A Network Approach for the Study of Drug Prescriptions: Analysis of Administrative Records from a Local Health Unit (ASL TO4, Regione Piemonte, Italy)
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
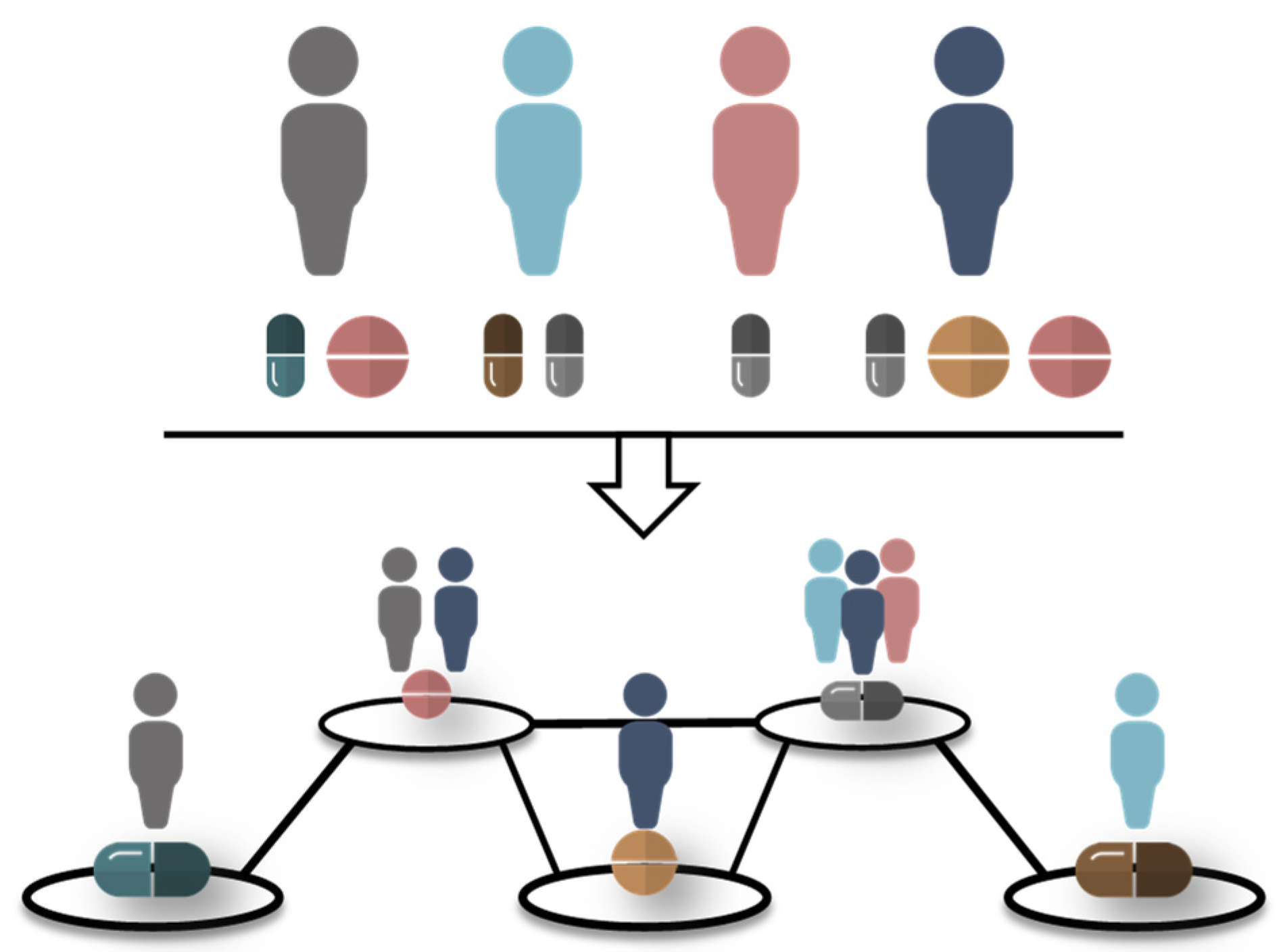
2.2. Drug Prescription Networks
2.3. Analysis of Drug Prescription Networks
2.4. Software
