer via mobile genetic elements through horizontal gene transfer, their abundance in water samples discharged from WWTPs into natural aquatic sources used by humans or animals suggests a potential risk of transmission resistance determinants into pathogenic and non-pathogenic bacteria and acquiring multidrug resistance as well as the participation of WWTPs in AMR transmission route and distribution into surrounding ecosystems and clinical settings. The growing problem of AMR and the spread of clinically relevant ARGs related to, i.e., β-lactams in the environment, indicate the need to improve and evaluate the procedures of wastewater treatment and disinfection; thus, ARB, ARGs, and factors influencing their selection and co-selection during the treatment process would be completely removed.
The development and improvement of techniques used in testing wastewater for antibiotic resistance has been very significant in recent years. There are more and more publications indicating the use of modern metagenomic assays, which enables broadening the knowledge of the complexity and structural and functional biodiversity of microbial communities—i.e., analysis of resistance genes; taxonomic assignment; functional genes characterization; the identification of the HGT mechanism and mobile elements involved in the gene transmission; and exploring relationships between pathogenic and non-pathogenic species and susceptible and resistant bacteria. Therefore metagenomic analysis seems to be a very useful tool to understand the process of AMR transmission. However, the clinical surveillance of resistant strains responsible for life-threatening infections and nosocomial outbreaks caused by β-lactam-resistant strains also involve molecular techniques, but still the gold standard are culture-based methods detecting the expression of genes and the resistance mechanism. Therefore, according to the One Health’s concept, collaborative approaches concerning AMR in the environment and clinical setting are indispensable and should combine new technology with standard microbiological methods. As WWTPs are the crucial points on the routes of ARB and ARGs’ spread, they should be deeply explored, which would help to understand the process and make it possible to introduce procedures to stop, or at least slow down, the spreading of antibiotic resistance.
Author Contributions
Conceptualization, I.W., writing—original draft preparation, I.W., preparing table and figures, I.W., A.K., E.K. and A.B.; writing—review and editing, I.W., A.K., E.K. and A.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financed by internal funding (DS 8/2022) from the National Medicines Institute.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO. Ten Threats to Global Health in 2019; World Health Organization: Geneva, Switzerland, 2019; pp. 1–18. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (accessed on 10 May 2022).
- CDC. Antibiotic Resistance Threats in the United States 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 1 April 2022).
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Abushaheen, M.A.; Muzaheed; Fatani, A.J.; Alosaimi, M.; Mansy, W.; George, M.; Acharya, S.; Rathod, S.; Divakar, D.D.; Jhugroo, C.; et al. Antimicrobial resistance, mechanisms and its clinical significance. Disease-a-Month 2020, 66, 100971. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Rasool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell. Infect. Microbiol. 2021, 11, 1–20. [Google Scholar] [CrossRef]
- Alexander, J.; Hembach, N.; Schwartz, T. Evaluation of antibiotic resistance dissemination by wastewater treatment plant effluents with different catchment areas in Germany. Sci. Rep. 2020, 10, 8952. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Harnisz, M.; Abreu-Silva, J.; Rolbiecki, D.; Korzeniewska, E.; Luczkiewicz, A.; Manaia, C.M.; Plaza, G. Antibiotic resistance in wastewater, does the context matter? Poland and Portugal as a case study. Crit. Rev. Environ. Sci. Technol. 2022, 52, 4194–4216. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 964. [Google Scholar] [CrossRef]
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—A review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef]
- Wang, J.; Chu, L.; Wojnárovits, L.; Takács, E. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: An overview. Sci. Total Environ. 2020, 744, 140997. [Google Scholar] [CrossRef]
- Felis, E.; Kalka, J.; Sochacki, A.; Kowalska, K.; Bajkacz, S.; Harnisz, M.; Korzeniewska, E. Antimicrobial pharmaceuticals in the aquatic environment—Occurrence and environmental implications. Eur. J. Pharm. 2020, 866, 172813. [Google Scholar] [CrossRef]
- Uluseker, C.; Kaster, K.M.; Thorsen, K.; Basiry, D.; Shobana, S.; Jain, M.; Kumar, G.; Kommedal, R.; Pala-Ozkok, I. A Review on Occurrence and Spread of Antibiotic Resistance in Wastewaters and in Wastewater Treatment Plants: Mechanisms and Perspectives. Front. Microbiol. 2021, 12, 717809. [Google Scholar] [CrossRef]
- Zieliński, W.; Korzeniewska, E.; Harnisz, M.; Drzymała, J.; Felis, E.; Bajkacz, S. Wastewater treatment plants as a reservoir of integrase and antibiotic resistance genes—An epidemiological threat to workers and environment. Environ. Int. 2021, 156, 106641. [Google Scholar] [CrossRef]
- Miłobedzka, A.; Ferreira, C.; Vaz-Moreira, I.; Calderón-Franco, D.; Gorecki, A.; Purkrtova, S.; Bartacek, J.; Dziewit, L.; Singleton, C.M.; Nielsen, P.H.; et al. Monitoring antibiotic resistance genes in wastewater environments: The challenges of filling a gap in the One-Health cycle. J. Hazard. Mater. 2022, 424, 127407. [Google Scholar] [CrossRef]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef]
- von Wintersdorff, C.J.H.; Penders, J.; van Niekerk, J.M.; Mills, N.D.; Majumder, S.; van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef]
- Czekalski, N.; Gascón Díez, E.; Bürgmann, H. Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J. 2014, 8, 1381–1390. [Google Scholar] [CrossRef]
- Kotlarska, E.; Łuczkiewicz, A.; Pisowacka, M.; Burzyński, A. Antibiotic resistance and prevalence of class 1 and 2 integrons in Escherichia coli isolated from two wastewater treatment plants, and their receiving waters (Gulf of Gdansk, Baltic Sea, Poland). Environ. Sci. Pollut. Res. 2015, 22, 2018–2030. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Chaurasia, D.; Pandey, A.; Gupta, P. Co-occurrence of multidrug resistance, β-lactamase and plasmid mediated AmpC genes in bacteria isolated from river Ganga, northern India. Environ. Pollut. 2020, 267, 115502. [Google Scholar] [CrossRef]
- Griffin, D.W.; Banks, K.; Gregg, K.; Shedler, S.; Walker, B.K. Antibiotic Resistance in Marine Microbial Communities Proximal to a Florida Sewage Outfall System. Antibiotics 2020, 9, 118. [Google Scholar] [CrossRef]
- Kvesić, M.; Kalinić, H.; Dželalija, M.; Šamanić, I.; Andričević, R.; Maravić, A. Microbiome and antibiotic resistance profiling in submarine effluent-receiving coastal waters in Croatia. Environ. Pollut. 2022, 292, 118282. [Google Scholar] [CrossRef] [PubMed]
- Sabri, N.A.; Schmitt, H.; Van Der Zaan, B.; Gerritsen, H.W.; Zuidema, T.; Rijnaarts, H.H.M.; Langenhoff, A.A.M. Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. J. Environ. Chem. Eng. 2020, 8, 102245. [Google Scholar] [CrossRef]
- Rodríguez-Molina, D.; Berglund, F.; Blaak, H.; Flach, C.-F.; Kemper, M.; Marutescu, L.; Gradisteanu, G.P.; Popa, M.; Spießberger, B.; Weinmann, T.; et al. Carriage of ESBL-producing Enterobacterales in wastewater treatment plant workers and surrounding residents—The AWARE Study. Eur. J. Clin. Microbiol. Infect. Dis. 2021. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.F.; Morris, D.; Schmitt, H.; Gaze, W.H. Natural recreational waters and the risk that exposure to antibiotic resistant bacteria poses to human health. Curr. Opin. Microbiol. 2022, 65, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Stanton, I.C.; Bethel, A.; Leonard, A.F.C.; Gaze, W.H.; Garside, R. Existing evidence on antibiotic resistance exposure and transmission to humans from the environment: A systematic map. Environ. Evid. 2022, 11, 1–24. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Zhang, T.; Fang, H.H.P. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Munk, P.; Njage, P.; van Bunnik, B.; McNally, L.; Lukjancenko, O.; Röder, T.; Nieuwenhuijse, D.; Pedersen, S.K.; Kjeldgaard, J.; et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019, 10, 1124. [Google Scholar] [CrossRef]
- Pärnänen, K.M.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef]
- Swami, O.C. Strategies to Combat Antimicrobial Resistance. J. Clin. Diagn. Res. 2014, 8, 8–11. [Google Scholar] [CrossRef]
- WHO. Global Action Plan on Antimicrobial Resistance 2015. Available online: https://apps.who.int/iris/handle/10665/193736 (accessed on 10 May 2022).
