Machine and Deep Learning towards COVID-19 Diagnosis and Treatment: Survey, Challenges, and Future Directions
Abstract
1. Introduction
2. Medical Image Inception Using AI-Based ML and DL for the Detection of COVID-19
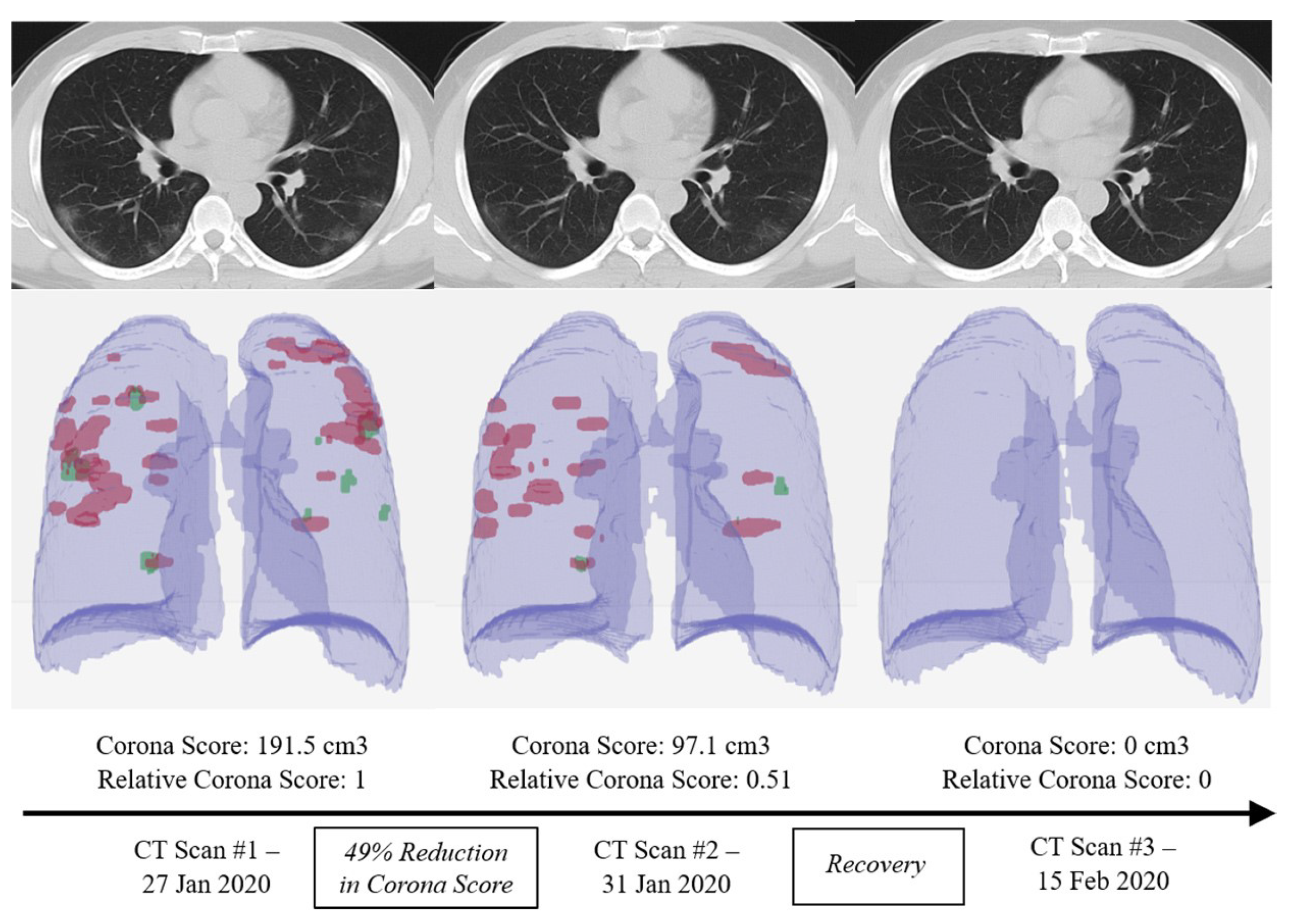
2.1. Chest CT Image Detection
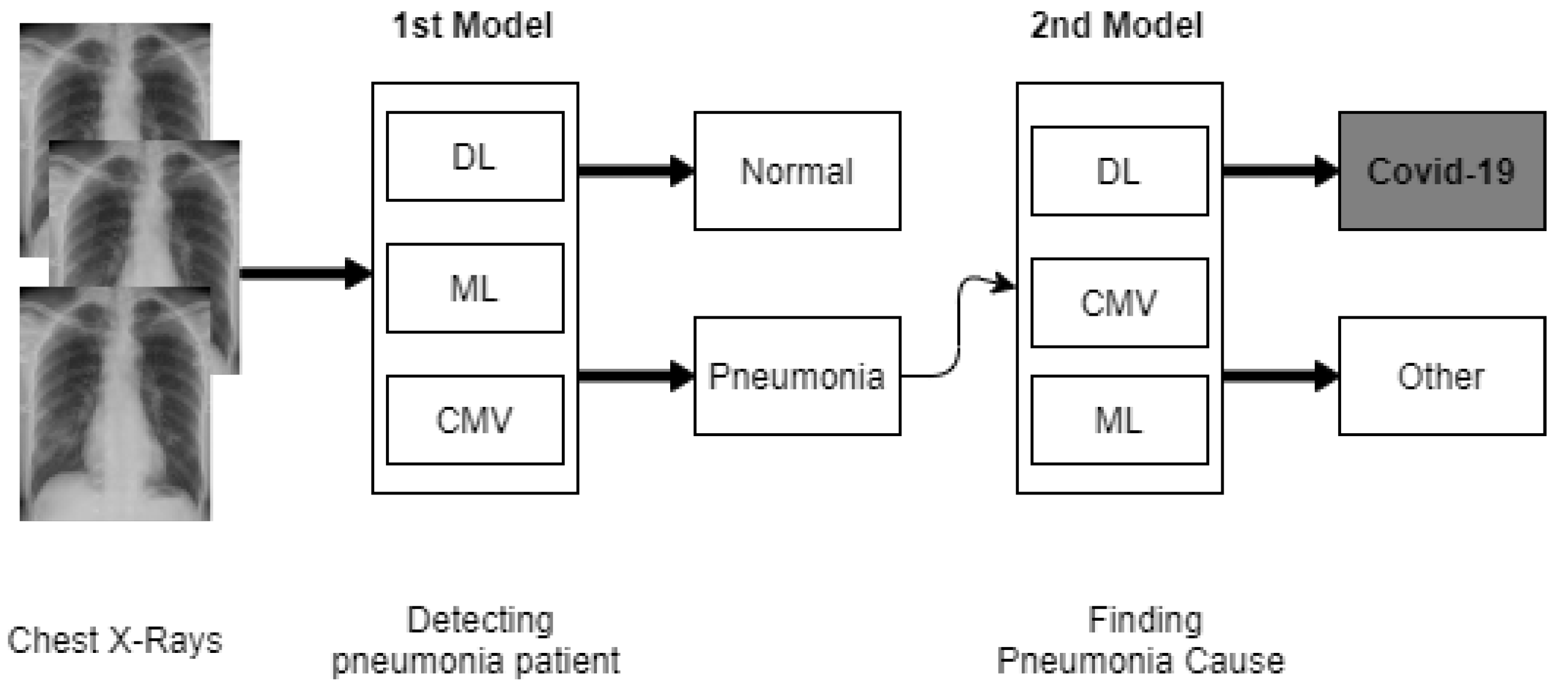