3. Results
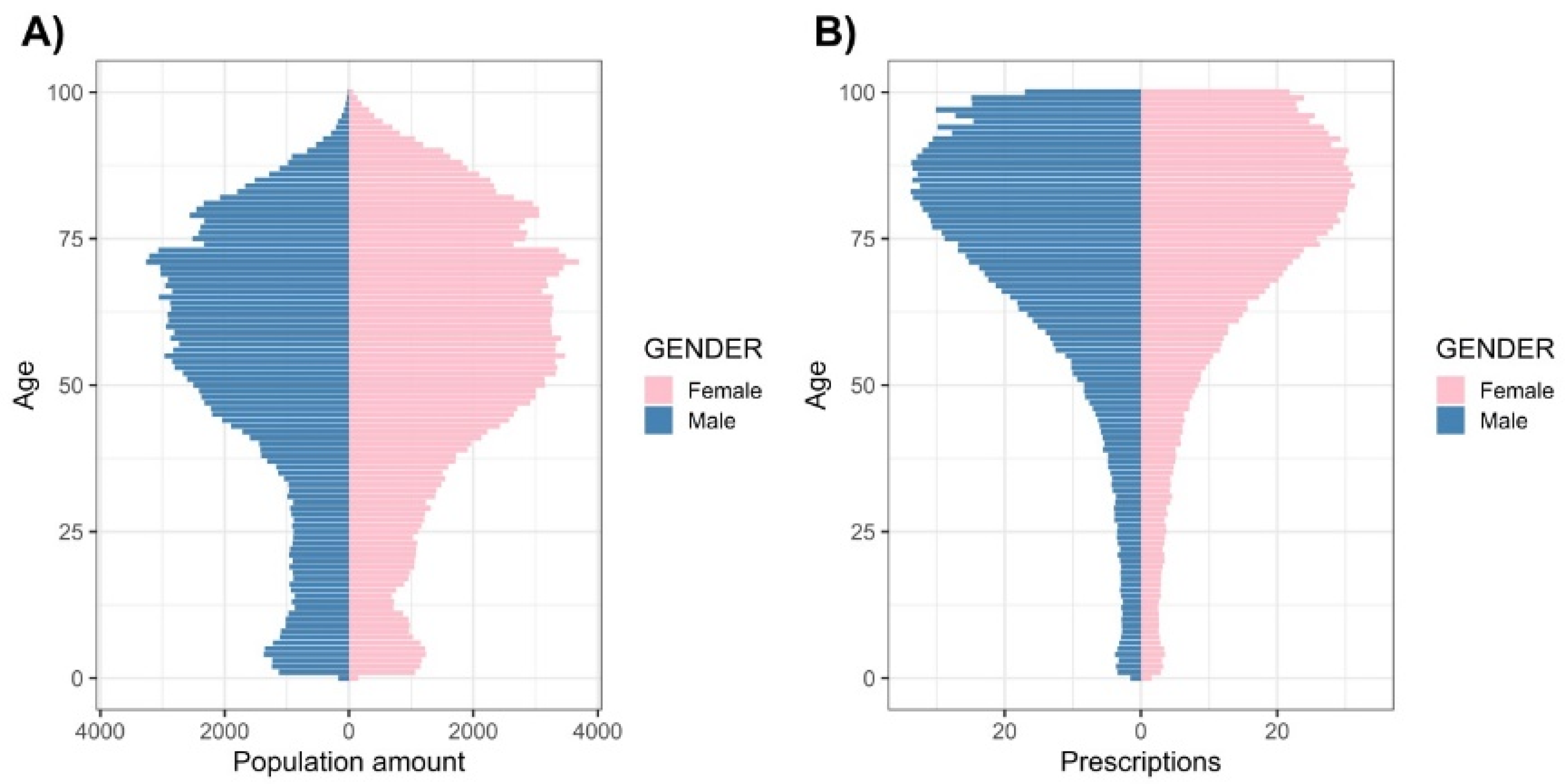
3.1. Study Population
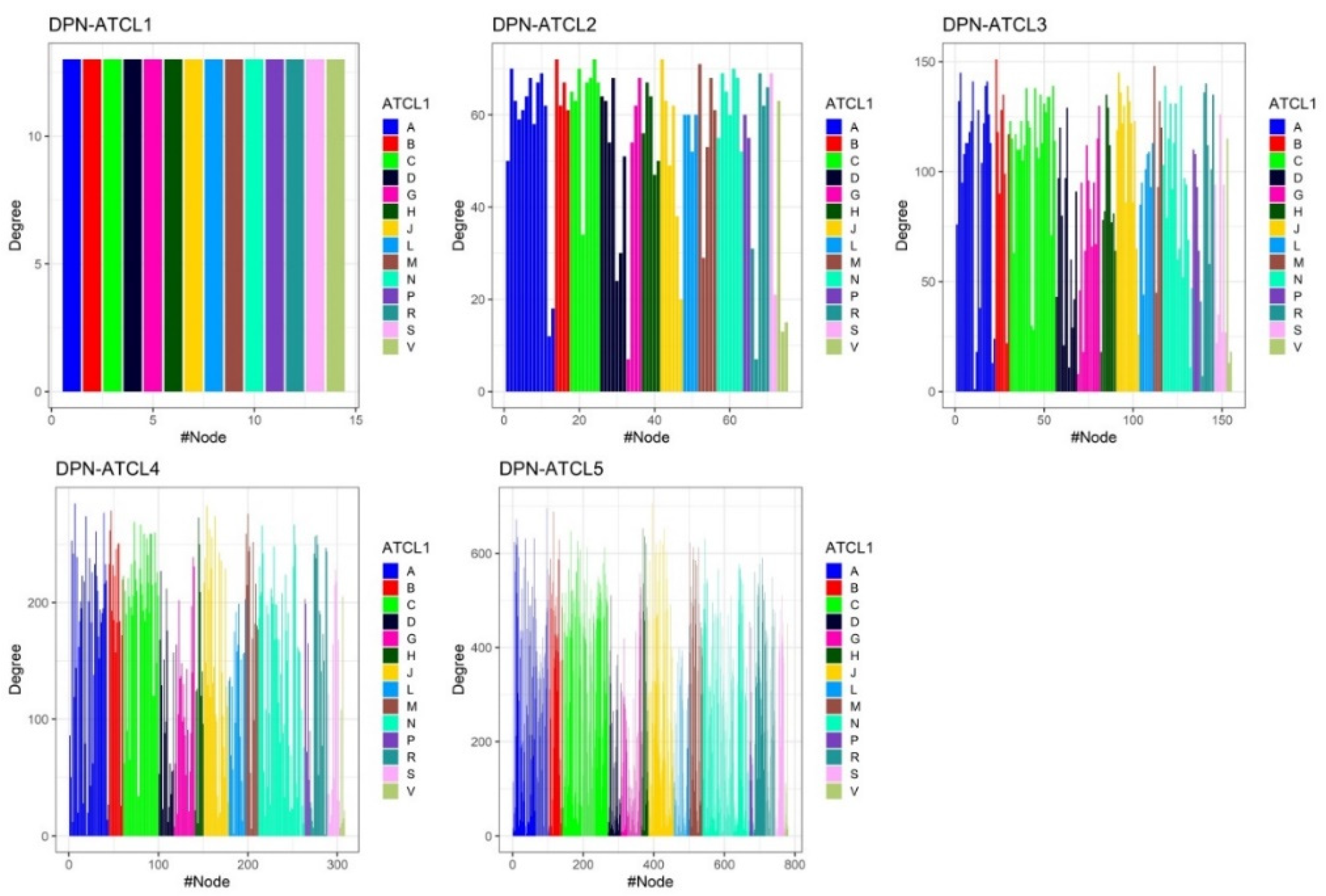
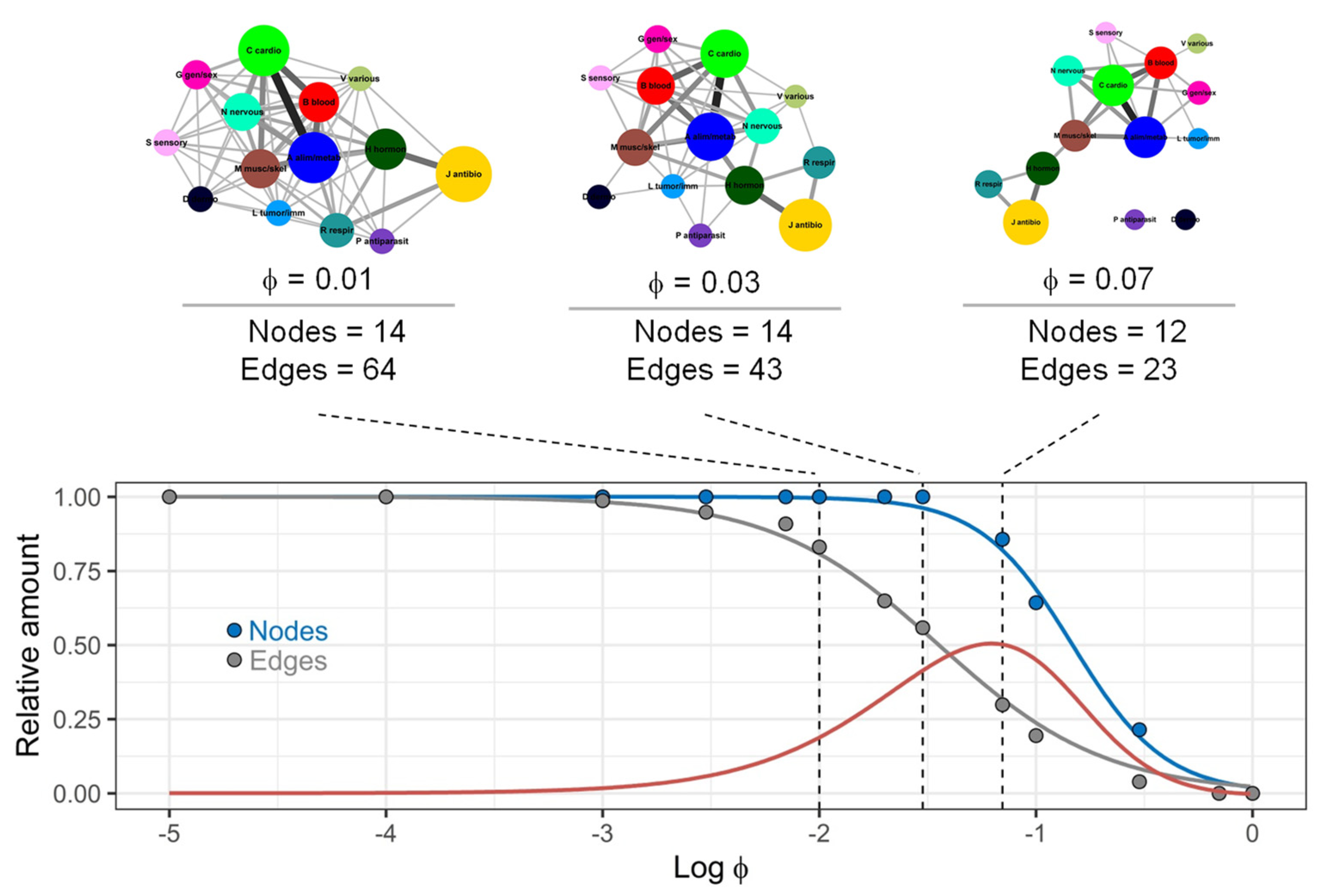
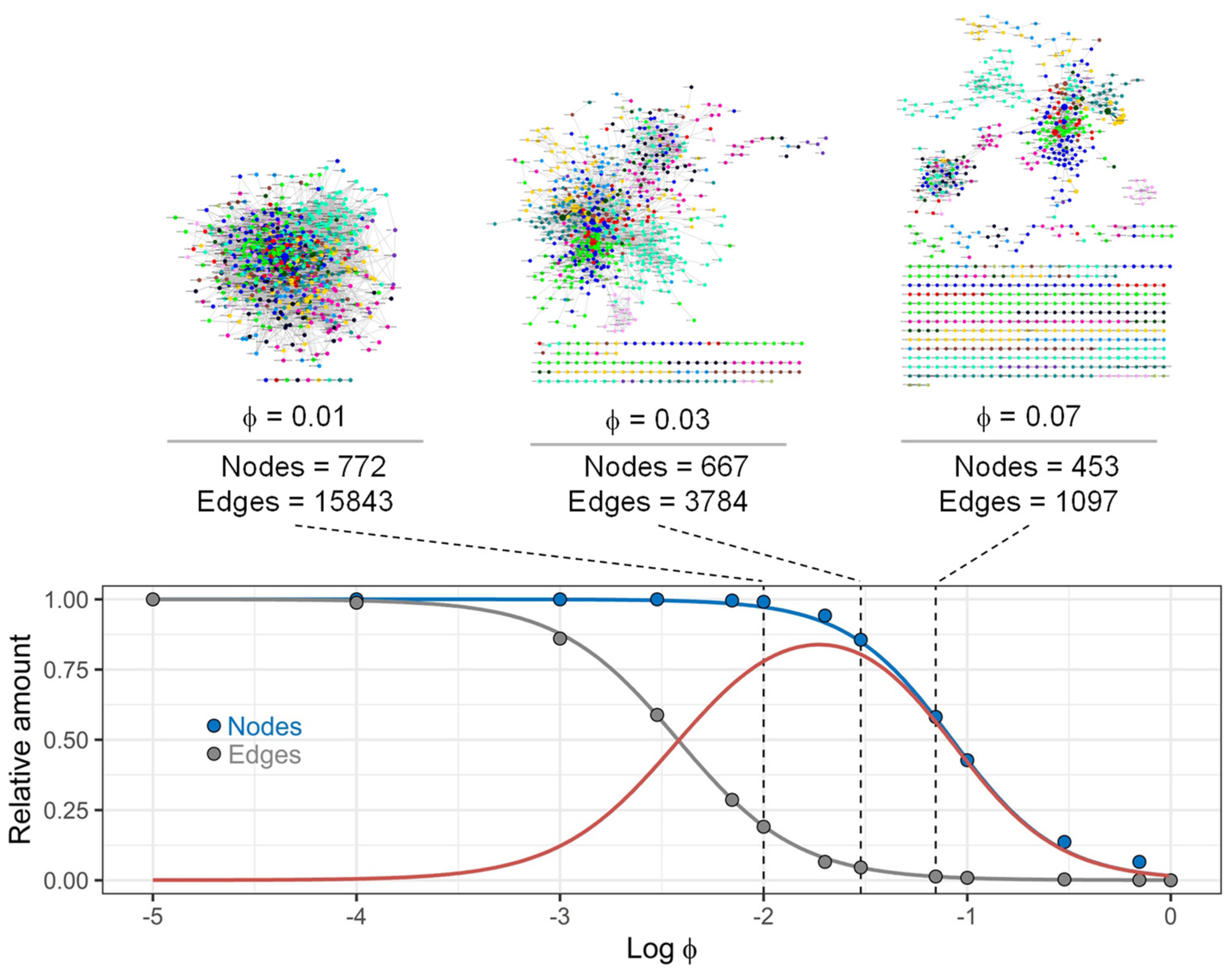
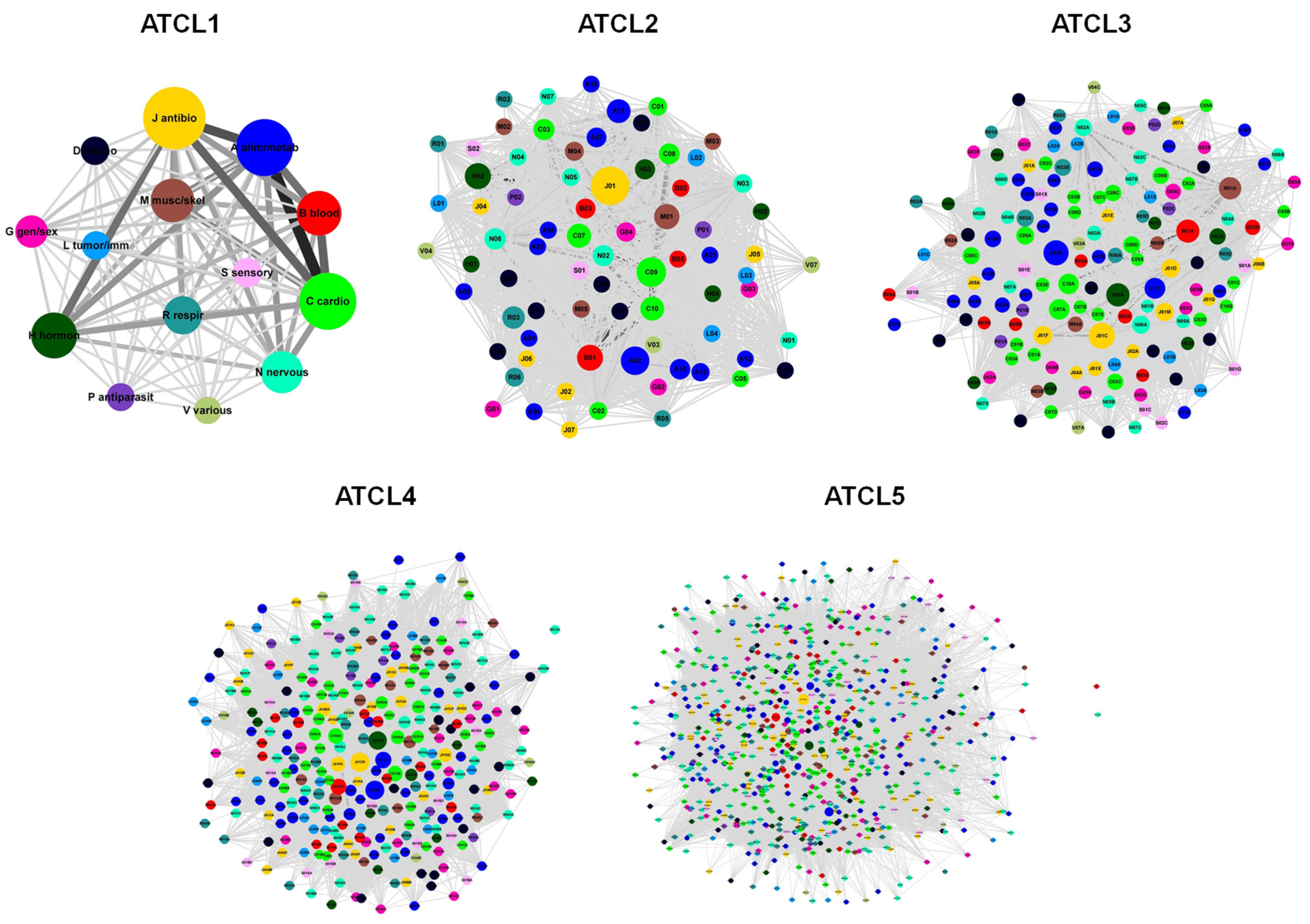
3.2. Structure of Drug Prescription Networks
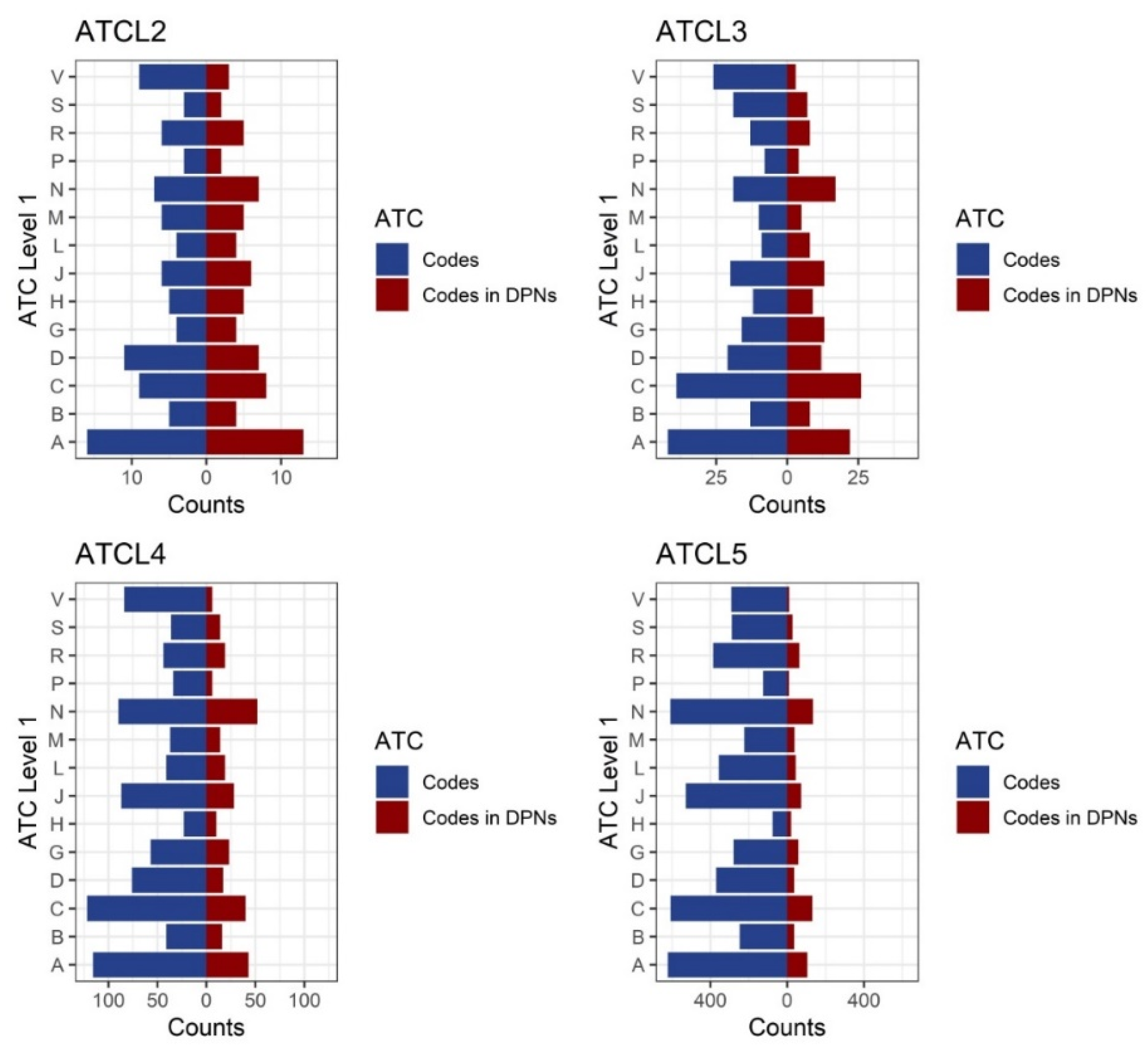