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.-N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Barancheshme, F.; Munir, M. Strategies to Combat Antibiotic Resistance in the Wastewater Treatment Plants. Front. Microbiol. 2018, 8, 2603. [Google Scholar] [CrossRef]
- Pruden, A.; Vikesland, P.J.; Davis, B.C.; de Roda Husman, A.M. Seizing the moment: Now is the time for integrated global surveillance of antimicrobial resistance in wastewater environments. Curr. Opin. Microbiol. 2021, 64, 91–99. [Google Scholar] [CrossRef]
- Robinson, T.P.; Bu, D.P.; Carrique-Mas, J.; Fèvre, E.M.; Gilbert, M.; Grace, D.; Hay, S.I.; Jiwakanon, J.; Kakkar, M.; Kariuki, S.; et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016, 110, 377–380. [Google Scholar] [CrossRef]
- Marano, R.B.M.; Fernandes, T.; Manaia, C.M.; Nunes, O.; Morrison, D.; Berendonk, T.U.; Kreuzinger, N.; Tenson, T.; Corno, G.; Fatta-Kassinos, D.; et al. A global multinational survey of cefotaxime-resistant coliforms in urban wastewater treatment plants. Environ. Int. 2020, 144, 106035. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.-F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Daoud, Z.; Rolain, J.-M. Editorial: “One Health” Approach for Revealing Reservoirs and Transmission of Antimicrobial Resistance. Front. Microbiol. 2022, 12, 2021–2023. [Google Scholar] [CrossRef]
- Zhuang, M.; Achmon, Y.; Cao, Y.; Liang, X.; Chen, L.; Wang, H.; Siame, B.A.; Leung, K.Y. Distribution of antibiotic resistance genes in the environment. Environ. Pollut. 2021, 285, 117402. [Google Scholar] [CrossRef]
- Australian Commision on Safety and Quality in Health Care. AURA 2019: Third Australian Report on Antimicrobial Use and Resistance in Human Health 2019. Available online: https://www.safetyandquality.gov.au/publications-and-resources/resource-library/aura-2019-third-australian-report-antimicrobial-use-and-resistance-human-health (accessed on 1 April 2022).
- Tang, S.S.; Apisarnthanarak, A.; Hsu, L.Y. Mechanisms of β-lactam antimicrobial resistance and epidemiology of major community- and healthcare-associated multidrug-resistant bacteria. Adv. Drug Deliv. Rev. 2014, 78, 3–13. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. Epidemiology of β-Lactamase-Producing Pathogens. Clin. Microbiol. Rev. 2020, 33, 1–37. [Google Scholar] [CrossRef]
- Li, X.-Z.; Nikaido, H. Efflux-Mediated Drug Resistance in Bacteria. Drugs 2009, 69, 1555–1623. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Jacoby, G.A.; Medeiros, A.A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 1995, 39, 1211–1233. [Google Scholar] [CrossRef] [PubMed]
- Bonomo, R.A. β-Lactamases: A Focus on Current Challenges. Cold Spring Harb. Perspect. Med. 2017, 7, a025239. [Google Scholar] [CrossRef] [PubMed]
- Frickmann, H.; Masanta, W.O.; Zautner, A.E. Emerging Rapid Resistance Testing Methods for Clinical Microbiology Laboratories and Their Potential Impact on Patient Management. Biomed Res. Int. 2014, 2014, 375681. [Google Scholar] [CrossRef] [PubMed]
- Karkman, A.; Do, T.T.; Walsh, F.; Virta, M.P.J. Antibiotic-Resistance Genes in Waste Water. Trends Microbiol. 2018, 26, 220–228. [Google Scholar] [CrossRef]
- Franklin, A.M.; Brinkman, N.E.; Jahne, M.A.; Keely, S.P. Twenty-first century molecular methods for analyzing antimicrobial resistance in surface waters to support One Health assessments. J. Microbiol. Methods 2021, 184, 106174. [Google Scholar] [CrossRef]
- Nguyen, A.Q.; Vu, H.P.; Nguyen, L.N.; Wang, Q.; Djordjevic, S.P.; Donner, E.; Yin, H.; Nghiem, L.D. Monitoring antibiotic resistance genes in wastewater treatment: Current strategies and future challenges. Sci. Total Environ. 2021, 783, 146964. [Google Scholar] [CrossRef]
- de Abreu, V.A.C.; Perdigão, J.; Almeida, S. Metagenomic Approaches to Analyze Antimicrobial Resistance: An Overview. Front. Genet. 2021, 11, 575592. [Google Scholar] [CrossRef]
- Manaia, C.M.; Rocha, J.; Scaccia, N.; Marano, R.; Radu, E.; Biancullo, F.; Cerqueira, F.; Fortunato, G.; Iakovides, I.C.; Zammit, I.; et al. Antibiotic resistance in wastewater treatment plants: Tackling the black box. Environ. Int. 2018, 115, 312–324. [Google Scholar] [CrossRef]
- Sawa, T.; Kooguchi, K.; Moriyama, K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J. Intensive Care 2020, 8, 13. [Google Scholar] [CrossRef]
- Baraniak, A.; Fiett, J.; Mrówka, A.; Walory, J.; Hryniewicz, W.; Gniadkowski, M. Evolution of TEM-Type Extended-Spectrum β-Lactamases in Clinical Enterobacteriaceae Strains in Poland. Antimicrob. Agents Chemother. 2005, 49, 1872–1880. [Google Scholar] [CrossRef]
- De Champs, C.; Sirot, D.; Chanal, C.; Bonnet, R.; Sirot, J. A 1998 Survey of Extended-Spectrum β-Lactamases in Enterobacteriaceae in France. Antimicrob. Agents Chemother. 2000, 44, 3177–3179. [Google Scholar] [CrossRef]
- Livermore, D.M.; Canton, R.; Gniadkowski, M.; Nordmann, P.; Rossolini, G.M.; Arlet, G.; Ayala, J.; Coque, T.M.; Kern-Zdanowicz, I.; Luzzaro, F.; et al. CTX-M: Changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 2006, 59, 165–174. [Google Scholar] [CrossRef]
- Doi, Y.; Iovleva, A.; Bonomo, R.A. The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J. Travel Med. 2017, 24, S44–S51. [Google Scholar] [CrossRef]
- Wiener, J. Multiple Antibiotic–Resistant Klebsiella and Escherichia coli in Nursing Homes. JAMA 1999, 281, 517. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Nordmann, P.; Laupland, K.B.; Poirel, L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 2005, 56, 52–59. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-Spectrum β-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Nordmann, P.; Cuzon, G.; Naas, T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect. Dis. 2009, 9, 228–236. [Google Scholar] [CrossRef]