2.2. Chest X-ray Image Detection
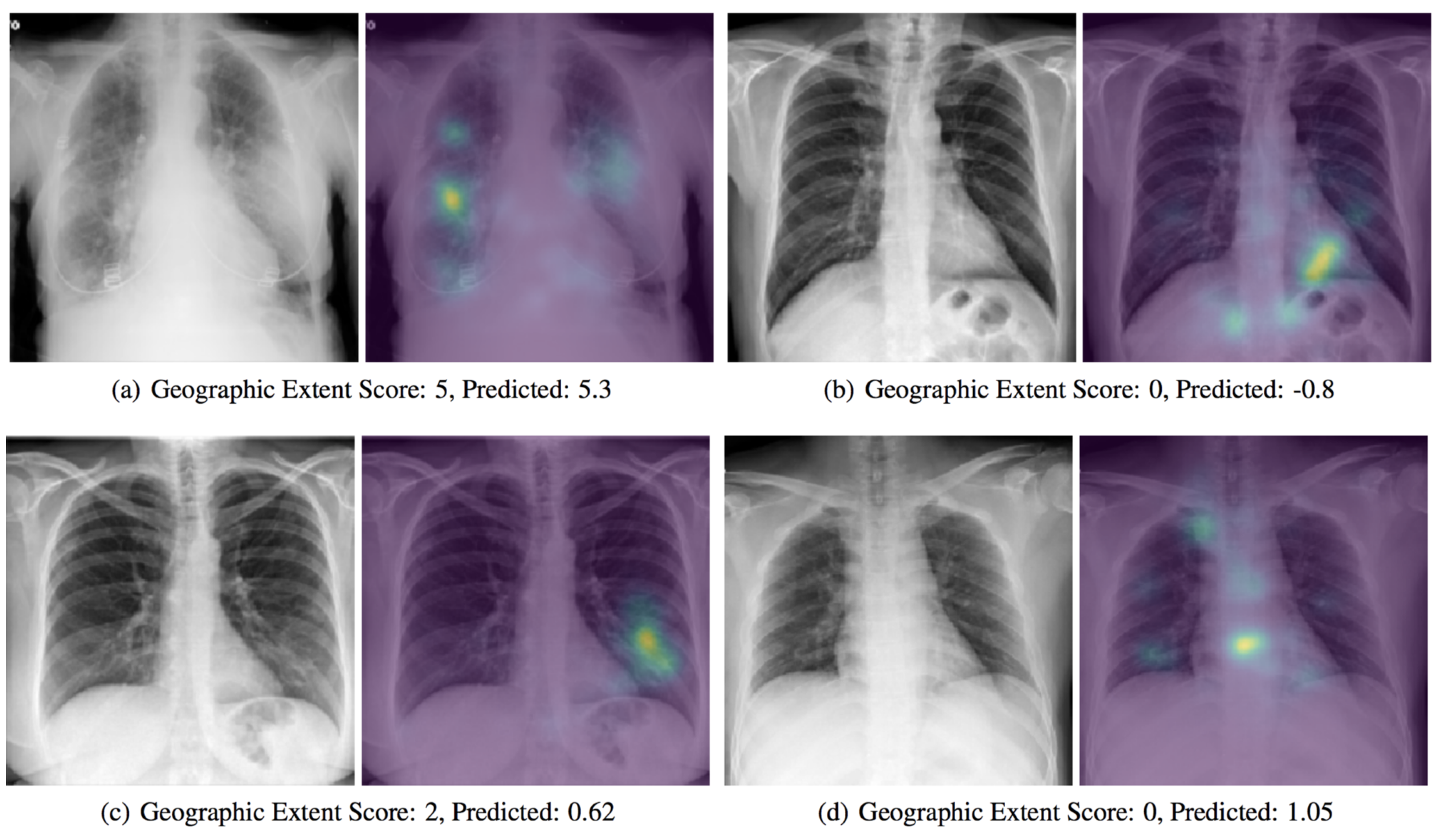
2.3. COVID-19 Severity Classification Using Chest X-ray with Deep Learning
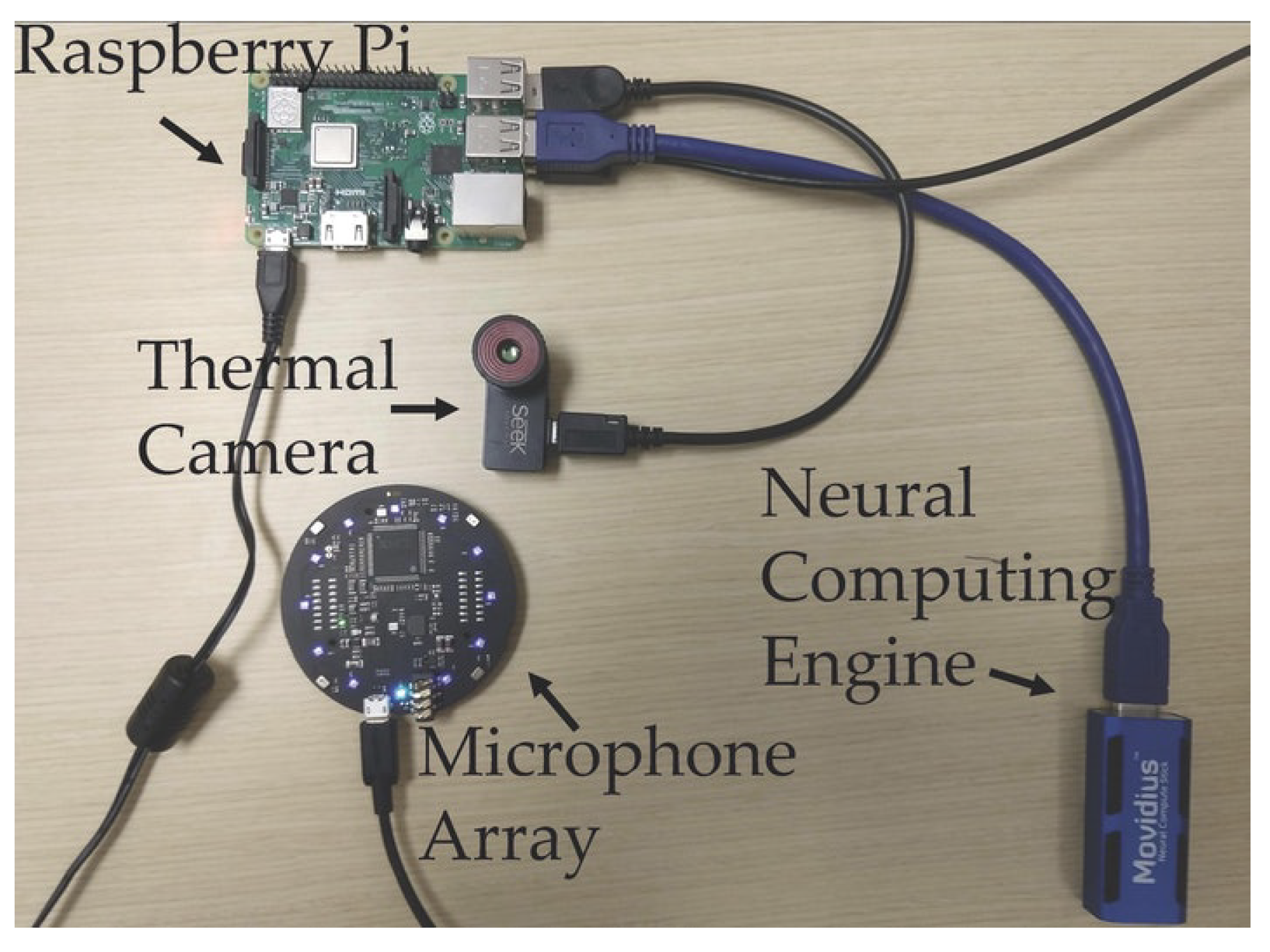
2.4. Observing COVID-19 Through AI-Based Cough Sound Analysis
- Separating positive users of COVID-19 from negative users.
