The Dual Roles of Nano Zero-Valent Iron and Zinc Oxide in Antibiotics Resistance Genes (ARGs) Spread in Sediment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sediment and Sampling
2.2. Basic Properties of Nanoparticles
2.3. Incubation Experiments
2.4. Quantitative-PCR and High-Throughput Sequencing
2.5. Data Analysis
3. Results
3.1. The Effect of NPs on Tet-ARGs/intI1 Abundance in Sediments
3.2. The Effect of NPs on the Inducing Role of TC in Tet-ARGs/intI1 Abundance in Sediment
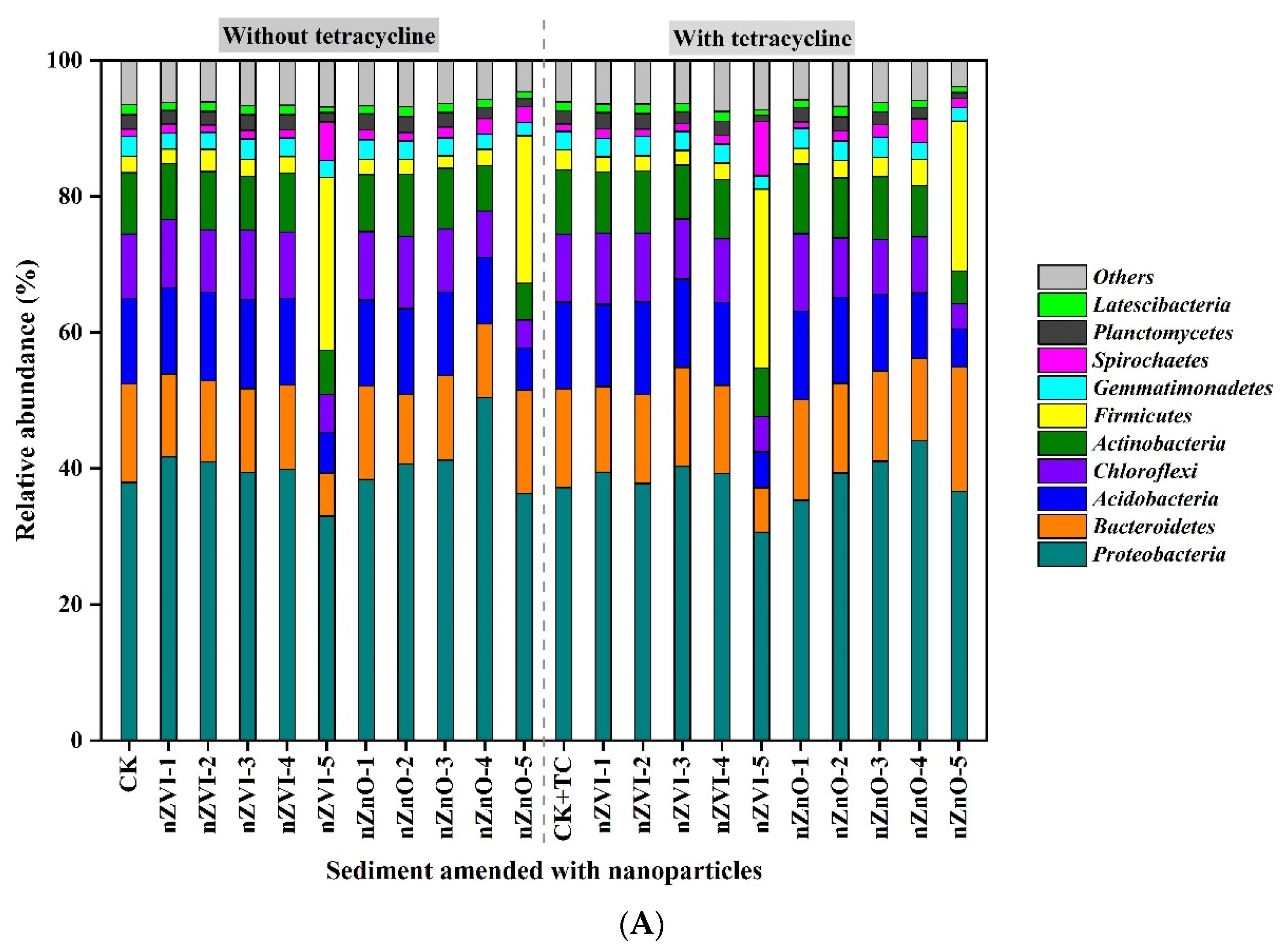
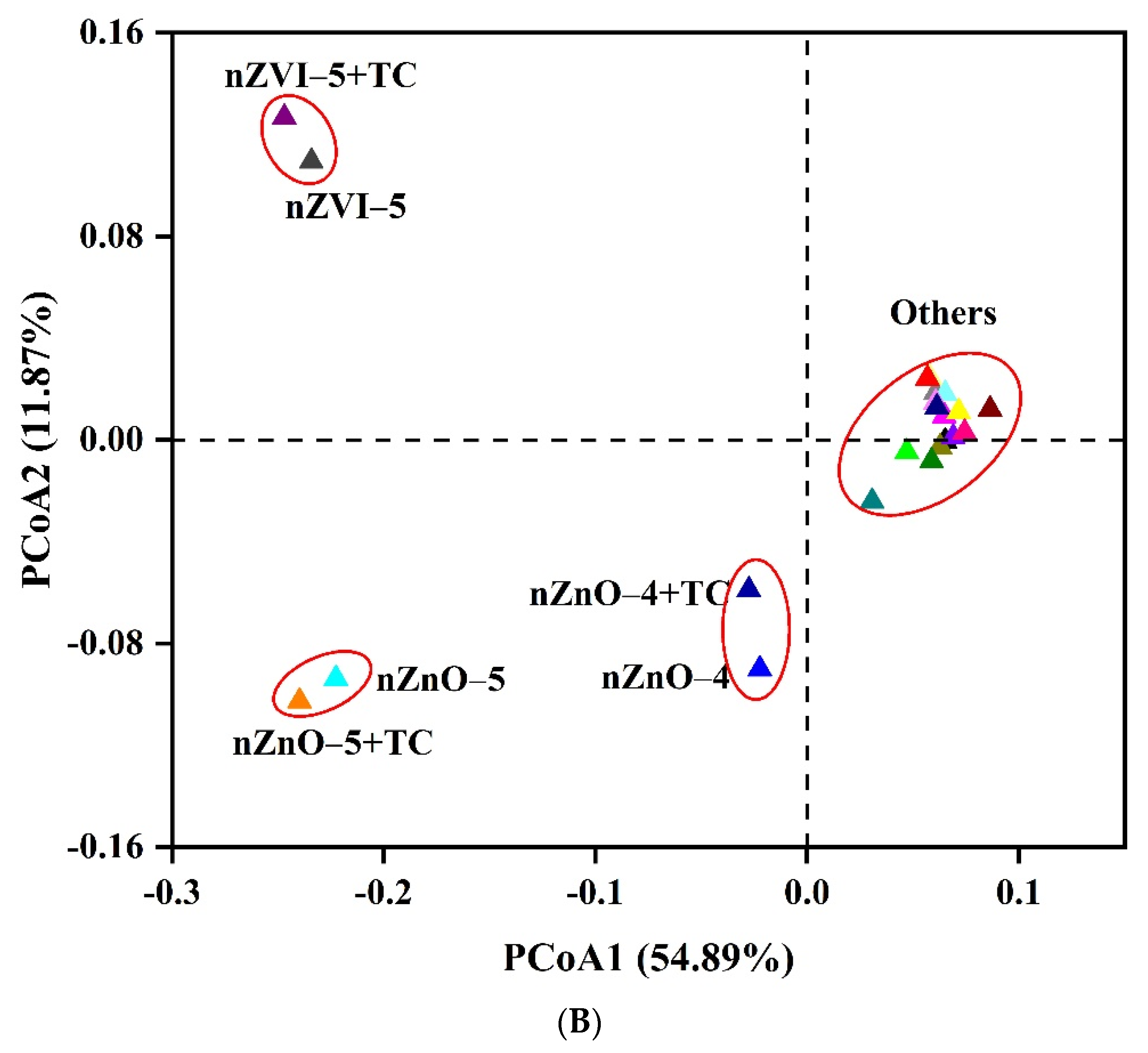
3.3. The Effect of nZVI and nZnO on Bacterial Diversity and Communities
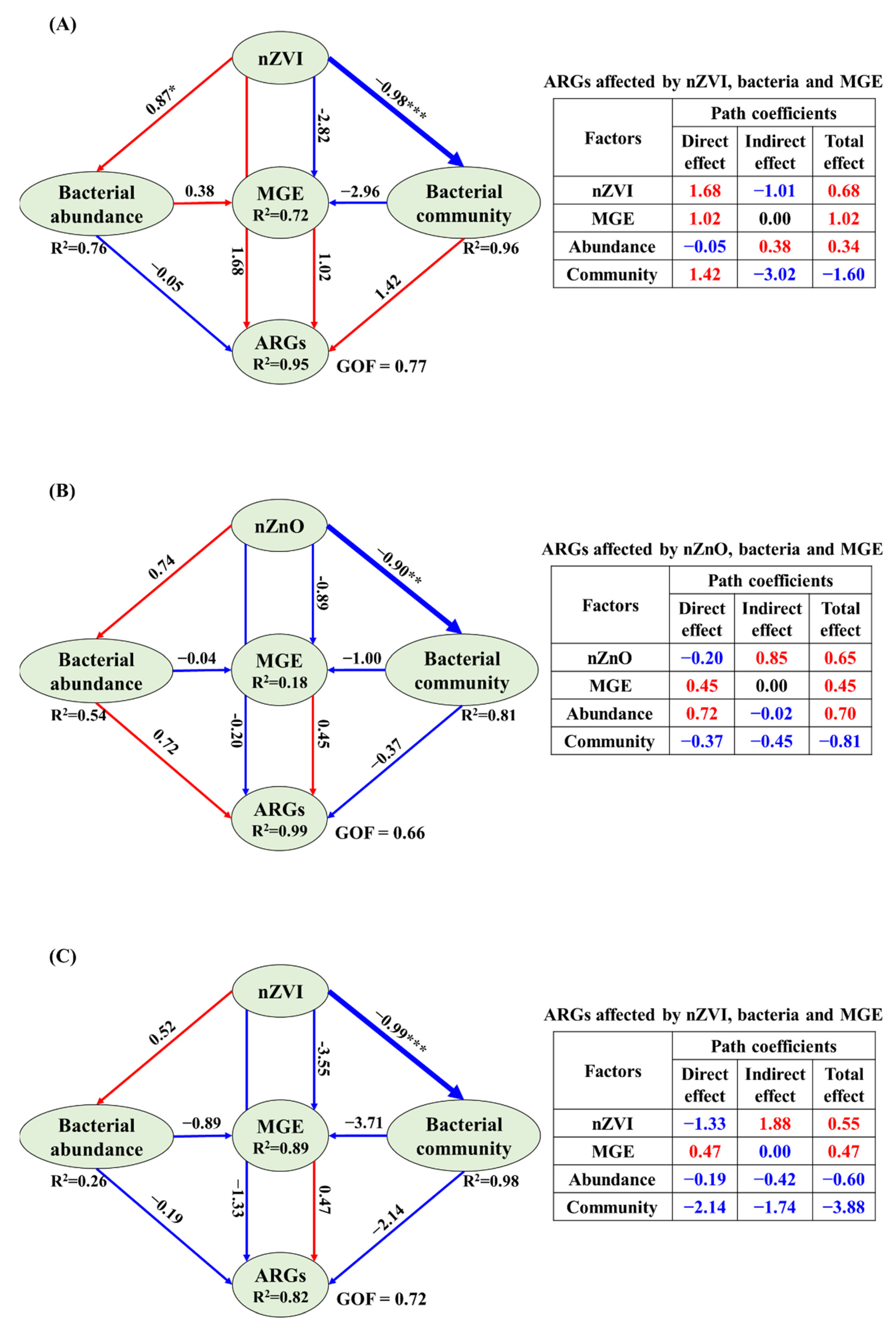
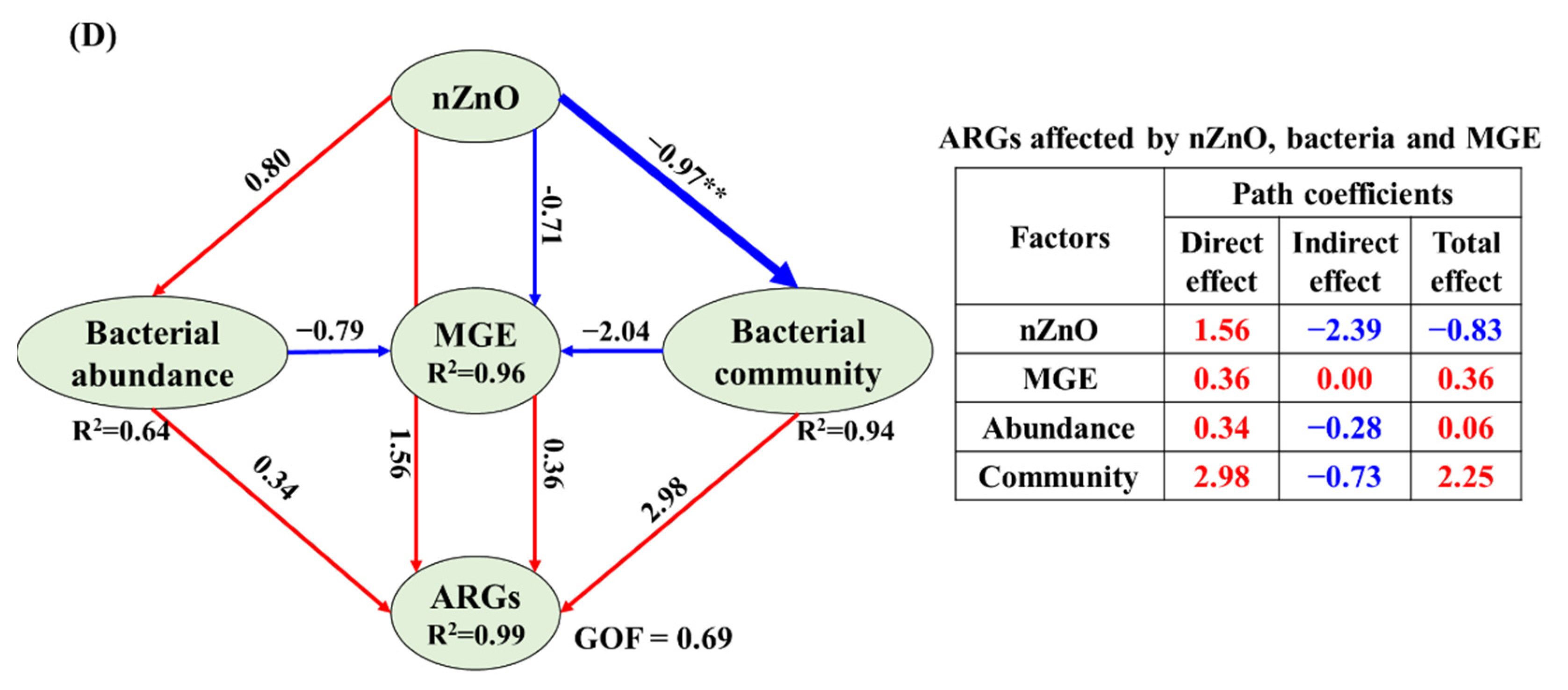
3.4. The Correlations Related Tet-ARGs to NPs, intI1 Abundance, Bacterial Abundance and Community
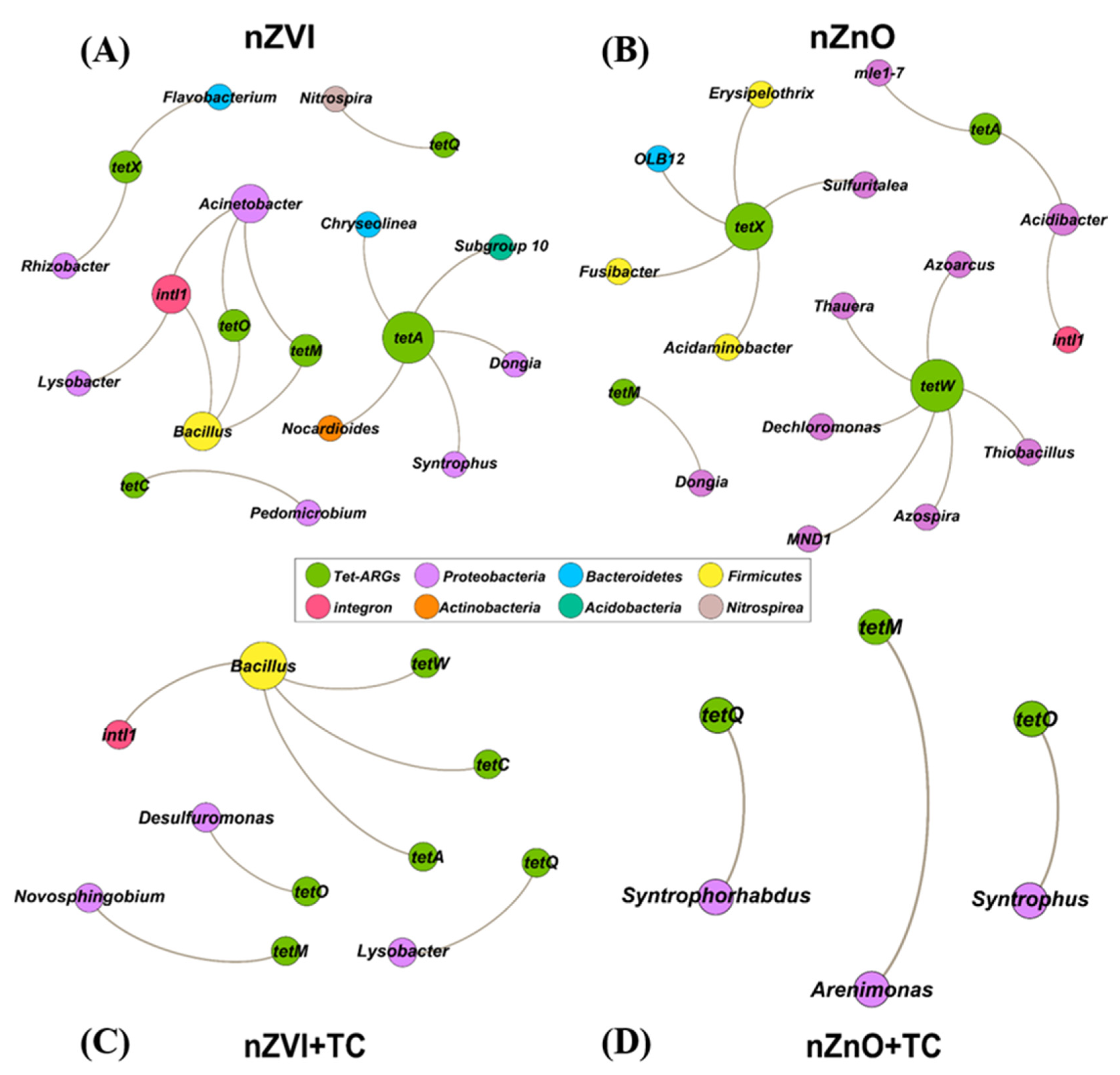
3.5. The Network Analysis between tet-ARGs/intI1 and the Bacterial Community at the Genus Level
4. Discussion
4.1. NPs Promote Tet-ARGs/intI1 Abundance in Sediment
4.2. NPs Reduce the Role of TC in Inducing tet-ARGs/intI1 in Sediment
4.3. NPs Alter Bacterial Diversity and Communities in Sediment
4.4. Potential Reasons for ARGs Spread Affected by NPs in Sediment
4.5. The Potential Hosts of Tet-ARGs in Different NPs Added Sediments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Feng, G.; Huang, H.; Chen, Y. Effects of emerging pollutants on the occurrence and transfer of antibiotic resistance genes: A review. J. Hazard. Mater. 2021, 420, 126602. [Google Scholar] [CrossRef]
- Zheng, H.; Feng, N.; Yang, T.; Shi, M.; Wang, X.; Zhang, Q.; Zhao, J.; Li, F.; Sun, K.; Xing, B. Individual and combined applications of biochar and pyroligneous acid mitigate dissemination of antibiotic resistance genes in agricultural soil. Sci. Total Environ. 2021, 796, 148962. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, J. Tackling Drug-Resistant Infections Globally: Final Report And Recommendations; Wellcome Trust/HM Government: London, UK, 2016.