- Nordmann, P.; Naas, T.; Poirel, L. Global Spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011, 17, 1791–1798. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Price, L.S.; Poirel, L.; Bonomo, R.A.; Schwaber, M.J.; Daikos, G.L.; Cormican, M.; Cornaglia, G.; Garau, J.; Gniadkowski, M.; Hayden, M.K.; et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 2013, 13, 785–796. [Google Scholar] [CrossRef]
- Li, L.; Nesme, J.; Quintela-Baluja, M.; Balboa, S.; Hashsham, S.; Williams, M.R.; Yu, Z.; Sørensen, S.J.; Graham, D.W.; Romalde, J.L.; et al. Extended-Spectrum β-Lactamase and Carbapenemase Genes are Substantially and Sequentially Reduced during Conveyance and Treatment of Urban Sewage. Environ. Sci. Technol. 2021, 55, 5939–5949. [Google Scholar] [CrossRef] [PubMed]
- Zaatout, N.; Bouras, S.; Slimani, N. Prevalence of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in wastewater: A systematic review and meta-analysis. J. Water Health 2021, 19, 705–723. [Google Scholar] [CrossRef] [PubMed]
- Gündoğdu, A.; Jennison, A.V.; Smith, H.V.; Stratton, H.; Katouli, M. Extended-spectrum β-lactamase producing Escherichia coli in hospital wastewaters and sewage treatment plants in Queensland, Australia. Can. J. Microbiol. 2013, 59, 737–745. [Google Scholar] [CrossRef]
- Korzeniewska, E.; Harnisz, M. Extended-spectrum beta-lactamase (ESBL)-positive Enterobacteriaceae in municipal sewage and their emission to the environment. J. Environ. Manag. 2013, 128, 904–911. [Google Scholar] [CrossRef]
- Makowska, N.; Philips, A.; Dabert, M.; Nowis, K.; Trzebny, A.; Koczura, R.; Mokracka, J. Metagenomic analysis of β-lactamase and carbapenemase genes in the wastewater resistome. Water Res. 2020, 170, 115277. [Google Scholar] [CrossRef]
- Yuan, W.; Tian, T.; Yang, Q.; Riaz, L. Transfer potentials of antibiotic resistance genes in Escherichia spp. strains from different sources. Chemosphere 2020, 246, 125736. [Google Scholar] [CrossRef]
- Ali, A.; Sultan, I.; Mondal, A.H.; Siddiqui, M.T.; Gogry, F.A.; Haq, Q.M.R. Lentic and effluent water of Delhi-NCR: A reservoir of multidrug-resistant bacteria harbouring bla CTX-M, bla TEM and bla SHV type ESBL genes. J. Water Health 2021, 19, 592–603. [Google Scholar] [CrossRef]
- Fadare, F.T.; Okoh, A.I. The Abundance of Genes Encoding ESBL, pAmpC and Non-β-Lactam Resistance in Multidrug-Resistant Enterobacteriaceae Recovered from Wastewater Effluents. Front. Environ. Sci. 2021, 9, 1–12. [Google Scholar] [CrossRef]
- Gumede, S.N.; Abia, A.L.K.; Amoako, D.G.; Essack, S.Y. Analysis of Wastewater Reveals the Spread of Diverse Extended-Spectrum β-Lactamase-Producing E. coli Strains in uMgungundlovu District, South Africa. Antibiotics 2021, 10, 860. [Google Scholar] [CrossRef]
- Alouache, S.; Estepa, V.; Messai, Y.; Ruiz, E.; Torres, C.; Bakour, R. Characterization of ESBLs and Associated Quinolone Resistance in Escherichia coli and Klebsiella pneumoniae Isolates from an Urban Wastewater Treatment Plant in Algeria. Microb. Drug Resist. 2014, 20, 30–38. [Google Scholar] [CrossRef]
- Bréchet, C.; Plantin, J.; Sauget, M.; Thouverez, M.; Talon, D.; Cholley, P.; Guyeux, C.; Hocquet, D.; Bertrand, X. Wastewater treatment plants release large amounts of extended-spectrum β-lactamase-producing escherichia coli into the environment. Clin. Infect. Dis. 2014, 58, 1658–1665. [Google Scholar] [CrossRef] [PubMed]
- Ojer-Usoz, E.; González, D.; García-Jalón, I.; Vitas, A.I. High dissemination of extended-spectrum β-lactamase-producing Enterobacteriaceae in effluents from wastewater treatment plants. Water Res. 2014, 56, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Akiba, M.; Sekizuka, T.; Yamashita, A.; Kuroda, M.; Fujii, Y.; Murata, M.; Lee, K.I.; Joshua, D.I.; Balakrishna, K.; Bairy, I.; et al. Distribution and relationships of antimicrobial resistance determinants among extended-spectrum-cephalosporin-resistant or carbapenem-resistant Escherichia coli isolates from rivers and sewage treatment plants in India. Antimicrob. Agents Chemother. 2016, 60, 2972–2980. [Google Scholar] [CrossRef]
- Caltagirone, M.; Nucleo, E.; Spalla, M.; Zara, F.; Novazzi, F.; Marchetti, V.M.; Piazza, A.; Bitar, I.; De Cicco, M.; Paolucci, S.; et al. Occurrence of Extended Spectrum β-Lactamases, KPC-Type, and MCR-1.2-Producing Enterobacteriaceae from Wells, River Water, and Wastewater Treatment Plants in Oltrepò Pavese Area, Northern Italy. Front. Microbiol. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Conte, D.; Palmeiro, J.K.; da Silva Nogueira, K.; de Lima, T.M.R.; Cardoso, M.A.; Pontarolo, R.; Degaut Pontes, F.L.; Dalla-Costa, L.M. Characterization of CTX-M enzymes, quinolone resistance determinants, and antimicrobial residues from hospital sewage, wastewater treatment plant, and river water. Ecotoxicol. Environ. Saf. 2017, 136, 62–69. [Google Scholar] [CrossRef]
- Guyomard-Rabenirina, S.; Dartron, C.; Falord, M.; Sadikalay, S.; Ducat, C.; Richard, V.; Breurec, S.; Gros, O.; Talarmin, A. Resistance to antimicrobial drugs in different surface waters and wastewaters of Guadeloupe. PLoS ONE 2017, 12, e0173155. [Google Scholar] [CrossRef]
- Galler, H.; Feierl, G.; Petternel, C.; Reinthaler, F.; Haas, D.; Habib, J.; Kittinger, C.; Luxner, J.; Zarfel, G. Multiresistant Multiresistant Bacteria Isolated from Activated Sludge in Austria. Int. J. Environ. Res. Public Health 2018, 15, 479. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, T.; Zhang, X.-X.; Liang, D.-W.; Zhang, M.; Gao, D.-W.; Zhu, H.-G.; Huang, Q.-G.; Fang, H.H.P. Quantification and characterization of β-lactam resistance genes in 15 sewage treatment plants from East Asia and North America. Appl. Microbiol. Biotechnol. 2012, 95, 1351–1358. [Google Scholar] [CrossRef]
- Khan, M.A.; Thurgood, N.E.; Faheem, S.M.; Rais, N.; Ansari, M.Z.; Kaleem, S.M.; Khan, S.T. Occurrence of Extended Spectrum Beta-Lactamase Gram-Negative Bacteria from Non-Clinical Sources in Dubai, United Arab Emirates. Water 2020, 12, 2562. [Google Scholar] [CrossRef]