- Separate COVID-19 cough users from healthy cough users.
- Separate COVID-19 cough users from asthma users who have declared coughing.
2.5. COVID-19 Diagnosis Based on Non-Invasive Measurements
3. ML and DL for Drug and Vaccine Development
3.1. ML and DL For Vaccine Development
3.2. Drug Development
- Drug discovery and development.
- Drug pre-clinical research.
- Drug clinical research.
- FDA drug review.
- FDA drug after-market safety control and development.
- Compound identification.
- Compound acquisition.
- Drug clinical research.
- FDA drug after-market safety control and development.
- Training only compound representation.
- Training only protein sequence representation.
- Training both protein representation and compound representations.
4. Challenges and Future Directions
4.1. Challenges
- Regulation. As the pandemic increases and maximizes the regular number of confirmed infected and deceased cases, numerous steps have been considered to contain this pandemic, e.g., Lockdown and social distancing. At the time of pandemic, authorities play a key part in identifying regulations and policies that can promote the participation of citizens, researchers, scientists, business owners, medical centers, technology giants, and major corporations to avert any barrier to COVID-19 prevention.
- Scarcity and unavailability of large-scale training data. Many AI-based DL techniques depend on large-scale training data, including medical imaging and different details of the environment. However, there are inadequate datasets available for AI, because of the rapid explosion of COVID-19. In reality, interpreting training samples is time-consuming and it may need trained medical professionals.
- Noisy data and online rumors. The challenges emerge from depending on portable online social media; without any significant changes, large audio information and false reports about COVID-19 have been reported in different online outlets. However, in judging and filtering audio and error data, AI-based ML and DL algorithms appear to be slow. Additionally, with the usage of noisy data, the results of the AI-based ML and DL algorithms become biased. The usage, functionality, and performance of AI-based methods are reduced by this issue, particularly in pandemic predictions and spread analysis.
- Lacking in the intersection of computer science and medicine fields. Most AI-based ML and DL researchers come from computer science major, but strong specialization in medical imaging, bioinformatics, virology, and many other related fields is also required to involve other medical knowledge for the use of ML and DL in the COVID-19 war. To deal with COVID-19, it is therefore important to organize the collaboration of experts from various fields and incorporate the information of multiple studies.
- Data privacy and protection. The cost of collecting personal privacy data in the age of big data and AI is very weak. Many governments aim to collect a range of personal information, including ID number, contact number, personal data, and medical data, in the face of public health issues, such as COVID-19. A problem worth addressing is how to efficiently preserve your privacy and human rights throughout AI-based discovery and processing.
- Incorrect structural and unstructural data (e.g., image, text, and numerical data). Having an ambiguous and incorrect information in text descriptions can be a challenge. Huge quantities of information from different sources can be incorrect. Additionally, excess data makes it impossible for valuable pieces of information to be extracted.
- COVID-19 early diagnosis using medical imaging e.g., Chest X-ray and CT scan. Dealing with unbalanced datasets due to inadequate medical imaging and long training period knowledge from COVID-19 and unable to clarify the challenges of the findings.
- Screen and triage patients, find functional therapies and cures, risk assessments, survival predictions, health care, and medical resource planning. The challenge is to collect physical attributes and treatment results for patients. Another difficulty is to deal with poor quality data that can lead to forecasts that are biased and unreliable.
4.2. Future Research Direction
- Detection of non-contact disease. The use of automated image classification in X-ray and CT imaging will effectively prevent the chance of disease transmissions from patients among radiologists during COVID-19 epidemics. For patient pose, the detection of X-ray and CT images, and camera facilities, AI-based ML and DL systems can be involved.
- Remote video diagnosis and consultations. It is possible to use the combination of AI and Natural Language Processing (NLP) techniques to build remote video diagnostic programs and robot systems in order to provide COVID-19 patient visits and first group diagnoses.
- Biological research. AI-based ML and DL systems can be used in the context of biological research to identify protein composition and viral factors via accurate biomedical knowledge analysis, such as significant protein structures, genetic sequences, and viral trajectories.
- Drug development and vaccination. AI-based ML and DL systems can not only be applied to identify possible drugs and vaccines, but they can also be used to mimic drug-protein and vaccine-receptor interactions, thus predicting future drug and vaccine reactions for people with various COVID-19 patients.
- Identification and screening of fake information. AI-based ML and DL systems can be applied to minimize and delete false news and audio data on internet forums in order to provide credible, factual, and scientific information about the COVID-19 outbreak.
- Impact assessment and evaluation. In order to analyse the effect of various social control modes on disease transmission, different types of simulations may use AI-based ML and DL systems. They can then be used to assess reliable and scientific approaches for disease control and prevention in the population.
- Patient contact tracking. AI-based ML and DL system can monitor and track the characteristics of people neighboring to patients with COVID-19 by creating social networks and knowledge graphs, thereby accurately predicting and monitoring the potential spread of the disease.
- Smart robots. In programs, such as sanitation in public areas, product delivery, and patient treatment without the need for human resources, intelligent robots are supposed to be used. This will stop the spreading of the virus from COVID-19.
- Future work with descriptive AI-based ML and DL techniques. The efficacy of deep learning models and graphical characteristics that lead to the differences between COVID-19 and other forms of pneumonia needs to be clarified. This will help radiologists and practitioners to gain an awareness of the virus and accurately analyse the possible X-ray and CT scans of the Coronavirus.
- COVID-19 diagnosis and treatment, which one is important? Both are essential, but finding a cure for COVID-19 is definitely more essential. We find that most of the existing AI-based ML and DL strategies are more centered on identifying COVID-19 from the existing literature in this survey. In order to find the COVID-19 treatment, more future research work based on ML and DL is required.
5. Final Comments
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Soghaier, M.A.; Saeed, K.M.; Zaman, K.K. Public Health Emergency of International Concern (PHEIC) has declared twice in 2014; polio and Ebola at the top. AIMS Public Health 2015, 2, 218. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Statistical modeling: The two cultures (with comments and a rejoinder by the author). Stat. Sci. 2001, 16, 199–231. [Google Scholar] [CrossRef]
- European Society of Medical Imaging Informatics. Automated Diagnosis and Quantitative Analysis of COVID-19 on Imaging. 2020. Available online: https://imagingcovid19ai.eu/ (accessed on 1 September 2020).