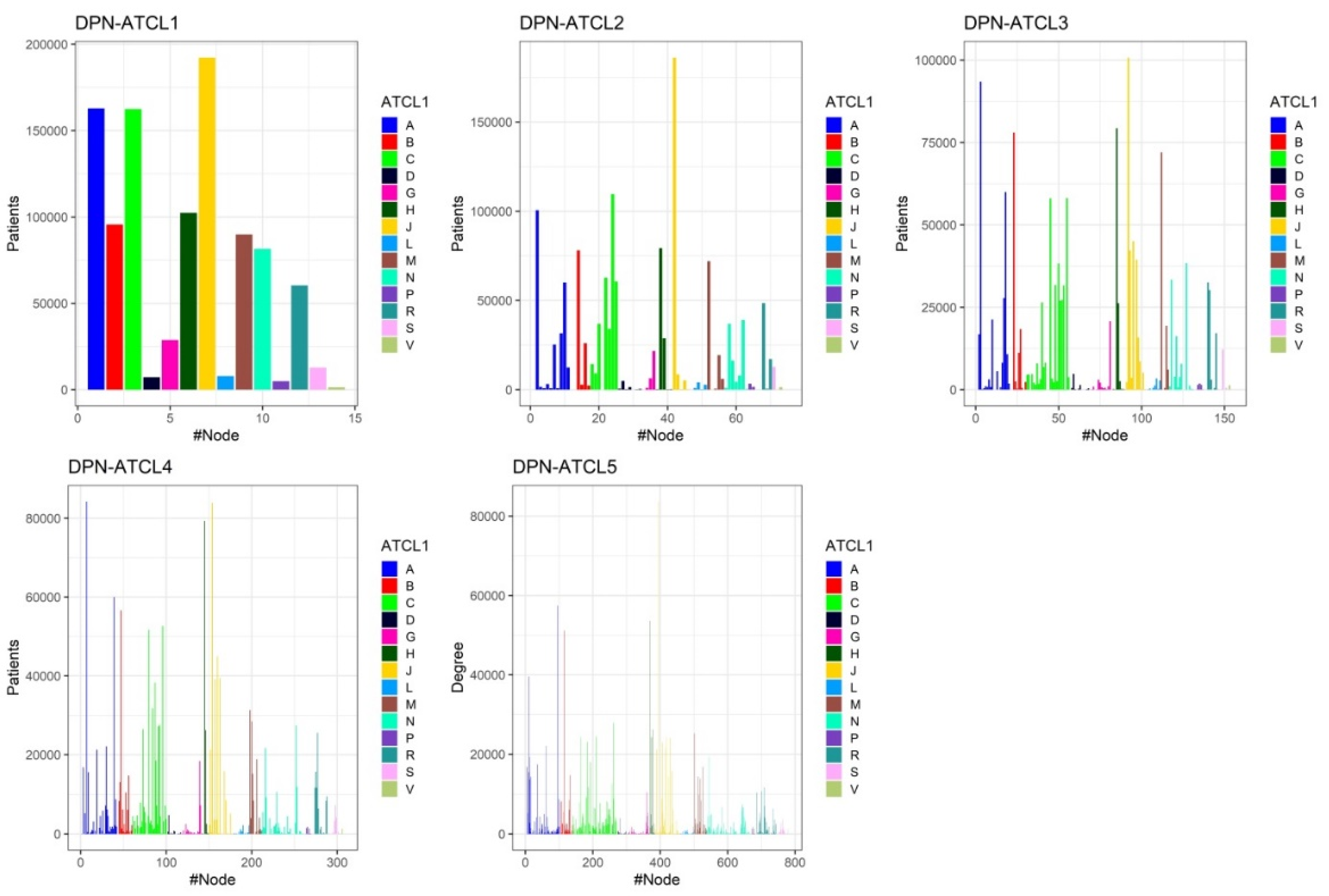
3.3. Structure of Drug Prescription Networks and Popularity of Drug (Co)Prescription
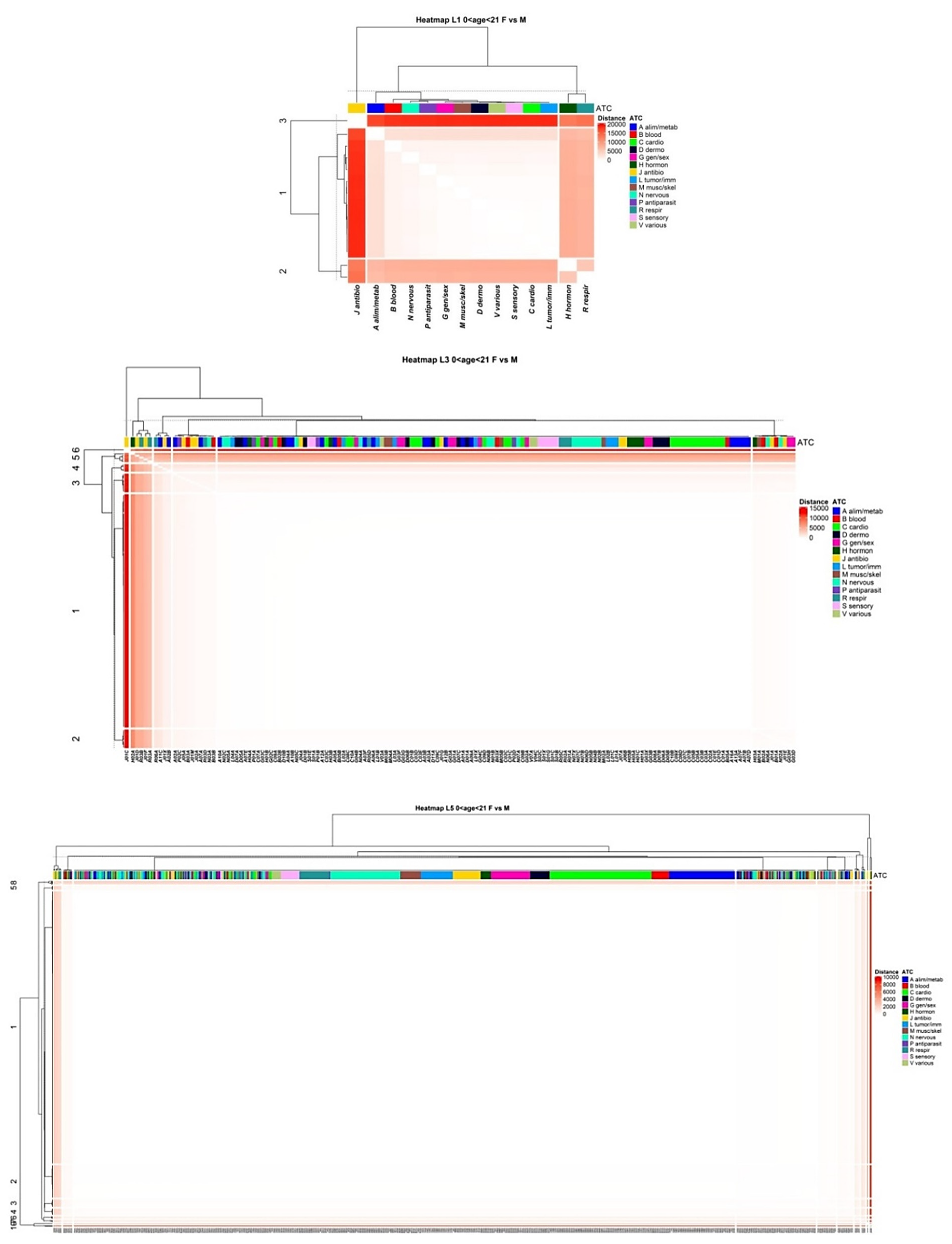
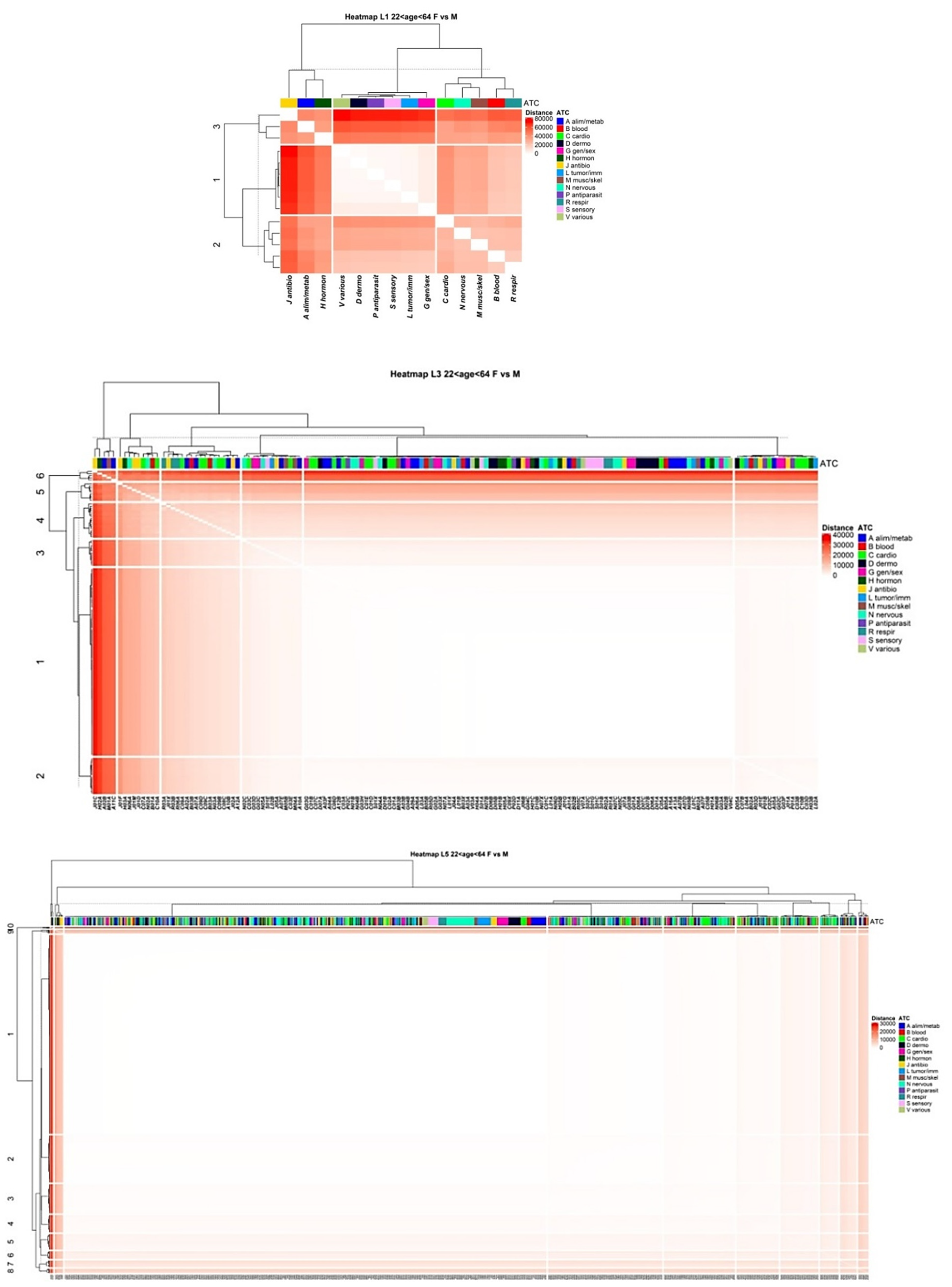
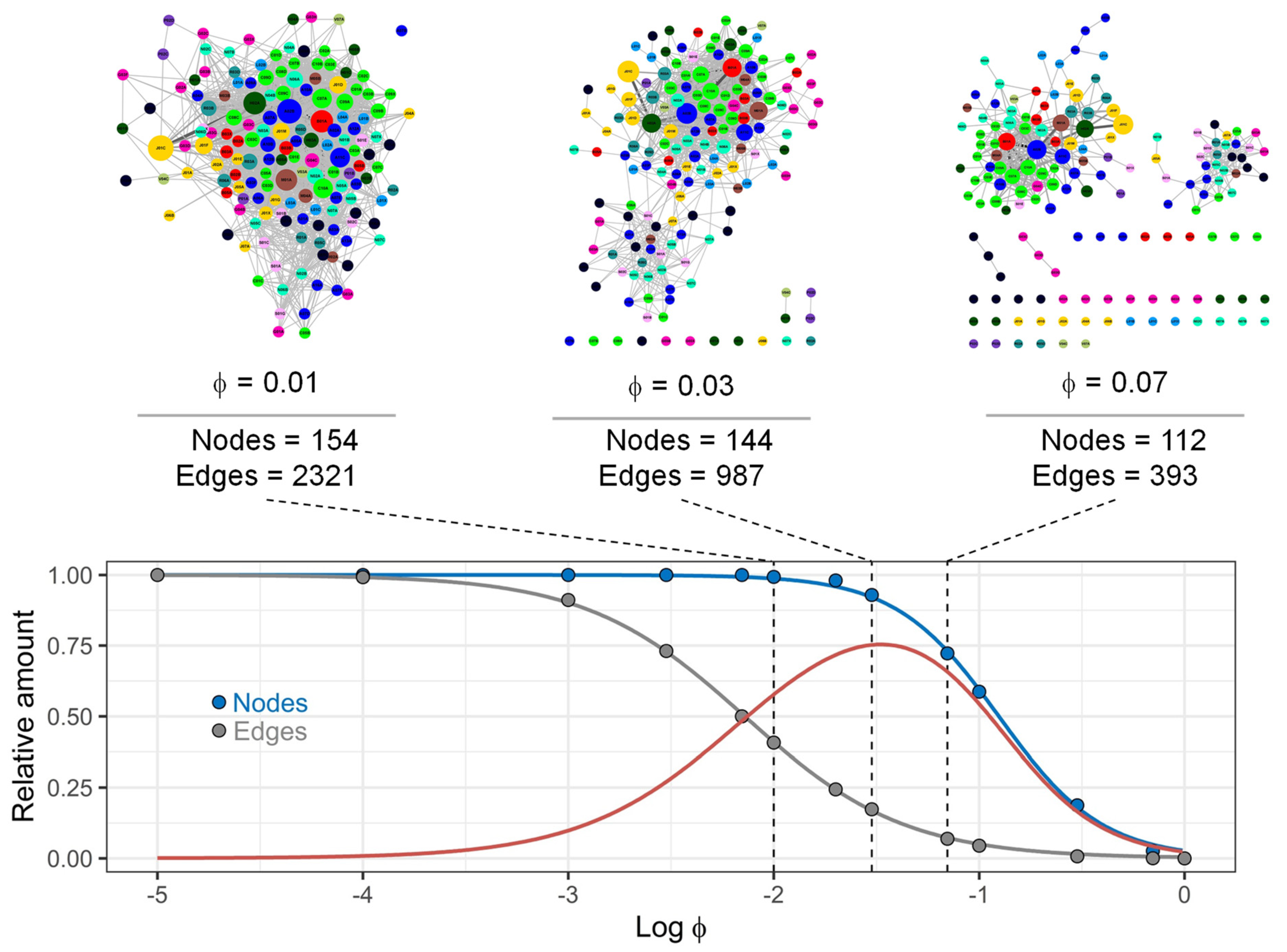
3.4. Comparison of Drug Prescription Networks
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Statistical and Network Measures Adopted in the Study
Appendix A.1.1. Degree
Appendix A.1.2. Betweenness Centrality
Appendix A.1.3. Closeness Centrality
Appendix A.1.4. Network Density
Appendix A.1.5. Network Assortativity
Appendix A.1.6. The Pearson’s Correlation Coefficient for Binary Variables
Appendix A.1.7. Difference of the Networks (Euclidean Distance)
Appendix A.2. The Anatomical Therapeutic Chemical (ATC) Classification System
- A—Alimentary tract and metabolism;
- B—Blood and blood forming organs;
- C—Cardiovascular system;
- D—Dermatologicals;
- G—Genito-urinary system and sex hormones;
- H—Systemic hormonal preparations, excluding sex hormones and insulins;
- J—Anti-infectives for systemic use;
- L—Antineoplastic and immunomodulating agents;
- M—Musculo-skeletal system;
- N—Nervous system;
- P—Antiparasitic products, insecticides and repellents;
- R—Respiratory system;
- S—Sensory organs;
- V—Various.
- (see https://www.whocc.no/atc/structure_and_principles/, accessed on 25 February 2021).