- Osińska, A.; Korzeniewska, E.; Harnisz, M.; Felis, E.; Bajkacz, S.; Jachimowicz, P.; Niestępski, S.; Konopka, I. Small-scale wastewater treatment plants as a source of the dissemination of antibiotic resistance genes in the aquatic environment. J. Hazard. Mater. 2020, 381, 121221. [Google Scholar] [CrossRef]
- Surleac, M.; Barbu, I.C.; Paraschiv, S.; Popa, L.I.; Gheorghe, I.; Marutescu, L.; Popa, M.; Sarbu, I.; Talapan, D.; Nita, M.; et al. Whole genome sequencing snapshot of multidrug resistant Klebsiella pneumoniae strains from hospitals and receiving wastewater treatment plants in Southern Romania. PLoS ONE 2020, 15, e0228079. [Google Scholar] [CrossRef] [PubMed]
- Smyth, C.; O’Flaherty, A.; Walsh, F.; Do, T.T. Antibiotic resistant and extended-spectrum β-lactamase producing faecal coliforms in wastewater treatment plant effluent. Environ. Pollut. 2020, 262, 114244. [Google Scholar] [CrossRef] [PubMed]
- Aristizábal-Hoyos, A.M.; Rodríguez, E.A.; Arias, L.; Jiménez, J.N. High clonal diversity of multidrug-resistant and extended spectrum beta-lactamase-producing Escherichia coli in a wastewater treatment plant. J. Environ. Manag. 2019, 245, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Amador, P.P.; Fernandes, R.M.; Prudêncio, M.C.; Barreto, M.P.; Duarte, I.M. Antibiotic resistance in wastewater: Occurrence and fate of Enterobacteriaceae producers of Class A and Class C β-lactamases. J. Environ. Sci. Health Part A 2015, 50, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Proia, L.; Anzil, A.; Borrego, C.; Farrè, M.; Llorca, M.; Sanchis, J.; Bogaerts, P.; Balcázar, J.L.; Servais, P. Occurrence and persistence of carbapenemases genes in hospital and wastewater treatment plants and propagation in the receiving river. J. Hazard. Mater. 2018, 358, 33–43. [Google Scholar] [CrossRef]
- Zhang, A.; Call, D.R.; Besser, T.E.; Liu, J.; Jones, L.; Wang, H.; Davis, M.A. β-lactam resistance genes in bacteriophage and bacterial DNA from wastewater, river water, and irrigation water in Washington State. Water Res. 2019, 161, 335–340. [Google Scholar] [CrossRef]
- Adegoke, A.A.; Madu, C.E.; Aiyegoro, O.A.; Stenström, T.A.; Okoh, A.I. Antibiogram and beta-lactamase genes among cefotaxime resistant E. coli from wastewater treatment plant. Antimicrob. Resist. Infect. Control 2020, 9, 46. [Google Scholar] [CrossRef]
- Piotrowska, M.; Kowalska, S.; Popowska, M. Diversity of β-lactam resistance genes in gram-negative rods isolated from a municipal wastewater treatment plant. Ann. Microbiol. 2019, 69, 591–601. [Google Scholar] [CrossRef]
- Hubeny, J.; Ciesielski, S.; Harnisz, M.; Korzeniewska, E.; Dulski, T.; Jałowiecki, Ł.; Płaza, G. Impact of Hospital Wastewater on the Occurrence and Diversity of Beta-Lactamase Genes During Wastewater Treatment with an Emphasis on Carbapenemase Genes: A Metagenomic Approach. Front. Environ. Sci. 2021, 9, 738158. [Google Scholar] [CrossRef]
- Teban-Man, A.; Farkas, A.; Baricz, A.; Hegedus, A.; Szekeres, E.; Pârvu, M.; Coman, C. Wastewaters, with or without Hospital Contribution, Harbour MDR, Carbapenemase-Producing, but Not Hypervirulent Klebsiella pneumoniae. Antibiotics 2021, 10, 361. [Google Scholar] [CrossRef]
- Galler, H.; Feierl, G.; Petternel, C.; Reinthaler, F.F.; Haas, D.; Grisold, A.J.; Luxner, J.; Zarfel, G. KPC-2 and OXA-48 carbapenemase-harbouring Enterobacteriaceae detected in an Austrian wastewater treatment plant. Clin. Microbiol. Infect. 2014, 20, O132–O134. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Hammarén, R.; Pal, C.; Östman, M.; Björlenius, B.; Flach, C.-F.; Fick, J.; Kristiansson, E.; Tysklind, M.; Larsson, D.G.J. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 2016, 572, 697–712. [Google Scholar] [CrossRef]
- Piotrowska, M.; Przygodzińska, D.; Matyjewicz, K.; Popowska, M. Occurrence and Variety of β-Lactamase Genes among Aeromonas spp. Isolated from Urban Wastewater Treatment Plant. Front. Microbiol. 2017, 8, 863. [Google Scholar] [CrossRef]
- Subirats, J.; Royo, E.; Balcázar, J.L.; Borrego, C.M. Real-time PCR assays for the detection and quantification of carbapenemase genes (bla KPC, bla NDM, and bla OXA-48) in environmental samples. Environ. Sci. Pollut. Res. 2017, 24, 6710–6714. [Google Scholar] [CrossRef]
- Zurfluh, K.; Bagutti, C.; Brodmann, P.; Alt, M.; Schulze, J.; Fanning, S.; Stephan, R.; Nüesch-Inderbinen, M. Wastewater is a reservoir for clinically relevant carbapenemase- and 16s rRNA methylase-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2017, 50, 436–440. [Google Scholar] [CrossRef]
- Müller, H.; Sib, E.; Gajdiss, M.; Klanke, U.; Lenz-Plet, F.; Barabasch, V.; Albert, C.; Schallenberg, A.; Timm, C.; Zacharias, N.; et al. Dissemination of multi-resistant Gram-negative bacteria into German wastewater and surface waters. FEMS Microbiol. Ecol. 2018, 94, fiy057. [Google Scholar] [CrossRef]
- Cacace, D.; Fatta-Kassinos, D.; Manaia, C.M.; Cytryn, E.; Kreuzinger, N.; Rizzo, L.; Karaolia, P.; Schwartz, T.; Alexander, J.; Merlin, C.; et al. Antibiotic resistance genes in treated wastewater and in the receiving water bodies: A pan-European survey of urban settings. Water Res. 2019, 162, 320–330. [Google Scholar] [CrossRef]
- Mathys, D.A.; Mollenkopf, D.F.; Feicht, S.M.; Adams, R.J.; Albers, A.L.; Stuever, D.M.; Grooters, S.V.; Ballash, G.A.; Daniels, J.B.; Wittum, T.E. Carbapenemase-producing Enterobacteriaceae and Aeromonas spp. Present in wastewater treatment plant effluent and nearby surface waters in the US. PLoS ONE 2018, 14, e0218650. [Google Scholar] [CrossRef]
- Hoelle, J.; Johnson, J.R.; Johnston, B.D.; Kinkle, B.; Boczek, L.; Ryu, H.; Hayes, S. Survey of US wastewater for carbapenem-resistant Enterobacteriaceae. J. Water Health 2019, 17, 219–226. [Google Scholar] [CrossRef]