- Mei, X.; Lee, H.C.; Diao, K.Y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Physics World. AI Checks CT Scans for COVID-19. 2020. Available online: https://physicsworld.com/a/ai-checks-ct-scans-for-covid-19/ (accessed on 1 September 2020).
- Wang, S.; Kang, B.; Ma, J.; Zeng, X.; Xiao, M.; Guo, J.; Cai, M.; Yang, J.; Li, Y.; Meng, X.; et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). medRxiv 2020. [Google Scholar] [CrossRef]
- ITN. CT Provides Best Diagnosis for Novel Coronavirus (COVID-19). 2020. Available online: https://www.itnonline.com/content/ct-provides-best-diagnosis-novel-coronavirus-covid-19 (accessed on 20 September 2020).
- Zhang, K.; Liu, X.; Shen, J.; Li, Z.; Sang, Y.; Wu, X.; Zha, Y.; Liang, W.; Wang, C.; Wang, K.; et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of covid-19 pneumonia using computed tomography. Cell 2020, 181, 1423–1433.e11. [Google Scholar] [CrossRef]
- VIDA. LungPrint Clinical Solutions. 2020. Available online: https://vidalung.ai/clinical_solutions/ (accessed on 14 October 2020).
- Analytics, H.I. Deep Learning Models Can Detect COVID-19 in Chest CT Scans. 2020. Available online: https://healthitanalytics.com/news/deep-learning-models-can-detect-covid-19-in-chest-ct-scans (accessed on 3 September 2020).
- Chen, J.; Wu, L.; Zhang, J.; Zhang, L.; Gong, D.; Zhao, Y.; Hu, S.; Wang, Y.; Hu, X.; Zheng, B.; et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: A prospective study. medRxiv 2020. [Google Scholar] [CrossRef]
- Huang, L.; Han, R.; Ai, T.; Yu, P.; Kang, H.; Tao, Q.; Xia, L. Serial quantitative chest ct assessment of covid-19: Deep-learning approach. Radiol. Cardiothorac. Imaging 2020, 2, e200075. [Google Scholar] [CrossRef]
- Qi, X.; Jiang, Z.; Yu, Q.; Shao, C.; Zhang, H.; Yue, H.; Ma, B.; Wang, Y.; Liu, C.; Meng, X.; et al. Machine learning-based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: A multicenter study. medRxiv 2020. [Google Scholar] [CrossRef]
- Pandit, M.K.; Banday, S.A. SARS n-CoV2-19 detection from chest X-ray images using deep neural networks. Int. J. Pervasive Comput. Commun. 2020, 16, 419–427. [Google Scholar] [CrossRef]
- Basu, S.; Mitra, S. Deep Learning for Screening COVID-19 using Chest X-ray Images. arXiv 2020, arXiv:2004.10507. [Google Scholar]
- Scudellari, M. Hospitals Deploy AI Tools to Detect COVID-19 on Chest Scans. 2020. Available online: https://spectrum.ieee.org/the-human-os/biomedical/imaging/hospitals-deploy-ai-tools-detect-covid19-chest-scans (accessed on 10 September 2020).
- BBVA. Artificial Intelligence to detect COVID-19 in Less than a Second Using X-rays. 2019. Available online: https://www.bbva.com/en/artificial-intelligence-to-detect-covid-19-in-less-than-a-second-using-x-rays/ (accessed on 6 September 2020).
- Cranfield University. Using Artificial Intelligence to Detect COVID-19 in X-rays. 2020. Available online: https://medicalxpress.com/news/2020-05-artificial-intelligence-covid-x-rays.html (accessed on 11 September 2020).
- Cranfield University. Using Machine Learning to Detect COVID-19 in X-rays. 2020. Available online: https://tectales.com/ai/using-ai-to-detect-covid-19-in-x-rays.html (accessed on 11 September 2020).
- Cranfield University. Artificial Intelligence Spots COVID-19 in Chest X-rays. 2020. Available online: https://pharmaphorum.com/news/artificialintelligence-spots-covid-19-from-chest-x-rays/ (accessed on 12 September 2020).
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Mangal, A.; Kalia, S.; Rajgopal, H.; Rangarajan, K.; Namboodiri, V.; Banerjee, S.; Arora, C. CovidAID: COVID-19 Detection Using Chest X-ray. arXiv 2020, arXiv:2004.09803. [Google Scholar]
- Das, D.; Santosh, K.; Pal, U. Truncated inception net: COVID-19 outbreak screening using chest X-rays. Phys. Eng. Sci. Med. 2020, 43, 915–925. [Google Scholar] [CrossRef]
- Minaee, S.; Kafieh, R.; Sonka, M.; Yazdani, S.; Soufi, G.J. Deep-covid: Predicting covid-19 from chest X-ray images using deep transfer learning. arXiv 2020, arXiv:2004.09363. [Google Scholar] [CrossRef]
- Yoo, S.H.; Geng, H.; Chiu, T.L.; Yu, S.K.; Cho, D.C.; Heo, J.; Choi, M.S.; Choi, I.H.; Cung Van, C.; Nhung, N.V.; et al. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Front. Med. 2020, 7, 427. [Google Scholar] [CrossRef]
- Loey, M.; Smarandache, F.; Khalifa, N.E.M. Within the Lack of Chest COVID-19 X-ray Dataset: A Novel Detection Model Based on GAN and Deep Transfer Learning. Symmetry 2020, 12, 651. [Google Scholar] [CrossRef]
- RadBoudumc. Evaluation of an AI System for Detection of COVID-19 on Chest X-ray Images. 2020. Available online: https://www.radboudumc.nl/en/news/2020/evaluation-of-an-ai-system-for-detection-of-covid-19-on-chest-x-ray-images (accessed on 13 September 2020).
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology 2020. [Google Scholar] [CrossRef]
- Butt, C.; Gill, J.; Chun, D.; Babu, B.A. Deep learning system to screen coronavirus disease 2019 pneumonia. Appl. Intell. 2020, 1. [Google Scholar]
- Ghoshal, B.; Tucker, A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv 2020, arXiv:2003.10769. [Google Scholar]
- Bai, X.; Fang, C.; Zhou, Y.; Bai, S.; Liu, Z.; Xia, L.; Chen, Q.; Xu, Y.; Xia, T.; Gong, S.; et al. Predicting COVID-19 malignant progression with AI techniques. 2020. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3557984 (accessed on 3 November 2020).