- Calero-Cáceres, W.; Méndez, J.; Martín-Díaz, J.; Muniesa, M. The Occurrence of antibiotic resistance genes in a Mediterranean river and their persistence in the riverbed sediment. Environ. Pollut. 2017, 223, 384–394. [Google Scholar] [CrossRef]
- Cheng, W.; Li, J.; Wu, Y.; Xu, L.; Su, C.; Qian, Y.; Zhu, Y.G.; Chen, H. Behavior of antibiotics and antibiotic resistance genes in eco-agricultural system: A case study. J. Hazard. Mater. 2016, 304, 18–25. [Google Scholar] [CrossRef] [PubMed]
- He, L.Y.; Ying, G.G.; Liu, Y.S.; Su, H.C.; Chen, J.; Liu, S.S.; Zhao, J.L. Discharge of swine wastes risks water quality and food safety: Antibiotics and antibiotic resistance genes from swine sources to the receiving environments. Environ. Int. 2016, 92–93, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: Studies in Northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Singh, A.P.; Kumar, S.; Giri, B.S.; Kim, K.H. Antibiotic resistance in major rivers in the world: A systematic review on occurrence, emergence, and management strategies. J. Clean. Prod. 2019, 234, 1484–1505. [Google Scholar] [CrossRef]
- Sun, J.; Jin, L.; He, T.; Wei, Z.; Liu, X.; Zhu, L.; Li, X. Antibiotic resistance genes (ARGs) in agricultural soils from the Yangtze river delta, China. Sci. Total Environ. 2020, 740, 140001. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, X.; Wang, M.; Chen, H.; Yang, Y.; Chen, Q.L.; Yao, M. Time-resolved spread of antibiotic resistance genes in highly polluted air. Environ. Int. 2019, 127, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Larsson, D.G.J.; Amézquita, A.; Collignon, P.; Brandt, K.K.; Graham, D.W.; Lazorchak, J.M.; Suzuki, S.; Silley, P.; Snape, J.R.; et al. Management of options for reducing the release of antibiotics. Environ. Health Perspect. 2013, 121, 878–885. [Google Scholar] [CrossRef]
- Li, N.; Sheng, G.P.; Lu, Y.Z.; Zeng, R.J.; Yu, H.Q. Removal of antibiotic resistance genes from wastewater treatment plant effluent by coagulation. Water Res. 2017, 111, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Xia, H.; Zhang, Y.; Li, J.; Cui, G.; Li, F.; Bai, W.; Jiang, Y.; Wu, N. Elimination of antibiotic resistance genes and human pathogenic bacteria by earthworms during vermicomposting of dewatered sludge by metagenomic analysis. Bioresour. Technol. 2020, 297, 122451. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Boo, C.; Guo, N.; Wang, S.; Elimelech, M.; Wang, Y. Photocatalytic reactive ultrafiltration membrane for removal of antibiotic resistant bacteria and antibiotic resistance genes from wastewater effluent. Environ. Sci. Technol. 2018, 52, 8666–8673. [Google Scholar] [CrossRef]
- Cui, P.; Bai, Y.; Li, X.; Peng, Z.; Chen, D.; Wu, Z.; Zhang, P.; Tan, Z.; Huang, K.; Chen, Z.; et al. Enhanced removal of antibiotic resistance genes and mobile genetic elements during sewage sludge composting covered with a semi-permeable membrane. J. Hazard. Mater. 2020, 396, 122738. [Google Scholar] [CrossRef]
- Wang, G.; Deng, D.; Hu, C.; Lou, L.; Luo, L.; He, J.; Tian, D.; Xiao, Y.; He, Y.; Zhang, S.; et al. More effective removal of antibiotic resistance genes from excess sludge by microwave integrated fenton treatment. Int. Biodeterior. Biodegrad. 2020, 149, 104920. [Google Scholar] [CrossRef]
- Luo, L.; Wang, G.; Wang, Z.; Ma, J.; He, Y.; He, J.; Wang, L.; Liu, Y.; Xiao, H.; Xiao, Y.; et al. Optimization of fenton process on removing antibiotic resistance genes from excess sludge by single-factor experiment and response surface methodology. Sci. Total Environ. 2021, 788, 147889. [Google Scholar] [CrossRef]
- Sekyere, J.O. Current state of resistance to antibiotics of last-resort in South Africa: A review from a public health perspective. Front. Public Health 2016, 4, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nnadozie, C.F.; Kumari, S.; Bux, F. Status of pathogens, antibiotic resistance genes and antibiotic residues in wastewater treatment systems. Rev. Environ. Sci. Biotechnol. 2017, 16, 491–515. [Google Scholar] [CrossRef]
- Liu, J.; Gefen, O.; Ronin, I.; Bar-Meir, M.; Balaban, N.Q. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 2020, 367, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, W.; Cai, Z.; Han, B.; Qian, T.; Zhao, D. An overview of preparation and applications of stabilized zero-valent iron nanoparticles for soil and groundwater remediation. Water Res. 2016, 100, 245–266. [Google Scholar] [CrossRef] [Green Version]
- Mohammed, A.A.; Al-Musawi, T.J.; Kareem, S.L.; Zarrabi, M.; Al-Ma’abreh, A.M. Simultaneous adsorption of tetracycline, amoxicillin, and ciprofloxacin by pistachio shell powder coated with zinc oxide nanoparticles. Arab. J. Chem. 2019, 13, 4629–4643. [Google Scholar] [CrossRef]