- Cooper, A.L.; Carter, C.; McLeod, H.; Wright, M.; Sritharan, P.; Tamber, S.; Wong, A.; Carrillo, C.D.; Blais, B.W. Detection of carbapenem-resistance genes in bacteria isolated from wastewater in Ontario. FACETS 2021, 6, 569–591. [Google Scholar] [CrossRef]
- Ebomah, K.E.; Okoh, A.I. Detection of Carbapenem-Resistance Genes in Klebsiella Species Recovered from Selected Environmental Niches in the Eastern Cape Province, South Africa. Antibiotics 2020, 9, 425. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Mao, D.; Zhou, H.; Luo, Y. Prevalence and Fate of Carbapenemase Genes in a Wastewater Treatment Plant in Northern China. PLoS ONE 2016, 11, e0156383. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.; Tan, B.; Jiang, X.-T.; Gu, X.; Chen, H.; Schmitz, B.W.; Haller, L.; Charles, F.R.; Zhang, T.; Gin, K. Metagenomic and Resistome Analysis of a Full-Scale Municipal Wastewater Treatment Plant in Singapore Containing Membrane Bioreactors. Front. Microbiol. 2019, 10, 172. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.; Tay, M.; Tan, B.; Le, T.-H.; Haller, L.; Chen, H.; Koh, T.H.; Barkham, T.M.S.; Thompson, J.R.; Gin, K.Y.H. Characterization of Metagenomes in Urban Aquatic Compartments Reveals High Prevalence of Clinically Relevant Antibiotic Resistance Genes in Wastewaters. Front. Microbiol. 2017, 8, 2200. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.A.; Söderquist, B.; Jass, J. Prevalence and Diversity of Antibiotic Resistance Genes in Swedish Aquatic Environments Impacted by Household and Hospital Wastewater. Front. Microbiol. 2019, 10, 688. [Google Scholar] [CrossRef]
- Araújo, S.; Sousa, M.; Tacão, M.; Baraúna, R.A.; Silva, A.; Ramos, R.; Alves, A.; Manaia, C.M.; Henriques, I. Carbapenem-resistant bacteria over a wastewater treatment process: Carbapenem-resistant Enterobacteriaceae in untreated wastewater and intrinsically-resistant bacteria in final effluent. Sci. Total Environ. 2021, 782, 146892. [Google Scholar] [CrossRef]
- Zhang, D.; Peng, Y.; Chan, C.-L.; On, H.; Wai, H.K.-F.; Shekhawat, S.S.; Gupta, A.B.; Varshney, A.K.; Chuanchuen, R.; Zhou, X.; et al. Metagenomic Survey Reveals More Diverse and Abundant Antibiotic Resistance Genes in Municipal Wastewater Than Hospital Wastewater. Front. Microbiol. 2021, 12, 712843. [Google Scholar] [CrossRef]
- Reinthaler, F.F.; Feierl, G.; Galler, H.; Haas, D.; Leitner, E.; Mascher, F.; Melkes, A.; Posch, J.; Winter, I.; Zarfel, G. ESBL-producing E. coli in Austrian sewage sludge. Water Res. 2010, 44, 1981–1985. [Google Scholar] [CrossRef]
- Dolejska, M.; Frolkova, P.; Florek, M.; Jamborova, I.; Purgertova, M.; Kutilova, I.; Cizek, A.; Guenther, S.; Literak, I. CTX-M-15-producing Escherichia coli clone B2-O25b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J. Antimicrob. Chemother. 2011, 66, 2784–2790. [Google Scholar] [CrossRef]
- Korzeniewska, E.; Korzeniewska, A.; Harnisz, M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013, 91, 96–102. [Google Scholar] [CrossRef]
- Čornejová, T.; Venglovsky, J.; Gregova, G.; Kmetova, M.; Kmet, V. Extended spectrum beta-lactamases in Escherichia coli from municipal wastewater. Ann. Agric. Environ. Med. 2015, 22, 447–450. [Google Scholar] [CrossRef]
- Kwak, Y.-K.; Colque, P.; Byfors, S.; Giske, C.G.; Möllby, R.; Kühn, I. Surveillance of antimicrobial resistance among Escherichia coli in wastewater in Stockholm during 1 year: Does it reflect the resistance trends in the society? Int. J. Antimicrob. Agents 2015, 45, 25–32. [Google Scholar] [CrossRef]
- Dropa, M.; Lincopan, N.; Balsalobre, L.C.; Oliveira, D.E.; Moura, R.A.; Fernandes, M.R.; da Silva, Q.M.; Matté, G.R.; Sato, M.I.Z.; Matté, M.H. Genetic background of novel sequence types of CTX-M-8- and CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae from public wastewater treatment plants in São Paulo, Brazil. Environ. Sci. Pollut. Res. 2016, 23, 4953–4958. [Google Scholar] [CrossRef]
- Silva, I.; Tacão, M.; Tavares, R.D.S.; Miranda, R.; Araújo, S.; Manaia, C.M.; Henriques, I. Fate of cefotaxime-resistant Enterobacteriaceae and ESBL-producers over a full-scale wastewater treatment process with UV disinfection. Sci. Total Environ. 2018, 639, 1028–1037. [Google Scholar] [CrossRef]
- Haberecht, H.B.; Nealon, N.J.; Gilliland, J.R.; Holder, A.V.; Runyan, C.; Oppel, R.C.; Ibrahim, H.M.; Mueller, L.; Schrupp, F.; Vilchez, S.; et al. Antimicrobial-Resistant Escherichia coli from Environmental Waters in Northern Colorado. J. Environ. Public Health 2019, 2019, 1–13. [Google Scholar] [CrossRef]
- Raven, K.E.; Ludden, C.; Gouliouris, T.; Blane, B.; Naydenova, P.; Brown, N.M.; Parkhill, J.; Peacock, S.J. Genomic surveillance of Escherichia coli in municipal wastewater treatment plants as an indicator of clinically relevant pathogens and their resistance genes. Microb. Genom. 2019, 5, e000267. [Google Scholar] [CrossRef]
- Sghaier, S.; Abbassi, M.S.; Pascual, A.; Serrano, L.; Díaz-De-Alba, P.; Said, M.B.; Hassen, B.; Ibrahim, C.; Hassen, A.; López-Cerero, L. Extended-spectrum β-lactamase-producing Enterobacteriaceae from animal origin and wastewater in Tunisia: First detection of O25b-B23-CTX-M-27-ST131 Escherichia coli and CTX-M-15/OXA-204-producing Citrobacter freundii from wastewater. J. Glob. Antimicrob. Resist. 2019, 17, 189–194. [Google Scholar] [CrossRef]
- Tanaka, H.; Hayashi, W.; Iimura, M.; Taniguchi, Y.; Soga, E.; Matsuo, N.; Kawamura, K.; Arakawa, Y.; Nagano, Y.; Nagano, N. Wastewater as a Probable Environmental Reservoir of Extended-Spectrum-β-Lactamase Genes: Detection of Chimeric β-Lactamases CTX-M-64 and CTX-M-123. Appl. Environ. Microbiol. 2019, 85, 1–12. [Google Scholar] [CrossRef]
- Schages, L.; Wichern, F.; Kalscheuer, R.; Bockmühl, D. Winter is coming—Impact of temperature on the variation of beta-lactamase and mcr genes in a wastewater treatment plant. Sci. Total Environ. 2020, 712, 136499. [Google Scholar] [CrossRef]