- Jin, C.; Chen, W.; Cao, Y.; Xu, Z.; Zhang, X.; Deng, L.; Zheng, C.; Zhou, J.; Shi, H.; Feng, J. Development and Evaluation of an AI System for COVID-19 Diagnosis. medRxiv 2020. [Google Scholar] [CrossRef]
- Jin, S.; Wang, B.; Xu, H.; Luo, C.; Wei, L.; Zhao, W.; Hou, X.; Ma, W.; Xu, Z.; Zheng, Z.; et al. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. medRxiv 2020. [Google Scholar] [CrossRef]
- Narin, A.; Kaya, C.; Pamuk, Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. arXiv 2020, arXiv:2003.10849. [Google Scholar]
- Wang, L.; Wong, A. COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-ray Images. arXiv 2020, arXiv:2003.09871. [Google Scholar]
- Gozes, O.; Frid-Adar, M.; Greenspan, H.; Browning, P.D.; Zhang, H.; Ji, W.; Bernheim, A.; Siegel, E. Rapid ai development cycle for the coronavirus (covid-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv 2020, arXiv:2003.05037. [Google Scholar]
- Chowdhury, M.E.; Rahman, T.; Khandakar, A.; Mazhar, R.; Kadir, M.A.; Mahbub, Z.B.; Islam, K.R.; Khan, M.S.; Iqbal, A.; Al-Emadi, N.; et al. Can AI help in screening viral and COVID-19 pneumonia? arXiv 2020, arXiv:2003.13145. [Google Scholar]
- Maghdid, H.S.; Asaad, A.T.; Ghafoor, K.Z.; Sadiq, A.S.; Khan, M.K. Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. arXiv 2020, arXiv:2004.00038. [Google Scholar]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef]
- Sethy, P.K.; Behera, S.K. Detection of coronavirus disease (covid-19) based on deep features. Preprints 2020. [Google Scholar] [CrossRef]
- Hemdan, E.E.D.; Shouman, M.A.; Karar, M.E. Covidx-net: A framework of deep learning classifiers to diagnose covid-19 in X-ray images. arXiv 2020, arXiv:2003.11055. [Google Scholar]
- Zheng, C.; Deng, X.; Fu, Q.; Zhou, Q.; Feng, J.; Ma, H.; Liu, W.; Wang, X. Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv 2020. [Google Scholar] [CrossRef]
- Hall, L.O.; Paul, R.; Goldgof, D.B.; Goldgof, G.M. Finding covid-19 from chest X-rays using deep learning on a small dataset. arXiv 2020, arXiv:2004.02060. [Google Scholar]
- Ni, Q.; Sun, Z.Y.; Qi, L.; Chen, W.; Yang, Y.; Wang, L.; Zhang, X.; Yang, L.; Fang, Y.; Xing, Z.; et al. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur. Radiol. 2020, 30, 6517–6527. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulos, I.D.; Aznaouridis, S.I.; Tzani, M.A. Extracting possibly representative COVID-19 Biomarkers from X-ray images with Deep Learning approach and image data related to Pulmonary Diseases. J. Med. Biol. Eng. 2020, 40, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Xia, L.; Shan, F.; Wu, D.; Wei, Y.; Yuan, H.; Jiang, H.; Gao, Y.; Sui, H.; Shen, D. Large-scale screening of covid-19 from community acquired pneumonia using infection size-aware classification. arXiv 2020, arXiv:2003.09860. [Google Scholar]
- Maia, M.; Pimentel, J.S.; Pereira, I.S.; Gondim, J.; Barreto, M.E.; Ara, A. Convolutional Support Vector Models: Prediction of Coronavirus Disease Using Chest X-rays. Information 2020, 11, 548. [Google Scholar] [CrossRef]
- Cohen, J.P.; Dao, L.; Morrison, P.; Roth, K.; Bengio, Y.; Shen, B.; Abbasi, A.; Hoshmand-Kochi, M.; Ghassemi, M.; Li, H.; et al. Predicting covid-19 pneumonia severity on chest X-ray with deep learning. arXiv 2020, arXiv:2005.11856. [Google Scholar] [CrossRef]
- Health EUROPA. Using Artificial Intelligence to Determine COVID-19 Severity. 2020. Available online: https://www.healtheuropa.eu/using-artificial-intelligence-to-determine-covid-19-severity/100501/ (accessed on 12 September 2020).
- New York University. App Determines COVID-19 Disease Severity Using Artificial Intelligence, Biomarkers. 2020. Available online: https://www.sciencedaily.com/releases/2020/06/200603132529.htm (accessed on 14 September 2020).
- Ridley, E. AI Can Assess COVID-19 Severity on Chest X-rays. 2020. Available online: https://www.auntminnie.com/index.aspx?sec=ser&sub=def&pag=dis&ItemID=129674 (accessed on 12 September 2020).
- Imran, A.; Posokhova, I.; Qureshi, H.N.; Masood, U.; Riaz, S.; Ali, K.; John, C.N.; Nabeel, M. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. arXiv 2020, arXiv:2004.01275. [Google Scholar]
- Stevens, S.; Volkmann, J.; Newman, E. The mel scale equates the magnitude of perceived differences in pitch at different frequencies. J. Acoust. Soc. Am. 1937, 8, 185–190. [Google Scholar] [CrossRef]
- HospiMedica. AI-Powered COVID-19 Cough Analyzer App Assesses Respiratory Health and Associated Risks. 2020. Available online: https://www.hospimedica.com/covid-19/articles/294784217/ai-powered-covid-19-cough-analyzer-app-assesses-respiratory-health-and-associated-risks.html (accessed on 14 September 2020).
- Schuller, B.W.; Schuller, D.M.; Qian, K.; Liu, J.; Zheng, H.; Li, X. Covid-19 and computer audition: An overview on what speech & sound analysis could contribute in the SARS-CoV-2 Corona crisis. arXiv 2020, arXiv:2003.11117. [Google Scholar]
- Wang, Y.; Hu, M.; Li, Q.; Zhang, X.P.; Zhai, G.; Yao, N. Abnormal respiratory patterns classifier may contribute to large-scale screening of people infected with COVID-19 in an accurate and unobtrusive manner. arXiv 2020, arXiv:2002.05534. [Google Scholar]
- Sharma, N.; Krishnan, P.; Kumar, R.; Ramoji, S.; Chetupalli, S.R.; Ghosh, P.K.; Ganapathy, S. Coswara—A Database of Breathing, Cough, and Voice Sounds for COVID-19 Diagnosis. arXiv 2020, arXiv:2005.10548. [Google Scholar]
- Al Hossain, F.; Lover, A.A.; Corey, G.A.; Reich, N.G.; Rahman, T. FluSense: A contactless syndromic surveillance platform for influenza-like illness in hospital waiting areas. Proc. ACM Interact. Mobile Wearable Ubiquitous Technol. 2020, 4, 1–28. [Google Scholar] [CrossRef]
- ET Health World. Scientists Develop Portable AI Device That Can Use Coughing Sounds to Monitor COVID-19 Trends. 2020. Available online: https://health.economictimes.indiatimes.com/news/diagnostics/scientists-develop-portable-ai-device-that-can-use-coughing-sounds-to-monitor-covid-19-trends/74730537 (accessed on 11 September 2020).
- Ravelo, J. This Nonprofit Needs Your Cough Sounds to Detect COVID-1. 2020. Available online: https://www.devex.com/news/this-nonprofit-needs-your-cough-sounds-to-detect-covid-19-97141 (accessed on 12 September 2020).
- Iqbal, M.Z.; Faiz, M.F.I. Active Surveillance for COVID-19 through artificial in-telligence using concept of real-time speech-recognition mobile application to analyse cough sound. 2020. Available online: https://www.researchgate.net/publication/340254891_Active_Surveillance_for_COVID-19_through_artificial_intelligence_using_concept_of_real-time_speech-recognition_mobile_application_to_analyse_cough_sound (accessed on 13 November 2020).