- Jeevanandam, J.; Barhoum, A.; Chan, Y.S.; Dufresne, A.; Danquah, M.K. Review on nanoparticles and nanostructured materials: History, sources, toxicity and regulations. Beilstein J. Nanotechnol. 2018, 9, 1050–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turan, N.B.; Erkan, H.S.; Engin, G.O.; Bilgili, M.S. Nanoparticles in the aquatic environment: Usage, properties, transformation and toxicity—A review. Process. Saf. Environ. Prot. 2019, 130, 238–249. [Google Scholar] [CrossRef]
- Pawlett, M.; Ritz, K.; Dorey, R.A.; Rocks, S.; Ramsden, J.; Harris, J.A. The impact of zero-valent iron nanoparticles upon soil microbial communities is context dependent. Environ. Sci. Pollut. Res. 2013, 20, 1041–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montes de Oca-Vásquez, G.; Solano-Campos, F.; Vega-Baudrit, J.R.; López-Mondéjar, R.; Odriozola, I.; Vera, A.; Moreno, J.L.; Bastida, F. Environmentally relevant concentrations of silver nanoparticles diminish soil microbial biomass but do not alter enzyme activities or microbial diversity. J. Hazard. Mater. 2020, 391, 122224. [Google Scholar] [CrossRef] [PubMed]
- Jośko, I.; Oleszczuk, P.; Dobrzyńska, J.; Futa, B.; Joniec, J.; Dobrowolski, R. Long-term effect of ZnO and CuO nanoparticles on soil microbial community in different types of soil. Geoderma 2019, 352, 204–212. [Google Scholar] [CrossRef]
- Zhang, C.; Sun, R.; Xia, T. Adaption/resistance to antimicrobial nanoparticles: Will it be a problem? Nano Today 2020, 34, 100909. [Google Scholar] [CrossRef]
- Chen, Y.; Li, P.; Huang, Y.; Yu, K.; Chen, H.; Cui, K.; Huang, Q.; Zhang, J.; Yew-Hoong Gin, K.; He, Y. Environmental media exert a bottleneck in driving the dynamics of antibiotic resistance genes in modern aquatic environment. Water Res. 2019, 162, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hou, L.; Liu, Y.; Liu, K.; Zhang, L.; Huang, F.; Wang, L.; Rashid, A.; Hu, A.; Yu, C. Horizontal and vertical gene transfer drive sediment antibiotic resistome in an urban lagoon system. J. Environ. Sci. 2021, 102, 11–23. [Google Scholar] [CrossRef]
- Wang, X.; Yang, F.; Zhao, J.; Xu, Y.; Mao, D.; Zhu, X.; Luo, Y.; Alvarez, P.J.J. Bacterial exposure to ZnO nanoparticles facilitates horizontal transfer of antibiotic resistance genes. NanoImpact 2018, 10, 61–67. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Song, H.; Lu, J.; Yuan, Z.; Guo, J. Copper nanoparticles and copper ions promote horizontal transfer of plasmid-mediated multi-antibiotic resistance genes across bacterial genera. Environ. Int. 2019, 129, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Wang, C.; Jiang, G.; Ma, J.; Li, Y.; Chen, H.; Guo, J. Bioaerosol is an important transmission route of antibiotic resistance genes in pig farms. Environ. Int. 2021, 154, 106559. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.C.; Liu, Y.; Lin, Z.J.; Shuai, X.Y.; Zhu, L.; Xu, L.; Meng, L.X.; Sun, Y.J.; Chen, H. Spread of antibiotic resistance genes and microbiota in airborne particulate aatter, dust, and human airways in the urban hospital. Environ. Int. 2021, 153, 106501. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Aydin, M.E.; Beduk, F.; Ulvi, A. Removal of antibiotics from aqueous solution by using magnetic Fe3O4/red mud-nanoparticles. Sci. Total Environ. 2019, 670, 539–546. [Google Scholar] [CrossRef]
- Dao, T.H.; Tran, T.T.; Nguyen, V.R.; Pham, T.N.M.; Vu, C.M.; Pham, T.D. Removal of antibiotic from aqueous solution using synthesized TiO2 nanoparticles: Characteristics and mechanisms. Environ. Earth Sci. 2018, 77, 359. [Google Scholar] [CrossRef]
- GB3838-02; Environmental Quality Standards for Surface Water. Ministry of Ecology and Environment of the People’s Republic of China: Beijing, China, 2002.
- Luo, L.; Ye, H.Y.; Zhang, D.H.; Gu, J.-D.; Deng, O.P. The dynamics of phosphorus fractions and the factors driving phosphorus cycle in Zoige Plateau peatland soil. Chemosphere 2021, 278, 130501. [Google Scholar] [CrossRef]
- Yao, M.; Rui, J.; Li, J.; Dai, Y.; Bai, Y.; Heděnec, P.; Wang, J.; Zhang, S.; Pei, K.; Liu, C.; et al. Rate-specific responses of prokaryotic diversity and structure to nitrogen deposition in the Leymus Chinensis Steppe. Soil Biol. Biochem. 2014, 79, 81–90. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Z.; Xu, T.; Feng, Z.; Liu, J.; Luo, L.; He, Y.; Xiao, Y.; Peng, H.; Zhang, Y.; et al. The fate of antibiotic resistance genes and their influential factors during excess sludge composting in a full-scale plant. Bioresour. Technol. 2021, 342, 126049. [Google Scholar] [CrossRef]
- Dinesh, R.; Anandaraj, M.; Srinivasan, V.; Hamza, S. Engineered nanoparticles in the soil and their potential implications to microbial activity. Geoderma 2012, 173–174, 19–27. [Google Scholar] [CrossRef]