- Urano, N.; Okai, M.; Tashiro, Y.; Takeuchi, A.; Endo, R.; Ishida, M.; Takashio, M. Behavior of Antibiotic-Resistant Fecal Coliforms in the Stream of a Sewage Treatment Plant in Tokyo. Adv. Microbiol. 2020, 10, 318–330. [Google Scholar] [CrossRef]
- Liedhegner, E.; Bojar, B.; Beattie, R.E.; Cahak, C.; Hristova, K.R.; Skwor, T. Similarities in Virulence and Extended Spectrum Beta-Lactamase Gene Profiles among Cefotaxime-Resistant Escherichia coli Wastewater and Clinical Isolates. Antibiotics 2022, 11, 260. [Google Scholar] [CrossRef] [PubMed]
- Igbinosa, I.H.; Okoh, A.I. Antibiotic Susceptibility Profile of Aeromonas Species Isolated from Wastewater Treatment Plant. Sci. World J. 2012, 2012, 1–6. [Google Scholar] [CrossRef]
- Ben Said, L.; Jouini, A.; Alonso, C.A.; Klibi, N.; Dziri, R.; Boudabous, A.; Ben Slama, K.; Torres, C. Characteristics of extended-spectrum β-lactamase (ESBL)- and pAmpC beta-lactamase-producing Enterobacteriaceae of water samples in Tunisia. Sci. Total Environ. 2016, 550, 1103–1109. [Google Scholar] [CrossRef] [PubMed]
- Ludden, C.; Reuter, S.; Judge, K.; Gouliouris, T.; Blane, B.; Coll, F.; Naydenova, P.; Hunt, M.; Tracey, A.; Hopkins, K.L.; et al. Sharing of carbapenemase-encoding plasmids between Enterobacteriaceae in UK sewage uncovered by MinION sequencing. Microb. Genom. 2017, 3, e000114. [Google Scholar] [CrossRef]
- Zhang, S.; Han, B.; Gu, J.; Wang, C.; Wang, P.; Ma, Y.; Cao, J.; He, Z. Fate of antibiotic resistant cultivable heterotrophic bacteria and antibiotic resistance genes in wastewater treatment processes. Chemosphere 2015, 135, 138–145. [Google Scholar] [CrossRef]
- Nzima, B.; Adegoke, A.A.; Ofon, U.A.; Al-Dahmoshi, H.O.M.; Saki, M.; Ndubuisi-Nnaji, U.U.; Inyang, C.U. Resistotyping and extended-spectrum beta-lactamase genes among Escherichia coli from wastewater treatment plants and recipient surface water for reuse in South Africa. New Microbes New Infect. 2020, 38, 100803. [Google Scholar] [CrossRef]
- Gomi, R.; Matsuda, T.; Matsumura, Y.; Yamamoto, M.; Tanaka, M.; Ichiyama, S.; Yoneda, M. Occurrence of clinically important lineages, including the sequence type 131 C1-M27 subclone, among extended-spectrum--lactamase-producing Escherichia coli in wastewater. Antimicrob. Agents Chemother. 2017, 61, e00564-17. [Google Scholar] [CrossRef]
- Finn, T.J.; Scriver, L.; Lam, L.; Duong, M.; Peirano, G.; Lynch, T.; Dong, T.; Pitout, J.D.D.; DeVinney, R. A comprehensive account of escherichia coli sequence type 131 in wastewater reveals an abundance of fluoroquinolone-resistant clade a strains. Appl. Environ. Microbiol. 2020, 86, 1–11. [Google Scholar] [CrossRef]
- Zhi, S.; Stothard, P.; Banting, G.; Scott, C.; Huntley, K.; Ryu, K.; Otto, S.; Ashbolt, N.; Checkley, S.; Dong, T.; et al. Characterization of water treatment-resistant and multidrug-resistant urinary pathogenic Escherichia coli in treated wastewater. Water Res. 2020, 182, 115827. [Google Scholar] [CrossRef]
- Mesquita, E.; Ribeiro, R.; Silva, C.J.C.; Alves, R.; Baptista, R.; Condinho, S.; Rosa, M.J.; Perdigão, J.; Caneiras, C.; Duarte, A. An update on wastewater multi-resistant bacteria: Identification of clinical pathogens such as escherichia coli o25b:H4-b2-st131-producing ctx-m-15 esbl and kpc-3 carbapenemase-producing klebsiella oxytoca. Microorganisms 2021, 9, 576. [Google Scholar] [CrossRef]
- Hembach, N.; Schmid, F.; Alexander, J.; Hiller, C.; Rogall, E.T.; Schwartz, T. Occurrence of the mcr-1 Colistin Resistance Gene and other Clinically Relevant Antibiotic Resistance Genes in Microbial Populations at Different Municipal Wastewater Treatment Plants in Germany. Front. Microbiol. 2017, 8, 1282. [Google Scholar] [CrossRef]
- Szczepanowski, R.; Linke, B.; Krahn, I.; Gartemann, K.-H.; Gützkow, T.; Eichler, W.; Pühler, A.; Schlüter, A. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 2009, 155, 2306–2319. [Google Scholar] [CrossRef]
- Wen, Q.; Yang, L.; Duan, R.; Chen, Z. Monitoring and evaluation of antibiotic resistance genes in four municipal wastewater treatment plants in Harbin, Northeast China. Environ. Pollut. 2016, 212, 34–40. [Google Scholar] [CrossRef]
- Narciso-da-Rocha, C.; Rocha, J.; Vaz-Moreira, I.; Lira, F.; Tamames, J.; Henriques, I.; Martinez, J.L.; Manaia, C.M. Bacterial lineages putatively associated with the dissemination of antibiotic resistance genes in a full-scale urban wastewater treatment plant. Environ. Int. 2018, 118, 179–188. [Google Scholar] [CrossRef]
- Neudorf, K.D.; Huang, Y.N.; Ragush, C.M.; Yost, C.K.; Jamieson, R.C.; Truelstrup Hansen, L. Antibiotic resistance genes in municipal wastewater treatment systems and receiving waters in Arctic Canada. Sci. Total Environ. 2017, 598, 1085–1094. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sànchez-Melsió, A.; Borrego, C.M.; Barceló, D.; Balcázar, J.L. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef]
- Laht, M.; Karkman, A.; Voolaid, V.; Ritz, C.; Tenson, T.; Virta, M.; Kisand, V. Abundances of Tetracycline, Sulphonamide and Beta-Lactam Antibiotic Resistance Genes in Conventional Wastewater Treatment Plants (WWTPs) with Different Waste Load. PLoS ONE 2014, 9, e103705. [Google Scholar] [CrossRef]
- Rafraf, I.D.; Lekunberri, I.; Sànchez-Melsió, A.; Aouni, M.; Borrego, C.M.; Balcázar, J.L. Abundance of antibiotic resistance genes in five municipal wastewater treatment plants in the Monastir Governorate, Tunisia. Environ. Pollut. 2016, 219, 353–358. [Google Scholar] [CrossRef]
- Kowalski, M.; Wolany, J.; Pastuszka, J.S.; Płaza, G.; Wlazło, A.; Ulfig, K.; Malina, A. Characteristics of airborne bacteria and fungi in some Polish wastewater treatment plants. Int. J. Environ. Sci. Technol. 2017, 14, 2181–2192. [Google Scholar] [CrossRef]
- Ginn, O.; Rocha-Melogno, L.; Bivins, A.; Lowry, S.; Cardelino, M.; Nichols, D.; Tripathi, S.N.; Soria, F.; Andrade, M.; Bergin, M.; et al. Detection and Quantification of Enteric Pathogens in Aerosols near Open Wastewater Canals in Cities with Poor Sanitation. Environ. Sci. Technol. 2021, 55, 14758–14771. [Google Scholar] [CrossRef]