- Brown, C.; Chauhan, J.; Grammenos, A.; Han, J.; Hasthanasombat, A.; Spathis, D.; Xia, T.; Cicuta, P.; Mascolo, C. Exploring Automatic Diagnosis of COVID-19 from Crowdsourced Respiratory Sound Data. arXiv 2020, arXiv:2006.05919. [Google Scholar]
- Miranda, I.D.; Diacon, A.H.; Niesler, T.R. A comparative study of features for acoustic cough detection using deep architectures. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 2601–2605. [Google Scholar]
- Yadav, S.; Keerthana, M.; Gope, D.; Ghosh, P.K. Analysis of Acoustic Features for Speech Sound Based Classification of Asthmatic and Healthy Subjects. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 6789–6793. [Google Scholar]
- Larson, E.C.; Lee, T.; Liu, S.; Rosenfeld, M.; Patel, S.N. Accurate and privacy preserving cough sensing using a low-cost microphone. In Proceedings of the 13th International Conference on Ubiquitous Computing, Beijing, China, 17–21 September 2011; pp. 375–384. [Google Scholar]
- Simply, R.M.; Dafna, E.; Zigel, Y. Obstructive sleep apnea (OSA) classification using analysis of breathing sounds during speech. In Proceedings of the 2018 26th European Signal Processing Conference (EUSIPCO), Eternal, Italy, 3–7 September 2018; pp. 1132–1136. [Google Scholar]
- Routray, A. Automatic Measurement of Speech Breathing Rate. In Proceedings of the 2019 27th European Signal Processing Conference (EUSIPCO), A Coruña, Spain, 2–6 September 2019; pp. 1–5. [Google Scholar]
- Nallanthighal, V.S.; Strik, H. Deep Sensing of Breathing Signal during Conversational Speech. 2019. Available online: https://repository.ubn.ru.nl/bitstream/handle/2066/214126/214126.pdf?sequence=1 (accessed on 13 November 2020).
- Partila, P.; Tovarek, J.; Rozhon, J.; Jalowiczor, J. Human stress detection from the speech in danger situation. Mobile Multimedia/Image Processing, Security, and Applications 2019. In Proceedings of the International Society for Optics and Photonics, San Diego, CA, USA, 11–15 August 2019; Volume 10993, p. 109930U. [Google Scholar]
- Maghdid, H.S.; Ghafoor, K.Z.; Sadiq, A.S.; Curran, K.; Rabie, K. A novel ai-enabled framework to diagnose coronavirus covid 19 using smartphone embedded sensors: Design study. arXiv 2020, arXiv:2003.07434. [Google Scholar]
- Etzioni, O.; Decario, N. AI Can Help Scientists Find a Covid-19 Vaccine. 2020. Available online: https://www.wired.com/story/opinion-ai-can-help-find-scientists-find-a-covid-19-vaccine/ (accessed on 16 September 2020).
- Senior, A.; Jumper, J.; Hassabis, D.; Kohli, P. AlphaFold: Using AI for Scientific Discovery. DeepMind. 2018. Available online: https://deepmind.com/blog/alphafold (accessed on 28 October 2020).
- HospiMedica. Scientists Use Cloud-Based Supercomputing and AI to Develop COVID-19 Treatments and Vaccine Models. 2020. Available online: https://www.hospimedica.com/covid-19/articles/294784537/scientists-use-cloud-based-supercomputing-and-ai-to-develop-covid-19-treatments-and-vaccine-models.html (accessed on 15 September 2020).
- Institute for Protein Design. Rosetta’s Role in Fighting Coronavirus. 2020. Available online: https://www.ipd.uw.edu/2020/02/rosettas-role-in-fighting-coronavirus/ (accessed on 10 September 2020).
- Rees, V. AI and Cloud Computing Used to Develop COVID-19 Vaccine. 2020. Available online: https://www.drugtargetreview.com/news/59650/ai-and-cloud-computing-used-to-develop-covid-19-vaccine/ (accessed on 17 September 2020).
- TABIP. Coronavirus Vaccine Candidate Developed In Adelaide Lab To Start Human Trials. 2020. Available online: https://covid19.tabipacademy.com/2020/07/03/coronavirus-vaccine-candidate-developed-in-adelaide-lab-to-start-human-trials/ (accessed on 20 September 2020).
- Flinders University. Microsoft’s AI for Health Supports COVID-19 Vaccine. 2020. Available online: https://medicalxpress.com/news/2020-08-microsoft-ai-health-covid-vaccine.html (accessed on 18 September 2020).
- Herst, C.V.; Burkholz, S.; Sidney, J.; Sette, A.; Harris, P.E.; Massey, S.; Brasel, T.; Cunha-Neto, E.; Rosa, D.S.; Chao, W.C.H.; et al. An effective CTL peptide vaccine for ebola zaire based on survivors’ CD8+ targeting of a particular nucleocapsid protein epitope with potential implications for COVID-19 vaccine design. Vaccine 2020, 38, 4464–4475. [Google Scholar] [CrossRef]
- Ong, E.; Wong, M.U.; Huffman, A.; He, Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. bioRxiv 2020. [Google Scholar] [CrossRef]
- Deoras, S. How ML Is Assisting In Development Of Covid-19 Vaccines. 2020. Available online: https://analyticsindiamag.com/how-ml-is-assisting-in-development-of-covid-19-vaccines/ (accessed on 19 September 2020).
- Rahman, M.S.; Hoque, M.N.; Islam, M.R.; Akter, S.; Rubayet-Ul-Alam, A.; Siddique, M.A.; Saha, O.; Rahaman, M.M.; Sultana, M.; Crandall, K.A.; et al. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2 etiologic agent of global pandemic COVID-19: An in silico approach. PeerJ 2020, 8, e9572. [Google Scholar] [CrossRef]
- Sarkar, B.; Ullah, M.A.; Johora, F.T.; Taniya, M.A.; Araf, Y. The Essential Facts of Wuhan Novel Coronavirus Outbreak in China and Epitope-based Vaccine Designing against 2019-nCoV. bioRxiv 2020. [Google Scholar] [CrossRef]
- Prachar, M.; Justesen, S.; Steen-Jensen, D.B.; Thorgrimsen, S.P.; Jurgons, E.; Winther, O.; Bagger, F.O. Covid-19 vaccine candidates: Prediction and validation of 174 sars-cov-2 epitopes. bioRxiv 2020. [Google Scholar] [CrossRef]
- Xue, H.; Li, J.; Xie, H.; Wang, Y. Review of drug repositioning approaches and resources. Int. J. Biol. Sci. 2018, 14, 1232. [Google Scholar] [CrossRef]
- Mohanty, S.; Rashid, M.H.A.; Mridul, M.; Mohanty, C.; Swayamsiddha, S. Application of Artificial Intelligence in COVID-19 drug repurposing. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Khuroo, M.S.; Sofi, A.A.; Khuroo, M. Chloroquine and Hydroxychloroquine in Coronavirus Disease 2019 (COVID-19). Facts, Fiction & the Hype. A Critical Appraisal. Int. J. Antimicrob. Agents 2020, 56, 106101. [Google Scholar] [PubMed]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Pahikkala, T.; Airola, A.; Pietilä, S.; Shakyawar, S.; Szwajda, A.; Tang, J.; Aittokallio, T. Toward more realistic drug–target interaction predictions. Brief. Bioinform. 2015, 16, 325–337. [Google Scholar] [CrossRef]