- Ge, Y.; Schimel, J.P.; Holden, P.A. Evidence for negative effects of TiO2 and ZnO nanoparticles on soil bacterial communities. Environ. Sci. Technol. 2011, 45, 1659–1664. [Google Scholar] [CrossRef]
- Hänsch, M.; Emmerling, C. Effects of silver nanoparticles on the microbiota and enzyme activity in soil. J. Plant Nutr. Soil Sci. 2010, 173, 554–558. [Google Scholar] [CrossRef]
- Sillen, W.M.A.; Thijs, S.; Abbamondi, G.R.; Janssen, J.; Weyens, N.; White, J.C.; Vangronsveld, J. Effects of silver nanoparticles on soil microorganisms and maize biomass are linked in the rhizosphere. Soil Biol. Biochem. 2015, 91, 14–22. [Google Scholar] [CrossRef]
- Xu, C.; Peng, C.; Sun, L.; Zhang, S.; Huang, H.; Chen, Y.; Shi, J. Distinctive effects of TiO2 and CuO nanoparticles on soil microbes and their community structures in flooded paddy soil. Soil Biol. Biochem. 2015, 86, 24–33. [Google Scholar] [CrossRef]
- Simonin, M.; Richaume, A. Impact of engineered aanoparticles on the activity, abundance, and diversity of soil microbial communities: A review. Environ. Sci. Pollut. Res. 2015, 22, 13710–13723. [Google Scholar] [CrossRef] [Green Version]
- Fajardo, C.; Ortíz, L.T.; Rodríguez-Membibre, M.L.; Nande, M.; Lobo, M.C.; Martin, M. Assessing the impact of zero-valent iron (ZVI) nanotechnology on soil microbial structure and functionality: A molecular approach. Chemosphere 2012, 86, 802–808. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Yu, Y.; Chen, Z.; Jin, M.; Yang, D.; Zhao, Z.; Wang, J.; Shen, Z.; Wang, X.; Qian, D.; et al. Nanoalumina promotes the horizontal transfer of multiresistance genes mediated by plasmids across genera. Proc. Natl. Acad. Sci. USA 2012, 109, 4944–4949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Wang, Y.; Jin, M.; Yuan, Z.; Bond, P.; Guo, J. Both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes. Water Res. 2020, 169, 115229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lu, J.; Wang, Y.; Verstraete, W.; Yuan, Z.; Guo, J. Insights of metallic nanoparticles and ions in accelerating the bacterial uptake of antibiotic resistance genes. J. Hazard. Mater. 2022, 421, 126728. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Zhang, T.; Fang, H.H.P. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Diehl, D.L.; Lapara, T.M. Effect of temperature on the fate of genes encoding tetracycline resistance and the integrase of Class 1 integrons within anaerobic and aerobic digesters treating municipal wastewater solids. Environ. Sci. Technol. 2010, 44, 9128–9133. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline antibiotics: Mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef] [Green Version]
- Auerbach, E.A.; Seyfried, E.E.; McMahon, K.D. Tetracycline resistance genes in activated sludge wastewater treatment plants. Water Res. 2007, 41, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Q.; Yuan, L.; Li, Z.H.; Zhang, H.C.; Sheng, G.P. Tetracycline exposure shifted microbial communities and enriched antibiotic resistance genes in the aerobic granular sludge. Environ. Int. 2019, 130, 104902. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Geng, J.; Ma, H.; Ren, H.; Xu, K.; Ding, L. Characterization of microbial community and antibiotic resistance genes in activated sludge under tetracycline and sulfamethoxazole selection pressure. Sci. Total Environ. 2016, 571, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Sun, M.; Ye, M.; Chao, H.; Zhao, Y.; Xia, B.; Jiao, W.; Feng, Y.; Zheng, X.; Liu, M.; et al. Coexistence and association between heavy metals, tetracycline and corresponding resistance genes in vermicomposts originating from different substrates. Environ. Pollut. 2019, 244, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhu, L.; Zhong, H.; Li, P.; Zhang, C.; Wei, D. Response of antibiotic and heavy metal resistance genes to tetracyclines and copper in substrate-free hydroponic microcosms with myriophyllum aquaticum. J. Hazard. Mater. 2021, 413, 125444. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Liu, G.; Dong, G.; Wu, X.; Wang, C.; Liu, Y. Reaction mechanism of zero-valent iron coupling with microbe to degrade tetracycline in permeable reactive barrier (PRB). Chem. Eng. J. 2017, 316, 525–533. [Google Scholar] [CrossRef]
- Cao, J.; Xiong, Z.; Lai, B. Effect of initial pH on the tetracycline (TC) removal by zero-valent iron: Adsorption, oxidation and reduction. Chem. Eng. J. 2018, 343, 492–499. [Google Scholar] [CrossRef]
- Auffan, M.; Rose, J.; Wiesner, M.R.; Bottero, J.Y. Chemical stability of metallic nanoparticles: A parameter controlling their potential cellular toxicity in vitro. Environ. Pollut. 2009, 157, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Chen, Z.; Hou, Z.; Li, T.; Lu, X. Ecotoxicological effect of zinc oxide nanoparticles on soil microorganisms. Front. Environ. Sci. Eng. 2015, 9, 912–918. [Google Scholar] [CrossRef]