- Wengenroth, L.; Berglund, F.; Blaak, H.; Chifiriuc, M.C.; Flach, C.-F.; Pircalabioru, G.G.; Larsson, D.G.J.; Marutescu, L.; van Passel, M.W.J.; Popa, M.; et al. Antibiotic Resistance in Wastewater Treatment Plants and Transmission Risks for Employees and Residents: The Concept of the AWARE Study. Antibiotics 2021, 10, 478. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Yoo, K. A review of the emergence of antibiotic resistance in bioaerosols and its monitoring methods. Rev. Environ. Sci. Bio/Technol. 2022, 21, 799–827. [Google Scholar] [CrossRef]
- Gaviria-Figueroa, A.; Preisner, E.C.; Hoque, S.; Feigley, C.E.; Norman, R.S. Emission and dispersal of antibiotic resistance genes through bioaerosols generated during the treatment of municipal sewage. Sci. Total Environ. 2019, 686, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Palzkill, T. Metallo-β-lactamase structure and function. Ann. N. Y. Acad. Sci. 2013, 1277, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Cui, X.; Zhang, H.; Du, H. Carbapenemases in Enterobacteriaceae: Detection and Antimicrobial Therapy. Front. Microbiol. 2019, 10, 1823. [Google Scholar] [CrossRef]
- Kazmierczak, K.M.; Rabine, S.; Hackel, M.; McLaughlin, R.E.; Biedenbach, D.J.; Bouchillon, S.K.; Sahm, D.F.; Bradford, P.A. Multiyear, Multinational Survey of the Incidence and Global Distribution of Metallo-β-Lactamase-Producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2016, 60, 1067–1078. [Google Scholar] [CrossRef]
- Matsumura, Y.; Peirano, G.; Motyl, M.R.; Adams, M.D.; Chen, L.; Kreiswirth, B.; DeVinney, R.; Pitout, J.D.D. Global Molecular Epidemiology of IMP-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2017, 61, e02729-16. [Google Scholar] [CrossRef]
- Matsumura, Y.; Peirano, G.; Bradford, P.A.; Motyl, M.R.; DeVinney, R.; Pitout, J.D.D. Genomic characterization of IMP and VIM carbapenemase-encoding transferable plasmids of Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 3034–3038. [Google Scholar] [CrossRef]
- Ghaith, D.M.; Zafer, M.M.; Ismail, D.K.; Al-Agamy, M.H.; Bohol, M.F.F.; Al-Qahtani, A.; Al-Ahdal, M.N.; Elnagdy, S.M.; Mostafa, I.Y. First reported nosocomial outbreak of Serratia marcescens harboring blaIMP-4 and blaVIM-2 in a neonatal intensive care unit in Cairo, Egypt. Infect. Drug Resist. 2018, 11, 2211–2217. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Mendes, R.E.; Canton, R.; Sader, H.S.; Jones, R.N. Variations in the Occurrence of Resistance Phenotypes and Carbapenemase Genes Among Enterobacteriaceae Isolates in 20 Years of the SENTRY Antimicrobial Surveillance Program. Open Forum Infect. Dis. 2019, 6, S23–S33. [Google Scholar] [CrossRef]
- Pournaras, S. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-beta-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 2003, 51, 1409–1414. [Google Scholar] [CrossRef]
- Lagatolla, C.; Edalucci, E.; Dolzani, L.; Riccio, M.L.; De Luca, F.; Medessi, E.; Rossolini, G.M.; Tonin, E.A. Molecular Evolution of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa in a Nosocomial Setting of High-Level Endemicity. J. Clin. Microbiol. 2006, 44, 2348–2353. [Google Scholar] [CrossRef]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a new metallo-β-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- van Loon, K.; Voor in ‘t holt, A.F.; Vos, M.C. A Systematic Review and Meta-analyses of the Clinical Epidemiology of Carbapenem-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2018, 62, 1–18. [Google Scholar] [CrossRef]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef]
- Hansen, G.T. Continuous Evolution: Perspective on the Epidemiology of Carbapenemase Resistance Among Enterobacterales and Other Gram-Negative Bacteria. Infect. Dis. Ther. 2021, 10, 75–92. [Google Scholar] [CrossRef]
- Alexander, J.; Bollmann, A.; Seitz, W.; Schwartz, T. Microbiological characterization of aquatic microbiomes targeting taxonomical marker genes and antibiotic resistance genes of opportunistic bacteria. Sci. Total Environ. 2015, 512–513, 316–325. [Google Scholar] [CrossRef]
- Hubeny, J.; Korzeniewska, E.; Buta-Hubeny, M.; Zieliński, W.; Rolbiecki, D.; Harnisz, M. Characterization of carbapenem resistance in environmental samples and Acinetobacter spp. isolates from wastewater and river water in Poland. Sci. Total Environ. 2022, 822, 153437. [Google Scholar] [CrossRef]
- Khan, F.A.; Hellmark, B.; Ehricht, R.; Söderquist, B.; Jass, J. Related carbapenemase-producing Klebsiella isolates detected in both a hospital and associated aquatic environment in Sweden. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 2241–2251. [Google Scholar] [CrossRef]
- Mills, M.C.; Lee, J. The threat of carbapenem-resistant bacteria in the environment: Evidence of widespread contamination of reservoirs at a global scale. Environ. Pollut. 2019, 255, 113143. [Google Scholar] [CrossRef] [PubMed]
- Cherak, Z.; Loucif, L.; Moussi, A.; Rolain, J.-M. Carbapenemase-producing Gram-negative bacteria in aquatic environments: A review. J. Glob. Antimicrob. Resist. 2021, 25, 287–309. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, R.; Thatikonda, S. β-Lactam Resistance Gene NDM-1 in the Aquatic Environment: A Review. Curr. Microbiol. 2021, 78, 3634–3643. [Google Scholar] [CrossRef] [PubMed]
- Lamba, M.; Graham, D.W.; Ahammad, S.Z. Hospital Wastewater Releases of Carbapenem-Resistance Pathogens and Genes in Urban India. Environ. Sci. Technol. 2017, 51, 13906–13912. [Google Scholar] [CrossRef] [PubMed]
- Parvez, S.; Khan, A.U. Hospital sewage water: A reservoir for variants of New Delhi metallo-β-lactamase (NDM)- and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2018, 51, 82–88. [Google Scholar] [CrossRef]