- He, T.; Heidemeyer, M.; Ban, F.; Cherkasov, A.; Ester, M. SimBoost: A read-across approach for predicting drug–target binding affinities using gradient boosting machines. J. Cheminform. 2017, 9, 1–14. [Google Scholar] [CrossRef]
- Öztürk, H.; Özgür, A.; Ozkirimli, E. DeepDTA: Deep drug–target binding affinity prediction. Bioinformatics 2018, 34, i821–i829. [Google Scholar] [CrossRef]
- Huang, K.; Fu, T.; Glass, L.; Zitnik, M.; Xiao, C.; Sun, J. DeepPurpose: A Deep Learning Library for Drug-Target Interaction Prediction and Applications to Repurposing and Screening. arXiv 2020, arXiv:cs.LG/2004.08919. [Google Scholar]
- Anwar, M.U.; Adnan, F.; Abro, A.; Khan, M.R.; Rehman, A.U.; Osama, M.; Javed, S.; Baig, A.; Shabbir, M.R.; Assir, M.Z.; et al. Combined Deep Learning and Molecular Docking Simulations Approach Identifies Potentially Effective FDA Approved Drugs for Repurposing Against SARS-CoV-2. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Zeng, X.; Zhu, S.; Liu, X.; Zhou, Y.; Nussinov, R.; Cheng, F. deepDR: A network-based deep learning approach to in silico drug repositioning. Bioinformatics 2019, 35, 5191–5198. [Google Scholar] [CrossRef]
- Kojima, R.; Ishida, S.; Ohta, M.; Iwata, H.; Honma, T.; Okuno, Y. kGCN: A graph-based deep learning framework for chemical structures. J. Cheminform. 2020, 12, 1–10. [Google Scholar] [CrossRef]
- Ramsundar, B.; Eastman, P.; Walters, P.; Pande, V. Deep Learning for the Life Sciences: Applying Deep Learning to Genomics, Microscopy, Drug Discovery, and More; O’Reilly Media, Inc.: Newton, MA, USA, 2019. [Google Scholar]
- Shi, Y.; Zhang, X.; Mu, K.; Peng, C.; Zhu, Z.; Wang, X.; Yang, Y.; Xu, Z.; Zhu, W. D3Targets-2019-nCoV: A webserver for predicting drug targets and for multi-target and multi-site based virtual screening against COVID-19. Acta Pharm. Sin. B 2020, 10, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Bung, N.; Krishnan, S.R.; Bulusu, G.; Roy, A. De novo design of new chemical entities (NCEs) for SARS-CoV-2 using artificial intelligence. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Tang, B.; He, F.; Liu, D.; Fang, M.; Wu, Z.; Xu, D. AI-aided design of novel targeted covalent inhibitors against SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Chen, J.; Li, K.; Zhang, Z.; Li, K.; Yu, P.S. A Survey on Applications of Artificial Intelligence in Fighting Against COVID-19. arXiv 2020, arXiv:2007.02202. [Google Scholar]
- Moskal, M.; Beker, W.; Roszak, R.; Gajewska, E.P.; Wołos, A.; Molga, K.; Szymkuć, S.; Grzybowski, B.A. Suggestions for second-pass anti-COVID-19 drugs based on the Artificial Intelligence measures of molecular similarity, shape and pharmacophore distribution. ChenRxiv 2020. [Google Scholar] [CrossRef]
- Hu, F.; Jiang, J.; Yin, P. Prediction of potential commercially inhibitors against SARS-CoV-2 by multi-task deep model. arXiv 2020, arXiv:2003.00728. [Google Scholar]
- Kadioglu, O.; Saeed, M.; Johannes Greten, H.; Efferth, T. Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Bull. WHO 2020. [Google Scholar] [CrossRef]
- Hofmarcher, M.; Mayr, A.; Rumetshofer, E.; Ruch, P.; Renz, P.; Schimunek, J.; Seidl, P.; Vall, A.; Widrich, M.; Hochreiter, S.; et al. Large-scale ligand-based virtual screening for SARS-CoV-2 inhibitors using deep neural networks. SSRN 2020. [Google Scholar] [CrossRef]
- Zhavoronkov, A.; Aladinskiy, V.; Zhebrak, A.; Zagribelnyy, B.; Terentiev, V.; Bezrukov, D.S.; Polykovskiy, D.; Shayakhmetov, R.; Filimonov, A.; Orekhov, P.; et al. Potential COVID-2019 3C-Like Protease Inhibitors Designed Using Generative Deep Learning Approaches; Insilico Medicine Hong Kong Ltd A: Hong Kong, China, 2020; Volume 307, p. E1. [Google Scholar]
| Author | Data | ML/DL Method | Accuracy | AUC | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| Li et al. [28] | A total of 4356 CT images from 3322 patients obtained from six clinics (1296 images for COVID-19, 1735 for CAP and 1325 for non-pneumonia). | COVNet using pre-traind ResNet-50 | N/A | 96% AUC | 87% | 92% |
| Butt et al. [29] | A total of 618 CT images (219 images from 110 COVID-19 patients, 224 CT images from 224 patients with influenza-A viral pneumonia, and 175 CT images from healthy people). | Location-attention network and ResNet-18 | 86.7% | N/A | N/A | N/A |
| Ghoshal and Tucker [30] | A total of 5941 CT images for four classes (1583 normal, 2786 bacterial pneumonia, 1504 viral pneumonia, and 68 COVID-19) | Drop-weights based Bayesian CNNs | 89.92% | N/A | N/A | N/A |
| Bai et al. [31] | Unknown number of chest CT images collected from 133 COVID-19 patients of which 54 patients had severe/critical cases. | MLP and LSTM | N/A | 95% | 95% | 95% |
| Jin et al. [32] | 496 chest CT scans from COVID-19 patients and 1385 non-COVID-19 CT scans. | CNN | N/A | 97.17% | 90.19% | 95.76% |
| Jin et al. [33] | A total of 1136 CT images (723 COVID-19 confirmed COVID-19 cases from five hospitals). | 3D UNet++ and ResNet-50 | N/A | N/A | 97% | 92% |
| Narin et al. [34] | Only 50 COVID-19 chest X-ray scans and 50 for non-COVID-19. | Pre-trained ResNet-50 | 98% | N/A | N/A | N/A |
| Wang and Wong [35] | 16,756 chest X-ray scans from 13,645 COVID-19 patients. | COVID-Net | 92.4% | N/A | 91.0% | N/A |
| Gozes et al. [36] | Unknown number of CT scans collected from 157 Chinese and U.S. COVID-19 patients. | ResNet-50 | N/A | 97% | 98.2% | 92.2% |
| Chowdhury et al. [37] | A total of 3487 chest CT scan images (1579 normal, 1485 viral pneumonia, and 423 COVID-19. | AlexNet, ResNet-18, DenseNet201, and SqueezeNet | 99.7% | N/A | 99.7% | 99.5% |
| Maghdid et al. [38] | A total of 170 COVID-19 X-ray scans and a total of 361 COVID-19 CT scans of collected from five different places. | CNN and pre-trained AlexNet models | 98% on X-ray scans and 94.1% on CT scans | N/A | 100% on X-ray scans and 90% on CT scans | N/A |
| Apostolopoulos and Mpesiana [39] | A total of 1428 chest X-ray scans (224 COVID-19, 700 pneumonia patient, and 504 normal). | Pre-trained VGG-19 | 93.48% | N/A | 92.85% | 98.75% |
| Sethy and Behera [40] | Chest X-ray scans of 25 COVID-19 positive and 25 negative. | ResNet-50 and SVM | 93.28% | N/A | 97% | 93% |
| Hemdan et al. [41] | Chest X-ray scans 25 COVID-19 positive and 25 Normal patient. | COVIDX-Net | 90% | N/A | N/A | N/A |
| Wang et al. [6] | Chest CT scans of 195 COVID-19 positive and 258 COVID-19 negative patients. | M-Inception | 89.5% | N/A | 87% | 88% |
| Zheng et al. [42] | Chest CT images of 313 COVID-19 and 229 non-COVID-19 patients. | U-Net and a 3D Deep Network | 90% | N/A | 95% | 95% |