- Hou, J.; Wu, Y.; Li, X.; Wei, B.; Li, S.; Wang, X. Toxic Effects of different types of zinc oxide nanoparticles on algae, plants, invertebrates, vertebrates and microorganisms. Chemosphere 2018, 193, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Wang, M.; Dai, J.; Sun, Y.; Zeng, Z. Application of manure containing tetracyclines slowed down the dissipation of tet resistance genes and caused changes in the composition of soil bacteria. Ecotoxicol. Environ. Saf. 2018, 147, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, W.; Xu, W. Effects of tetracycline antibiotics in chicken manure on soil microbes and antibiotic resistance genes (ARGs). Environ. Geochem. Health 2022, 44, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Jin, M.; Ma, J.; Chen, Z.; Shen, Z.; Yang, D.; Shi, D.; Liu, W.; Kang, M.; Wang, J.; et al. Nano-Al2O3 can mediate transduction-like transformation of antibiotic resistance genes in water. J. Hazard. Mater. 2021, 405, 124224. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Lou, L.P.; Cui, X.Y.; Wu, B.B.; Hou, J.A.; Xun, B.; Xu, X.H.; Chen, Y.X. Sorption and desorption of pentachlorophenol to black carbon of three different origins. J. Hazard. Mater. 2011, 185, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Markley, J.L.; Wencewicz, T.A. Tetracycline-inactivating enzymes. Front. Microbiol. 2018, 9, 1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, B.W.; Liang, X.M.; Huang, X.P.; Zhang, T.; Li, X.D. Differentiating anthropogenic impacts on ARGs in the Pearl River Estuary by using suitable gene indicators. Water Res. 2013, 47, 2811–2820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Chen, M.; Feng, F.; Zhang, J.; Sui, Q.; Tong, J.; Wei, Y.; Wei, D. Effects of chlortetracycline and copper on tetracyclines and copper resistance genes and microbial community during swine manure anaerobic digestion. Bioresour. Technol. 2017, 238, 57–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| NPs | Concentration (mmol kg−1) | Scenario 1: Sediment Only | Scenario 2: Sediment Overlaid with Tetracycline Solution (10 mg L−1) |
|---|---|---|---|
| 0 | CK | CK+TC | |
| nZVI | 0.0035 | nZVI-1 | nZVI-1+TC |
| 0.035 | nZVI-2 | nZVI-2+TC | |
| 0.35 | nZVI-3 | nZVI-3+TC | |
| 3.5 | nZVI-4 | nZVI-4+TC | |
| 35 | nZVI-5 | nZVI-5+TC | |
| nZnO | 0.0035 | nZnO-1 | nZnO-1+TC |
| 0.035 | nZnO-2 | nZnO-2+TC | |
| 0.35 | nZnO-3 | nZnO-3+TC | |
| 3.5 | nZnO-4 | nZnO-4+TC | |
| 35 | nZnO-5 | nZnO-5+TC |
| Scenario 1: Sediment Only | Scenario 2: Sediment with Tetracycline (+TC) | |||||
|---|---|---|---|---|---|---|
| OTUs | Shannon | Chao1 | OTUs | Shannon | Chao1 | |
| CK | 10,364 | 12.59 | 17,814 | 10,019 | 12.51 | 17,181 |
| nZVI-1 | 8230 | 11.90 | 12,665 | 10,304 | 12.59 | 17,840 |
| nZVI-2 | 7913 | 11.85 | 12,280 | 9820 | 12.49 | 15,962 |
| nZVI-3 | 9473 | 12.31 | 15,975 | 9496 | 12.40 | 15,428 |
| nZVI-4 | 9790 | 12.49 | 15,969 | 10,418 | 12.61 | 17,982 |
| nZVI-5 | 7547 | 10.93 | 12,584 | 7306 | 10.70 | 12,177 |
| nZnO-1 | 9691 | 12.47 | 15,823 | 10,122 | 12.56 | 16,829 |
| nZnO-2 | 10,247 | 12.64 | 15,614 | 8630 | 12.17 | 12,814 |
| nZnO-3 | 10,260 | 12.56 | 17,784 | 9936 | 12.47 | 17,027 |
| nZnO-4 | 8164 | 11.89 | 12,500 | 9190 | 12.20 | 15,385 |
| nZnO-5 | 7127 | 10.33 | 13,105 | 6542 | 10.22 | 10,689 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, L.; Deng, D.; Zhao, X.; Hu, H.; Li, X.; Gu, J.; He, Y.; Yang, G.; Deng, O.; Xiao, Y. The Dual Roles of Nano Zero-Valent Iron and Zinc Oxide in Antibiotics Resistance Genes (ARGs) Spread in Sediment. Int. J. Environ. Res. Public Health 2022, 19, 9405. https://doi.org/10.3390/ijerph19159405
Luo L, Deng D, Zhao X, Hu H, Li X, Gu J, He Y, Yang G, Deng O, Xiao Y. The Dual Roles of Nano Zero-Valent Iron and Zinc Oxide in Antibiotics Resistance Genes (ARGs) Spread in Sediment. International Journal of Environmental Research and Public Health. 2022; 19(15):9405. https://doi.org/10.3390/ijerph19159405
Chicago/Turabian StyleLuo, Ling, Dahang Deng, Xin Zhao, Hairong Hu, Xinyi Li, Jidong Gu, Yan He, Gang Yang, Ouping Deng, and Yinlong Xiao. 2022. "The Dual Roles of Nano Zero-Valent Iron and Zinc Oxide in Antibiotics Resistance Genes (ARGs) Spread in Sediment" International Journal of Environmental Research and Public Health 19, no. 15: 9405. https://doi.org/10.3390/ijerph19159405
APA StyleLuo, L., Deng, D., Zhao, X., Hu, H., Li, X., Gu, J., He, Y., Yang, G., Deng, O., & Xiao, Y. (2022). The Dual Roles of Nano Zero-Valent Iron and Zinc Oxide in Antibiotics Resistance Genes (ARGs) Spread in Sediment. International Journal of Environmental Research and Public Health, 19(15), 9405. https://doi.org/10.3390/ijerph19159405