- Bardhan, T.; Chakraborty, M.; Bhattacharjee, B. Prevalence of colistin-resistant, carbapenem-hydrolyzing proteobacteria in hospital water bodies and out-falls of West Bengal, India. Int. J. Environ. Res. Public Health 2020, 17, 1007. [Google Scholar] [CrossRef]
- Hwang, S.H.; Kim, Y.J. Meropenem-resistant bacteria in hospital effluents in Seoul, Korea. Environ. Monit. Assess. 2018, 190, 673. [Google Scholar] [CrossRef]
- Haller, L.; Chen, H.; Ng, C.; Le, T.H.; Koh, T.H.; Barkham, T.; Sobsey, M.; Gin, K.Y.H. Occurrence and characteristics of extended-spectrum β-lactamase- and carbapenemase- producing bacteria from hospital effluents in Singapore. Sci. Total Environ. 2018, 615, 1119–1125. [Google Scholar] [CrossRef]
- Suzuki, Y.; Nazareno, P.J.; Nakano, R.; Mondoy, M.; Nakano, A.; Bugayong, M.P.; Bilar, J.; Perez, M.; Medina, E.J.; Saito-Obata, M.; et al. Environmental Presence and Genetic Characteristics of Carbapenemase-Producing Enterobacteriaceae from Hospital Sewage and River Water in the Philippines. Appl. Environ. Microbiol. 2020, 86, 1–10. [Google Scholar] [CrossRef]
- Nasri, E.; Subirats, J.; Sànchez-Melsió, A.; Ben Mansour, H.; Borrego, C.M.; Balcázar, J.L. Abundance of carbapenemase genes (blaKPC, blaNDM and blaOXA-48) in wastewater effluents from Tunisian hospitals. Environ. Pollut. 2017, 229, 371–374. [Google Scholar] [CrossRef]
- Marathe, N.P.; Berglund, F.; Razavi, M.; Pal, C.; Dröge, J.; Samant, S.; Kristiansson, E.; Larsson, D.G.J. Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 2019, 7, 97. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, X.; Luo, L.; Hu, N.; Duan, J.; Tang, Z.; Zhong, R.; Li, Y. The Prevalence and Characterization of Extended-Spectrum β-Lactamase- and Carbapenemase-Producing Bacteria from Hospital Sewage, Treated Effluents and Receiving Rivers. Int. J. Environ. Res. Public Health 2020, 17, 1183. [Google Scholar] [CrossRef]
- Luo, Y.; Yang, F.; Mathieu, J.; Mao, D.; Wang, Q.; Alvarez, P.J.J. Proliferation of Multidrug-Resistant New Delhi Metallo-β-lactamase Genes in Municipal Wastewater Treatment Plants in Northern China. Environ. Sci. Technol. Lett. 2014, 1, 26–30. [Google Scholar] [CrossRef]
- Yang, F.; Mao, D.; Zhou, H.; Wang, X.; Luo, Y. Propagation of New Delhi Metallo-β-lactamase Genes (bla NDM-1) from a Wastewater Treatment Plant to Its Receiving River. Environ. Sci. Technol. Lett. 2016, 3, 138–143. [Google Scholar] [CrossRef]
- Mantilla-Calderon, D.; Jumat, M.R.; Wang, T.; Ganesan, P.; Al-Jassim, N.; Hong, P.Y. Isolation and characterization of NDM-positive Escherichia coli from municipal wastewater in Jeddah, Saudi Arabia. Antimicrob. Agents Chemother. 2016, 60, 5223–5231. [Google Scholar] [CrossRef]
- Stachurová, T.; Piková, H.; Bartas, M.; Semerád, J.; Svobodová, K.; Malachová, K. Beta-lactam resistance development during the treatment processes of municipal wastewater treatment plants. Chemosphere 2021, 280, 130749. [Google Scholar] [CrossRef]
- Mahon, B.M.; Brehony, C.; McGrath, E.; Killeen, J.; Cormican, M.; Hickey, P.; Keane, S.; Hanahoe, B.; Dolan, A.; Morris, D. Indistinguishable NDM-producing Escherichia coli isolated from recreational waters, sewage, and a clinical specimen in Ireland, 2016 to 2017. Eurosurveillance 2017, 22, 30513. [Google Scholar] [CrossRef]
- Walsh, T.R.; Weeks, J.; Livermore, D.M.; Toleman, M.A. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect. Dis. 2011, 11, 355–362. [Google Scholar] [CrossRef]
- Lamba, M.; Gupta, S.; Shukla, R.; Graham, D.W.; Sreekrishnan, T.R.; Ahammad, S.Z. Carbapenem resistance exposures via wastewaters across New Delhi. Environ. Int. 2018, 119, 302–308. [Google Scholar] [CrossRef]
- Divyashree, M.; Mani, M.K.; Shama Prakash, K.; Vijaya Kumar, D.; Veena Shetty, A.; Shetty, A.K.; Karunasagar, I. Hospital wastewater treatment reduces NDM-positive bacteria being discharged into water bodies. Water Environ. Res. 2020, 92, 562–568. [Google Scholar] [CrossRef]
- Bauernfeind, A.; Stemplinger, I.; Jungwirth, R.; Wilhelm, R.; Chong, Y. Comparative characterization of the cephamycinase blaCMY-1 gene and its relationship with other beta-lactamase genes. Antimicrob. Agents Chemother. 1996, 40, 1926–1930. [Google Scholar] [CrossRef] [PubMed]
- Philippon, A.; Arlet, G.; Jacoby, G.A. Plasmid-Determined AmpC-Type β-Lactamases. Antimicrob. Agents Chemother. 2002, 46, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A. AmpC β-Lactamases. Clin. Microbiol. Rev. 2009, 22, 161–182. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Etayo, L.; González, D.; Leiva, J.; Vitas, A.I. Multidrug-Resistant Bacteria Isolated from Different Aquatic Environments in the North of Spain and South of France. Microorganisms 2020, 8, 1425. [Google Scholar] [CrossRef] [PubMed]
- Yim, G.; Kwong, W.; Davies, J.; Miao, V. Complex integrons containing qnrB4-ampC (bla DHA-1) in plasmids of multidrug-resistant Citrobacter freundii from wastewater. Can. J. Microbiol. 2013, 59, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Su, H.-C.; Ying, G.-G.; He, L.-Y.; Liu, Y.-S.; Zhang, R.-Q.; Tao, R. Antibiotic resistance, plasmid-mediated quinolone resistance (PMQR) genes and ampC gene in two typical municipal wastewater treatment plants. Environ. Sci. Process. Impacts 2014, 16, 324. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Peirano, G.; Kock, M.M.; Strydom, K.; Matsumura, Y. The Global Ascendency of OXA-48-Type Carbapenemases. Clin. Microbiol. Rev. 2019, 33, 1–48. [Google Scholar] [CrossRef]
- Evans, B.A.; Amyes, S.G.B. OXA β-Lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef]
- Mairi, A.; Pantel, A.; Sotto, A.; Lavigne, J.-P.; Touati, A. OXA-48-like carbapenemases producing Enterobacteriaceae in different niches. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 587–604. [Google Scholar] [CrossRef]
- Poirel, L.; Héritier, C.; Tolün, V.; Nordmann, P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).