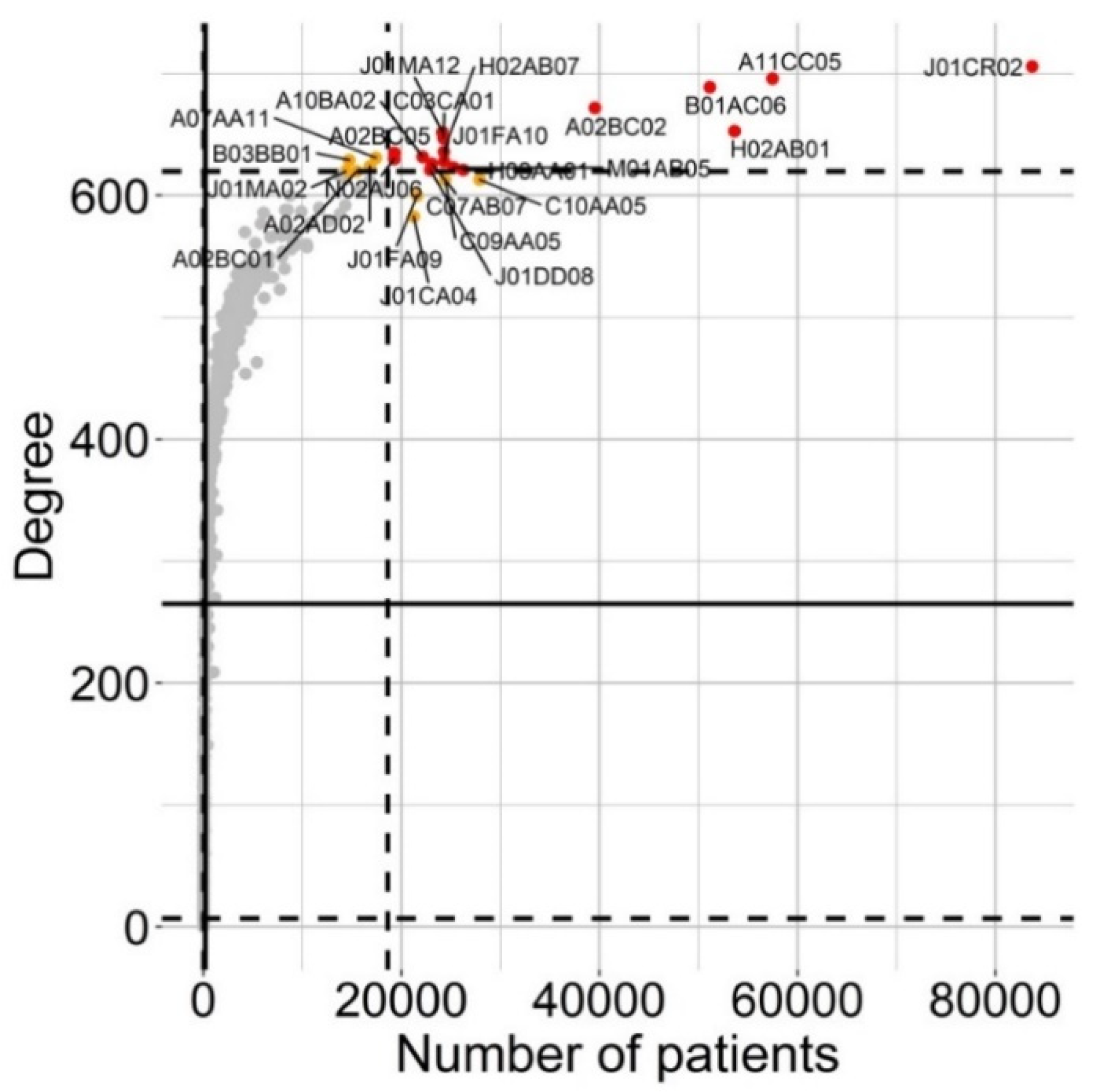
| ATC | Age Strata (Years) | ||
|---|---|---|---|
| 0–21 | 22–64 | 65–100 | |
| L1 | J | J, A and H | A and C |
| L3 | J01C | J01C, H02A, A02B, M01A and A11C | A02B, B01A, A11C, C07A, C10A and M01A |
| L5 | J01CR02 and H02AB01 | J01CR02, A11CC05 and H02AB01 | A11CC05 and B01AC06 |
References
- Nielsen, C.R.; Halling, A.; Andersen-Ranberg, K. Disparities in multimorbidity across Europe—Findings from the SHARE Survey. Eur. Geriatr. Med. 2017, 8, 16–21. [Google Scholar] [CrossRef]
- OECD. Health at a Glance 2019; OECD: Paris, France, 2019. [Google Scholar] [CrossRef]
- Ma, C.; Smith, H.W.; Chu, C.; Juarez, D.T. Big data in pharmacy practice: Current use, challenges, and the future. Integr. Pharm. Res. Pract. 2015, 4, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Dalton, K.; Byrne, S. Role of the pharmacist in reducing healthcare costs: Current insights. Integr. Pharm. Res. Pract. 2017, 6, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Hallas, J. Drug utilization statistics for individual-level pharmacy dispensing data. Pharmacoepidemiol. Drug Saf. 2005, 14, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Euler, L. Solutio problematis ad geometriam situs pertinentis. Comment. Acad. Sci. Petropolitanae 1736, 8, 128–140. [Google Scholar]
- Estrada, E. The Structure of Complex. Networks: Theory and Applications; Oxford University Press: Oxford, UK, 2011. [Google Scholar] [CrossRef]
- Barabási, A.L. Network Science; Cambridge University Press: Cambridge, UK, 2016. [Google Scholar]
- Latora, V.; Nicosia, V.; Russo, G. Complex Networks; Cambridge University Press: Cambridge, MA, USA, 2017. [Google Scholar] [CrossRef]
- Cavallo, P.; Pagano, S.; Boccia, G.; De Caro, F.; De Santis, M.; Capunzo, M. Network analysis of drug prescriptions. Pharmacoepidemiol. Drug Saf. 2013, 22, 130–137. [Google Scholar] [CrossRef]
- Bazzoni, G.; Marengoni, A.; Tettamanti, M.; Franchi, C.; Pasina, L.; Djade, C.D.; Fortino, I.; Bortolotti, A.; Merlino, L.; Nobili, A. The drug prescription network: A system-level view of drug co-prescription in community-dwelling elderly people. Rejuvenation Res. 2015, 18, 153–161. [Google Scholar] [CrossRef]
- WHO Collaborating Centre for Drug Statistics Methodology. Guidelines for ATC Classification and DDD Assignment 2020. Available online: https://www.whocc.no/filearchive/publications/2020_guidelines_web.pdf (accessed on 25 February 2021).