| Hall et al. [43] | A total of 455 chest X-ray (135 of COVID-19 and 320 of viral and bacterial pneumonia. | Pre-trained ResNet-50 | 89.2% | 95% | N/A | N/A |
| Ni et al. [44] | CT images of 96 COVID-19 positive collected from three hospitals in China. | Deep CNN | 73% | N/A | 95% | 95% |
| Apostolopoulos et al. [45] | A total of 3905 X-ray scans for several classes of disease including COVID-19. | MobileNet-v2 | 99.18% | N/A | 97.36% | 99.42% |
| Shi et al. [46] | CT images of 2685 collected from three hospitals, 1658 of COVID-19 positive cases and 1027 cases were CAP patients. | RF | 87.9% | N/A | 90.7% | 83.3% |
| Loey et al. [26] | Chest X-ray scans of 307 for COVID-19, normal, pneumonia, bacterial, and pneumonia virus. | GAN with AlexNet, GoogLeNet, and ResNet18 pre-trained models. | 80.56% | N/A | N/A | N/A |
| Maia et al. [47] | A total of 437 X-ray images (217 COVID-19, 108 other diseases, and 112 healthy). | Different Convolutional Support Vector Machines (CSVMs) | 98.14% | N/A | N/A | N/A |
| Parameters | Severity Score | ||||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |
| Extent of Lung Involvement | No | <25% | 25–50% | 50–75% | >75% |
| Degree of ambiguity | No | Ground glass ambiguity | Unification | White-out | - |
| Author | Usage | ML/DL Method | Dataset | Predictive Performance |
|---|---|---|---|---|
| Miranda et al. [63] | TB Diagnosis | MFB, MFCC, STFT with CNN | Google audio dataset which includes 1.8 million Youtube Freesound videos and audio database | 94.6% AUC |
| Yadav et al. [64] | Asthmatic Discovery | INTERSPEECH 2013 (The basis of the Computational Paralinguistics challenge acoustic features) | Speech from 47 asthmatic and 48 healthy controls | 48% Accuracy |
| Larson et al. [65] | Avoid recording of speech while hearing cough | PCA on audio spectrograms, FFT coefficients and random separation | Acted cough from 17 patients having 8 people cough due to cold weather, 3 patients because of Asthama, 1 patient because of allergies and 5 patients who cough due to chronic cough condition | 92% of True positive rate and 50% of false positive rate |
| Simply et al. [66] | Obstructive sleep apnea (OSA) acquisition from sounds breathing in speech | MFCCs have a single layer neural network for respiration and MFCCs, strength, tone, kurtosis and ZCR with OV SVM separation | 90 Male subjects’ speech and sleep quality measures using WatchPAT | 50% for breathing detection using Cohen’s kappa coefficient and 54% using OSA detection |
| Routray et al. [67] | Automatic speech breathing average rating | Cepstrogram and support Vector machine with radial base work | 16 recording of speeches 21-year-old group participants age means | 89% F1-measure |
| Nallanthighal and Strik [68] | Receiving a breathing signal in conversational speech | Pectrogram with CNN and RNN | 20 healthy speeches recordings using a microphone and a breathing signal using two respirator transducers belts | 91.2% respiratory sensitivity |
| Partila et al. [69] | Pressure detection in speech in cases such as a car accident, domestic violence, and conditions near death | LLD and functional characteristics are obtained using openSMILE with SVM and CNN separators | 312 emergency call recording of 112 Emergency Cable Emergency Plan from the Czech Republic | 87.9% accuracy using SVM and 87.5% Accuracy using CNN |
| Author | Data | Method | Role of ML/DL Method | COVID-19 Target | Potent In Drugs |
|---|---|---|---|---|---|
| Moskal et al. [100] | A random subset of 6000 small molecules from the ZINC database and divided it randomly into the training and test sets in 5:1 proportion | VAE, CNN, LSTM, and MLP | Help in generating SMILES strings and molecules | N/A | 110 Drugs |
| Beck et al. [87] | Drug Target Common (DTC) and BindingDB datasets | MT-DTI | Predict binding affinity between drugs and protein targets | 3CLpro, RdRp, Helicase | 6 Drugs |
| Hu et al. [101] | A dataset contains 4895 commercially drugs | DL | Predict binding between drugs and protein targets | 3CLpro, RdRp | 10 drugs |
| Kadioglu et al. [102] | FDA-approved drugs for drug repositioning, Natural compound dataset from literature mining, and ZINC dataset to select compounds interacting with SARS-CoV-2 target proteins. | NN and Naive Bayes | Construct drug likelihood prediction model | spike protein and more | 3 drugs |
| Hofmarcher et al. [103] | Binary ECFP4 fingerprints folded to a length of 1024 and 75 output units of ChemAI | ChemAI | Classify the moecules effects on COVID-19 proteases | 3CLpro, PLpro | 30,000 molecules |
| Bung et al. [97] | A dataset of 1.6 million drug-like small molecules from the ChEMBL database | RNN and RL | Classify protease inhibitor molecules | 3CLpro | 31 compounds |
| Zhavoronkov et al. [104] | Crystal structure of 2019-nCoV 3C-like protease | 28 ML | Generate new molecular structures for 3CLpro | 3CLpro | 100 molecules |
| Tang et al. [98] | SARS-CoV 3CLpro inhibitors (284 molecules) | RL and DQN | Predict molecules and lead compounds for each target fragment | 3CLpro | 47 compounds |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alafif, T.; Tehame, A.M.; Bajaba, S.; Barnawi, A.; Zia, S. Machine and Deep Learning towards COVID-19 Diagnosis and Treatment: Survey, Challenges, and Future Directions. Int. J. Environ. Res. Public Health 2021, 18, 1117. https://doi.org/10.3390/ijerph18031117
Alafif T, Tehame AM, Bajaba S, Barnawi A, Zia S. Machine and Deep Learning towards COVID-19 Diagnosis and Treatment: Survey, Challenges, and Future Directions. International Journal of Environmental Research and Public Health. 2021; 18(3):1117. https://doi.org/10.3390/ijerph18031117
Chicago/Turabian StyleAlafif, Tarik, Abdul Muneeim Tehame, Saleh Bajaba, Ahmed Barnawi, and Saad Zia. 2021. "Machine and Deep Learning towards COVID-19 Diagnosis and Treatment: Survey, Challenges, and Future Directions" International Journal of Environmental Research and Public Health 18, no. 3: 1117. https://doi.org/10.3390/ijerph18031117
APA StyleAlafif, T., Tehame, A. M., Bajaba, S., Barnawi, A., & Zia, S. (2021). Machine and Deep Learning towards COVID-19 Diagnosis and Treatment: Survey, Challenges, and Future Directions. International Journal of Environmental Research and Public Health, 18(3), 1117. https://doi.org/10.3390/ijerph18031117