- Newman, M.E.J. Assortative Mixing in Networks. Phys. Rev. Lett. 2002, 89, 208701. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.E.J. Mixing patterns in networks. Phys. Rev. E 2003, 67, 26126. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, C.A.; Blumm, N.; Barabási, A.L.; Christakis, N.A. A dynamic network approach for the study of human phenotypes. PLoS Comput. Biol. 2009, 5, e1000353. [Google Scholar] [CrossRef] [PubMed]
- Tantardini, M.; Ieva, F.; Tajoli, L.; Piccardi, C. Comparing methods for comparing networks. Sci. Rep. 2019, 9, 17557. [Google Scholar] [CrossRef] [PubMed]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Gustavsen, J.A.; Pai, S.; Isserlin, R.; Demchak, B.; Pico, A.R. RCy3: Network biology using Cytoscape from within R. F1000Research 2019, 8, 1774. [Google Scholar] [CrossRef] [PubMed]
- Landon, B.E.; Keating, N.L.; Barnett, M.L.; Onnela, J.-P.; Paul, S.; O’Malley, A.J.; Keegan, T.; Christakis, N.A. Variation in patient-sharing networks of physicians across the United States. JAMA 2012, 308, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, K.; Currim, F.; Ram, S. Predicting high-cost patients at point of admission using network science. IEEE J. Biomed. Health Inform. 2018, 22, 1970–1977. [Google Scholar] [CrossRef] [PubMed]
- Brunson, J.C.; Laubenbacher, R.C. Applications of network analysis to routinely collected health care data: A systematic review. J. Am. Med. Inform. Assoc. 2018, 25, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Geissler, K.H.; Lubin, B.; Ericson, K.M.M. The association between patient sharing network structure and healthcare costs. PLoS ONE 2020, 15, e0234990. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Gallagher, M.; Loveday, W.; Dev, A.; Connor, J.P. Network analysis and visualisation of opioid prescribing data. IEEE J. Biomed. Health Inform. 2020, 24, 1447–1455. [Google Scholar] [CrossRef] [PubMed]
- Bianconi, G. Multilayer Networks: Structure and Function; Oxford University Press: Oxford UK, 2018. [Google Scholar] [CrossRef]

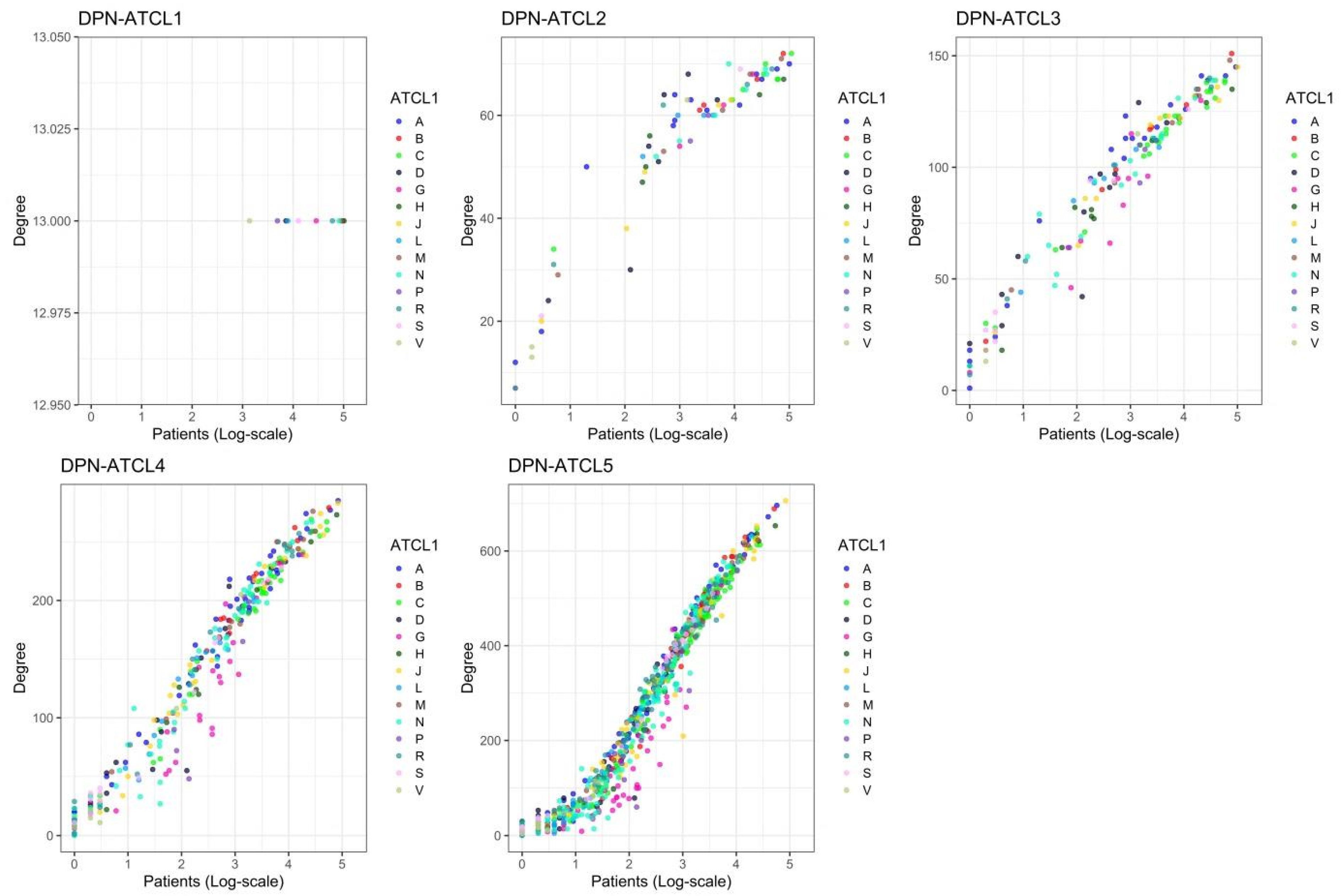

| ATCL 1 | 2 | 3 | 4 | [CV%] 5 | [CV%] 6 | [CV%] | 7 | |
|---|---|---|---|---|---|---|---|---|
| 1 | 14 | 2.639 | 91 | 1.000 | 13.000 [0.0] | 5.571 [374.2] | 0.876 [41.5] | NA 8 |
| 2 | 75 | 4.317 | 2053 | 0.740 | 54.747 [32.2] | 51.839 [140.4] | 50.444 [11.7] | −0.194 |
| 3 | 155 | 5.043 | 7191 | 0.603 | 92.787 [42.9] | 91.999 [104.4] | 26.907 [7.9] | −0.189 |
| 4 | 308 | 5.731 | 22,747 | 0.481 | 147.708 [55.9] | 173.130 [85.3] | 9.712 [7.2] | −0.264 |
| 5 | 781 | 6.661 | 104,063 | 0.342 | 266.487 [72.9] | 423.840 [74.0] | 2.984 [6.4] | −0.332 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miglio, G.; Basso, L.; Armando, L.G.; Traina, S.; Benetti, E.; Diarassouba, A.; Baroetto Parisi, R.; Esiliato, M.; Rolando, C.; Remani, E.; et al. A Network Approach for the Study of Drug Prescriptions: Analysis of Administrative Records from a Local Health Unit (ASL TO4, Regione Piemonte, Italy). Int. J. Environ. Res. Public Health 2021, 18, 4859. https://doi.org/10.3390/ijerph18094859
Miglio G, Basso L, Armando LG, Traina S, Benetti E, Diarassouba A, Baroetto Parisi R, Esiliato M, Rolando C, Remani E, et al. A Network Approach for the Study of Drug Prescriptions: Analysis of Administrative Records from a Local Health Unit (ASL TO4, Regione Piemonte, Italy). International Journal of Environmental Research and Public Health. 2021; 18(9):4859. https://doi.org/10.3390/ijerph18094859
Chicago/Turabian StyleMiglio, Gianluca, Lara Basso, Lucrezia G. Armando, Sara Traina, Elisa Benetti, Abdoulaye Diarassouba, Raffaella Baroetto Parisi, Mariangela Esiliato, Cristina Rolando, Elisa Remani, and et al. 2021. "A Network Approach for the Study of Drug Prescriptions: Analysis of Administrative Records from a Local Health Unit (ASL TO4, Regione Piemonte, Italy)" International Journal of Environmental Research and Public Health 18, no. 9: 4859. https://doi.org/10.3390/ijerph18094859
APA StyleMiglio, G., Basso, L., Armando, L. G., Traina, S., Benetti, E., Diarassouba, A., Baroetto Parisi, R., Esiliato, M., Rolando, C., Remani, E., & Cena, C. (2021). A Network Approach for the Study of Drug Prescriptions: Analysis of Administrative Records from a Local Health Unit (ASL TO4, Regione Piemonte, Italy). International Journal of Environmental Research and Public Health, 18(9), 4859. https://doi.org/10.3390/ijerph18094859







